Abstract
We isolated two Candida pseudointermedia strains from the Atlantic rain forest in Brazil, and analyzed cellobiose metabolization in their cells. After growth in cellobiose medium, both strains had high intracellular β-glucosidase activity [~ 200 U (g cells)−1 for 200 mM cellobiose and ~ 100 U (g cells)−1 for 2 mM pNPβG] and negligible periplasmic cellobiase activity. During batch fermentation, the strain with the best performance consumed all the available cellobiose in the first 18 h of the assay, producing 2.7 g L−1 of ethanol. Kinetics of its cellobiase activity demonstrated a high-affinity hydrolytic system inside cells, with Km of 12.4 mM. Our data suggest that, unlike other fungal species that hydrolyze cellobiose extracellularly, both analyzed strains transport it to the cytoplasm, where it is then hydrolyzed by high-affinity intracellular β-glucosidases. We believe this study increases the fund of knowledge regarding yeasts from Brazilian microbiomes.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Yeasts have been used by humans since the Neolithic Revolution. This was long before their role as fermenting microorganisms was discovered in the nineteenth century with the work of Louis Pasteur. Over these millennia human activity has primarily selected Saccharomyces spp. as industrial yeasts; nevertheless, several wild species have come to be known in the last century (Eliodório et al. 2019). Despite the fact that many of them have already been taxonomically described, the number of biochemically characterized species is much smaller. This is the case of Candida pseudointermedia (family Metschnikowiaceae, order Saccharomycetales, class Saccharomycetes subphylum Saccharomycotina, phylum Ascomycota), first described by Nakase et al. (1976), but hitherto poorly characterized in terms of carbohydrate metabolism.
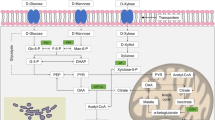
The lasting relationship of yeasts with humans is mainly due to the broad spectrum of carbohydrates that they metabolize (Lagunas 1993; Gancedo 1998; Marques et al. 2016). Cellobiose, one of these sugars, is a disaccharide formed by two glucose molecules joined by a β-1,4 glycosidic bond, with one glucose unit rotated 180° with respect to the other. This disaccharide is mostly found in partially hydrolyzed cellulosic material (Parisutham et al. 2017). Cellobiose hydrolysis depends on the activity of β-glucosidases (also called cellobiases), enzymes primarily produced by representatives of the fungal kingdom (Srivastava et al. 2018; Alves et al. 2019). Both hydrolysis and fermentation of this sugar are interesting to the biofuel industry.
In yeasts, cellobiose metabolism also depends on β-glucosidases that can be secreted by cells in the medium in which they are found (Barbosa et al. 2010; Baffi et al. 2011; Molnárová et al. 2014), or, they may be located either in the periplasm (Guo et al. 2015) or in the cytoplasm (Santos et al. 2011; Reis et al. 2014; Parisutham et al. 2017). With the exception of the species Sporidiobolus pararoseus (Baffi et al. 2011), Naganishia albida (Cryptococcus albidus) and Aureobasidium pullulans (Molnárová et al. 2014), which, respectively, belong to the orders Sporidiobolales (class Microbotryomycetes, subphylum Pucciniomycotina, phylum Basidiomycota), Filobasidiales (class Tremellomycetes, subphylum Agaricomycotina, phylum Basidiomycota) and Dothideales (class Dothideomycetes, subphylum Pezizomycotina, phylum Ascomycota), all the other yeasts analyzed by those authors belong to the order Saccharomycetales (class Saccharomycetes, subphylum Saccharomycotina, phylum Ascomycota), being distributed in five different families: Metschnikowiaceae, Debaryomycetaceae, Phaffomycetaceae, Pichiaceae and Saccharomycodaceae. While the ability to extracellularly hydrolyze cellobiose is observed in the first three orders, the presence of β-glucosidases in the periplasm or in the cytoplasm of cells seems to be restricted to the order Saccharomycetales. If cellobiose hydrolysis occurs either extracellularly or in the periplasm, yeast cells only need to uptake the glucose molecules generated by this breakdown, then metabolize these hexoses through fermentative or respiratory metabolism. On the other hand, for cellobiose hydrolysis to occur in the cytoplasm, the yeast cell must have membrane permeases capable of transporting the molecule into the cell, where it can then be hydrolyzed by intracellular β-glucosidases, releasing glucose molecules that can be directly fermented or respired (Parisutham et al. 2017; Ríos-Fránquez et al. 2017; Eliodório et al. 2019).
Although several studies have already observed C. pseudointermedia on different substrates, including fish-paste (Nakase et al. 1976), lake (Rosa et al. 1995), coffee (Mausoud et al. 2004), human body (Putigani et al. 2011), coconut water (Maciel et al. 2016), rice phylloplane (Limtong and Kaewwichian 2015), tap water (Babič et al. 2016) and spoiled ready-to-eat meals (Feliciano et al. 2019), these studies have focused on ecological, taxonomic and phylogenetic aspects; there are no detailed metabolic characterizations of this yeast. Through simple physiological tests, used for taxonomic description of this species, it was discovered that this yeast assimilates cellobiose, among other sugars (Nakase et al. 1976). Nevertheless, the mechanism of this metabolism by yeast cells has not been detailed, nor has the role of their permeases and hydrolytic enzymes been analyzed. Therefore, with the purpose of broadening the base of knowledge regarding still poorly-characterized yeast species, we analyzed the cellobiose catabolism in two C. pseudointermedia strains isolated from rotting wood samples collected in the Atlantic rain forest of Southern Brazil. To the best of our knowledge, this is the first work to detail the metabolism of a disaccharide in C. pseudointermedia.
Materials and methods
Yeast strain isolation and identification
The yeast strains used in this work were isolated from rotting wood samples collected from the Chapecó National Forest, 27° 5′ S and 52° 46′ W (strain FLONA-CE-3.4), and in the woods surrounding the Chapecó campus of the Federal University of the Southern Border, 27° 7′ S and 52° 42′ W (strain UFFS-CE-3.6), following the methodology described by Bazoti et al. (2017). Samples were stored in sterile plastic bags and were transported in isothermal boxes under refrigeration. One gram of each sample was inoculated in flaks with 20 mL minimal YNB media (yeast nitrogen base 6.7 g L−1, pH 5.0) containing 10 g L−1 of cellobiose and 0.2 g L−1 of chloramphenicol. Flasks were incubated at 25 °C on a shaker at 145 rpm until growth was detected by turbidity. Subsequently, one loopful of each tube was streaked on plates containing the same media described above with addition of 20 g L−1 agar. Plates were incubated at 25 °C, and the various yeast morphotypes were purified by repeated streaking on YMA plates (d-glucose 10 g L−1, peptone 5 g L−1, yeast extract 3 g L−1, malt extract 3 g L−1, agar 20 g L−1) and preserved at − 80 °C for later identification. The yeasts were characterized using standard methods (Kurtzman et al. 2011). Species identification was performed using analysis of the D1/D2 variable domains of the large subunit of rRNA (White et al. 1990; O'Donnell 1993; Kurtzman and Robnett 1998; Lachance et al. 1999). The amplified DNA was concentrated, cleaned and sequenced in an ABI 3130 Genetic Analyzer automated sequencing system (Life Technologies, California, USA) using BigDye v3.1 and POP7 polymer. The sequences were assembled, edited, and aligned using the program MEGA6 (Tamura et al. 2013). The sequences obtained were deposited in Genbank (MT007799 and MT007800 for FLONA-CE-3.4 and UFFS-CE-3.6, respectively) and compared with those included in that database using the Basic Local Alignment Search Tool (BLAST, at https://www.ncbi.nlm.nih.gov). Furthermore, a phylogenetic tree was assembled with sequences from type material of the order Saccharomycetales, obtained through this BLAST. To this end, the sequences were first aligned using the Multiple Sequence Comparison by Log-Expectation (MUSCLE, https://www.ebi.ac.uk/Tools/msa/muscle/), and then the alignment file were Gblock-curated on the Phylogeny.fr platform (Castresana 2000; Dereeper et al. 2008, 2010; https://www.phylogeny.fr). After that, the cured alignment file was used to generate the phylogenetic tree using Akaike Information Criterion (AIC) in Smart Model Selection (Lefort et al. 2017) implemented in the PhyML environment (Guindon et al. 2010—https://www.atgc-montpellier.fr/phyml-sms), with a bootstrap analysis of 1000 replicates. For the management of the phylogenetic tree, we used the online tool Interactive Tree of Life (iTOL, https://itol.embl.de).
Media and culture conditions
Two culture media were used in this study: rich YP medium (10 g L−1 yeast extract and 20 g L−1 Bacto peptone), and minimal YNB medium, containing 6.7 g L−1 yeast nitrogen base without amino acids. Both media were supplemented with 20 g L−1 of the indicated carbon source (glucose or cellobiose), and the pH of the media was adjusted to pH 5.0 using HCl. Cells were grown aerobically at 25 °C with shaking (145 rpm) in cotton-plugged Erlenmeyer flasks filled to 1/5 of the volume with medium. Cellular growth was followed by turbidity measurements at 570 nm, and culture samples were harvested regularly, centrifuged (3500g, 3 min), and their supernatants were used for determination of sugars and ethanol.
For batch fermentations, yeasts were pre-grown on YP with 20 g L−1 of glucose until the exponential phase (~ 1 mg of dry yeast mL−1), centrifuged (3500g, 3 min) and washed twice with cold water, and inoculated at high cell density (10 mg of dry yeast mL−1) into minimal YNB medium containing 20 g L−1 of glucose or cellobiose. Samples were collected regularly and processed as described above. The data were expressed as averages and standard error from two completely independent experiments.
Sugars and ethanol determination
Samples of the supernatants were filtered through 0.45-μm filters before transfer to chromatographic vials. The concentrations of glucose, cellobiose and ethanol were determined using high performance liquid chromatography (HPLC; LCMS-2020, Shimadzu), with a refractive index detector (RID-10, Shimadzu) and a column for organic acids (Aminex HPX-87H, Bio-Rad). The mobile phase used 5 mM sulfuric acid at 50 °C with a flow rate of 0.6 mL min−1. Calibration curves were established for all samples using seven concentrations ranging from 0.25 to 20 g L−1 for carbohydrates and 0.125 to 10 g L−1 for ethanol.
β-Glucosidase assays
Periplasmic and intracellular β-Glucosidase activity was determined in cells grown up to the beginning of the exponential growth phase (OD570nm ~ 3.5) in rich YP medium containing 20 g L−1 of glucose or cellobiose as carbon sources. Cells were initially centrifuged, washed twice with water at 4 °C, and resuspended to reach 20 g L−1. Periplasmic cellobiose hydrolysis by β-glucosidase was determined in vivo with whole cells previously incubated with 50 mM sodium fluoride to block glycolysis (Silveira et al. 1996; Batista et al. 2004), using 100 mM cellobiose in 50 mM succinate-Tris buffer, pH 5.0. Intracellular β-glucosidase activity was determined in situ with permeabilized yeast cells (Stambuk 1999) using 200 mM cellobiose or 2 mM p-nitrophenyl-β-d-glucopyranoside (pNPβG) in 100 mM MOPS-NaOH buffer, pH 6.8. For the kinetic analysis, cellobiose concentration ranged from 12.5 to 400 mM. After incubation, the tubes were quickly boiled for 10 min and then cooled in an ice bath. The tubes were centrifuged for 5 min at 3500 rpm and the supernatants were used to quantify the glucose released from cellobiose hydrolysis or the p-nitrophenol released from pNPβG hydrolysis. Glucose was measured by the glucose oxidase and peroxidase method using a commercial kit (BioDiagnostica-Laborclin), and p-nitrophenol was measured on a spectrophotometer at 400 nm. Negative controls using previously boiled cells were used. Cellobiase activity was expressed as U (g dry yeast cells)−1, where one unit corresponds to 1 μmol of glucose or p-nitrophenol produced per min at 30 °C. The presented data were averages and standard error from two independent experiments. The kinetic parameters Km and Vmax were calculated according to Lineaweaver and Burk (1934).
Additionally, β-glucosidase activity was also analyzed in the media, to verify if there was any secreted cellobiase by yeast cells. To this end, strains FLONA-CE-3.4 and UFFS-CE-3.6 were cultured in rich YP medium containing 20 g L−1 of cellobiose, and media was harvested by centrifugation at the beginning of the exponential growth phase (OD570nm ~ 3.5) or after 48 h of culture. The harvested medium was mixed (1:3) with cellobiose (300 mM cellobiose in 150 mM succinate-Tris buffer, pH 5.0), and then incubated for 30 min at 30 °C or 50 °C. Extracellular β-glucosidase activity was assessed by glucose release as described above. For negative controls, the harvested medium was previously boiled.
Transport assays
β-Glucoside transport activity was determined using a colorimetric transport assay using p-nitrophenyl-β-d-glucopyranoside (pNPβG), as previously described for the determination of α-glucoside transport activity in yeasts (Hollatz and Stambuk 2001; Alves et al. 2007, 2008). Cells collected at the exponential phase of growth (OD570nm ~ 3.5) in rich YP medium containing 20 g L−1 of glucose or cellobiose as carbon source were initially centrifuged, washed twice with water at 4 °C, and resuspended to reach 20 g L−1. The cell suspension was incubated with 10 mM pNPβG in 100 mM succinate-Tris buffer, pH 5.0. After 5 min incubation at room temperature, the reaction was interrupted with 1.0 mL of 2 M sodium bicarbonate, and the p-nitrophenol released was measured using a spectrophotometer at 400 nm. All assays were performed in triplicate, and controls with previously boiled yeast cells were used. Transport activity was expressed as U (g dry yeast cells)−1, where one unit corresponds to 1 μmol of pNPβG transported per min. The data were expressed as averages and standard error from two completely independent experiments.
Results and discussion
The two Candida pseudointermedia yeast strains (strains FLONA-CE-3.4 and UFFS-CE-3.6) were isolated from rotting wood samples collected from the Atlantic rain forests of southern Brazil. The sequencing of the D1/D2 variable domains (see Materials and Methods) of both strains revealed an identity of 99.80% between them. When compared to a type strain of C. pseudointermedia (NCBI accession number MK394147.1), the identity was 99.80% for FLONA-CE-3.4 and 100.00% for UFFS-CE-3.6. Considering the proximity between the species C. pseudointermedia and C. intermedia, we also compared the D1/D2 variable domains of these strains with a type strain of C. intermedia (NCBI accession number NG_055404.1) and we found 99.39% and 99.60% of identity for FLONA-CE-3.4 and UFFS-CE-3.6, respectively. It is worth mentioning that the D1/D2 variable domains of C. pseudointermedia and C. intermedia type strains also share 99.60% of identity with each other. Thus, that led us to classify both FLONA-CE-3.4 and UFFS-CE-3.6 as members of the species Candida pseudointermedia.
Initially, we cultivated both strains in rich YP media and minimal YNB media containing cellobiose or glucose as carbon source. The two strains showed distinct profiles of cell growth and sugar consumption in the two types of medium employed (Fig. 1). Strain UFFS-CE-3.6 presented higher cell growth and sugar (glucose or cellobiose) consumption rate in rich YP medium (Fig. 1a, b), when compared with strain FLONA-CE-3.4; however, surprisingly, this last strain showed better performance in minimal YNB medium (Fig. 1c, d), with a 70% higher cellobiose consumption rate by the cells when cultivated in this medium (Table 1).
Cellular growth (a, c) and sugar consumption (b, d) in rich YP medium (a, b) or minimal YNB medium (c, d) containing 20 g L−1 of cellobiose (open symbols) or glucose (closed symbols) by strains FLONA-CE-3.4 (circles) and UFFS-CE-3.6 (squares). Data represent averages and standard error from two independent experiments
Previous studies reported that some sugars may not be completely consumed if the cells are grown in minimal media (Hahn-Hagerdal et al. 2005; Alves et al. 2007), and the ethanol yield in media containing ammonia as nitrogen source (as in the minimal media used in the present work) is roughly half of the yield observed in media whose source of nitrogen comes from yeast extract and peptone (Hande et al. 2013). However, in the present study, this was true only for cellobiose consumption by strain UFFS-CE-3.6 that produced 1.1 g L−1 of ethanol from cellobiose in rich media, but not in YNB media (Table 1). During growth on glucose, both strains consumed this sugar in the first 10–20 h of incubation in rich YP media; however, they needed ~ 24 h to totally consume glucose from minimal YNB medium (Fig. 1). Consequently, the ethanol yield (Ye/s) from glucose in rich media was around 0.38 g g−1, while in YNB media the ethanol yield did not exceed 0.19 g g−1 (Table 1). The lower cellobiose consumption by FLONA-CE-3.4 in rich YP media may be explained by the presence of other sugars in yeast extract such as trehalose (Iwahara et al. 1993) that may interfere with cellobiose consumption in the rich YP media.
The two analyzed strains showed high intracellular β-glucosidase activity against cellobiose, and negligible periplasmic activity (Fig. 2). No cellobiase activity was detected in the medium, neither in the beginning of exponential growth nor at the end of cultivation, after 48 h (data not shown). Taking together, these results, therefore, suggest that the analyzed strains harbor only intracellular isoforms of β-glucosidase. Although cellobiose-grown cells showed high intracellular cellobiose hydrolysis rates, when the cells were pre-cultured in media with glucose, the expression of cellobiase was strongly inhibited in both strains. Indeed, glucose promotes the catabolic repression of a series of enzymes involved in the metabolism of alternative sugars (Santangelo 2006; Alves et al. 2014; Cadete and Rosa 2018).
Similar β-glucosidase activity was shown by Santos et al. (2011) in two strains of the cellobiose-fermenting yeast Scheffersomyces (Candida) queiroziae, although in that case, yeasts had higher ethanol yields than did FLONA-CE-3.4 and UFFS-CE-3.6, suggesting that cellobiose transport through the plasma membrane of this strains might limit the fermentation of this sugar. Indeed, Casa-Villegas et al. (2018) recently showed that a genetically-modified Saccharomyces cerevisiae strain heterologously expressing intracellular β-glucosidases had its growth in cellobiose limited by low sugar intake. In our study, this was also corroborated by the fact that cellobiose consumption was slower than glucose consumption in both FLONA-CE-3.4 and UFFS-CE-3.6 strains (Table 1). When sugars are taken up slowly by yeast cells, their metabolism is essentially respiratory (Duval et al. 2010). Moreover, when the pNPβG (a cellobiose synthetic analog) transport and hydrolysis activities were analyzed in cells cultured in media with cellobiose (Fig. 3), we found that the transport system may indeed be the main bottleneck for cellobiose fermentation, because transport activity (Fig. 3a) reached no more than 12.5% of pNPβG hydrolysis rate (Fig. 3b). Regarding these analyses with pNPβG, it is also worth noting that not only the transport activity, but also hydrolytic activity was negligible when both strains were pre-grown in glucose. This corroborates cellobiase activity data (Fig. 2), suggesting that cellobiose metabolization is impaired in the presence of this hexose.
Some new yeast species have been described as cellobiose-fermenting yeasts, despite the fact that, in most studies, investigators have only performed assays in Durham tubes, and/or the fermentation was considered weak or slow in relation to that observed with glucose (Santos et al. 2011; Wang et al. 2015, 2016; Peter et al. 2017; Cadete and Rosa 2018). Nevertheless, Lopes et al. (2018) recently showed that yeast strains from the species Candida jaroonii, Scheffersomyces queiroziae as well as from a new species of Yamadozyma fermented 50 g L−1 cellobiose in rich YP medium with efficiencies ranging from 82.0 to 94.4% of the maximum theoretical ethanol yield (0.538 g of ethanol per g of cellobiose consumed). Therefore, in addition to the growth experiments in full aerobic condition with a low initial cell concentrations inoculum, we also carried out batch fermentations with a high cell concentration (~ 10 g L−1) in minimal YNB media containing cellobiose or glucose as carbon source. Both strains consumed all available glucose in the first two hours of the experiment; however, it took 18 h for UFFS-CE-3.6 to deplete all the cellobiose from the medium, and 24 h of incubation was not sufficient to allow total cellobiose consumption by the strain FLONA-CE-3.4 (Fig. 4a). Even so, strain UFFS-CE-3.6 fermented cellobiose with an ethanol yield (Ye/s) of 0.11 g g−1, representing 57% more than that obtained during growth in this carbon source and one-third of the ethanol yield achieved from glucose (see Fig. 4b and Table 1). One might say that this delayed cellobiose consumption observed in batch fermentations can be explained by the repressive effect exerted by glucose, because the strains were pre-grown on media with glucose; nevertheless, when the strains were pre-grown in cellobiose, the fermentation of this β-glucoside was not improved at all (data not shown).
Because strain UFFS-CE-3.6 showed the best performance during fully aerobic growth and batch fermentation (Table 1) and showed slightly higher β-glucosidase activities than strain FLONA-CE-3.4 (Figs. 2, 3), the kinetics of its cellobiase activity were analyzed against 12.5–400 mM cellobiose (Fig. 5). We found that β-glucosidases in this yeast have high affinity (Km = 12.4 mM) for cellobiose. This is approximately half of the Michaelis–Menten constant observed for the β-glucosidase BGLII from the thermo-tolerant yeast Pichia etchellsii (Wallecha and Mishra 2003) and one-sixth of the observed Km for the C. peltate cellobiase (Saha and Bothast 1996). Therefore, this represents an affinity for cellobiose two and six times higher, respectively, than the affinities observed by those authors. In an extracellular β-glucosidase from the wine-associated yeast Debaromyces pseudopolymorphus, Villena et al. (2006) measured a Km value (11.9 mM) similar to what we obtained for C. pseudointermedia UFFS-CE-3.6, although we found a Vmax 2.6 times higher than the one found for that winery yeast. More recently, a thermostable β-glucosidase from Aspergillus fumigatus was shown to have a very high affinity for cellobiose (Km = 1.75 mM), but a low Vmax (Liu et al. 2012), with only 28.2% of the Vmax we found for UFFS-CE-3.6.
Our results thus depict the metabolism of cellobiose in yet another non-domesticated yeast species. The assimilation of this disaccharide seems to be a constant in yeasts associated with processes of degradation of lignocellulosic material in nature (Riley et al. 2016). However, the data available in the literature do not seem to indicate close phylogenetic relationship among species capable of assimilating cellobiose, i.e., this characteristic is not restricted to closely related taxa, not even to a single phylum within the kingdom Fungi (Molnárová et al. 2014). Indeed, when comparing the D1/D2 variable domains of 28S rRNA gene of cellobiose-assimilating species with those of yeasts unable to metabolize this sugar, we see that positive-phenotype yeasts do not necessarily cluster together, and that the cellobiose-assimilation capacity is randomly spread among the members of the order Saccharomycetales (Fig. 6). On the other hand, recently Shen et al. (2018) showed that this trait is inferred as being from the budding yeast common ancestor (BYCA) and that those budding yeasts which are not able to metabolize this disaccharide probably have lost such trait.
Phylogenetic tree assembled from the D1/D2 domains of 49 related yeast species, belonging to the order Saccharomycetales. Besides the two strains of the present study (UFFS-CE-3.6 and FLONA-CE-3.4) and the Candida pseudointermedia type strain, the following species were evaluated: Geotrichum cucujoidarum, Galactomyces reessii, Metschnikowia bicuspidate, Metschnikowia pulcherrima, Candida intermedia, Brettanomyces nanus, Brettanomyces bruxellensis, Magnusiomyces clavatus, Saturnispora zaruensis, Pichia fermentans, Pichia membranifaciens, Pichia manshurica, Groenewaldozyma salmanticensis, Sugiyamaella ligni, Trigonopsis vinaria, Lipomyces starkeyi, Lipomyces oligophage, Saccharomycopsis amapae, Saccharomycopsis synnaedendra, Spathaspora arborariae, Spathaspora passalidarum, Ambrosiozyma kamigamensis, Candida tropicalis, Meyerozyma guilliermondii, Meyerozyma caribbica, Scheffersomyces stipites, Nakazawaea wyomingensis, Cephaloascus fragrans, Peterozyma xylosa, Debaromyces hansenii, Candida queiroziae, Pachysolen tannophilus, Schwanniomyces pseudopolymorphus, Ascoidea tarda, Ascoidea rubescens, Saccharomyces cerevisiae, Hanseniaspora guilliermondii, Nadsonia fulvescens, Alloascoidea hylecoeti, Alloascoidea africana, Naumovozyma dairenensis, Tetrapisispora blattae, Tortispora caseinolytica, Kazachstania naganishii, Torulaspora delbrueckii, Saccharomycodes ludwigii, Wickerhamomyces anomalus and Wickerhamomyces queroliae. Accession numbers of the sequences used are shown to the right of species name. The numbers above each branch represent the branch lengths; the numbers below represent the bootstraps values. Bootstrap values were based on 1000 replications. In the assembled tree, yeasts capable of assimilating cellobiose are in bold. This feature was verified in the Westerdijk Fungal Biodiversity Institute (https://www.wi.knaw.nl/), with the exception of the following species, which have been checked in the literature: Geotrichum cucujoidarum (Suh and Blackwell 2006); Galactomyces reessii (Hoog et al. 1998); Metschnikowia bicuspidata (Miller and Phaff 1998); Magnusiomyces clavatus (Buchta et al. 2019); Pichia manshurica (Amoikon et al. 2018); Spathaspora arborariae (Cadete et al. 2009); Cephaloascus fragrans (Kurtzman and Hoog 2011); Debaromyces hansenii (Prillinger et al. 1999); Candida queiroziae (Santos et al. 2011); Ascoidea tarda, Ascoidea rubescens, Alloascoidea hylecoeti and Alloascoidea africana (Kurtzman and Robnett, 2013); and, Wickerhamomyces queroliae (Rosa et al. 2009). Yeasts labeled with *, ** and ***belong to CUG-Ser1, CUG-Ser2 and CUG-Ala clades, respectively (Shen et al. 2018)
Conclusion
The present study showed that, to ferment cellobiose, the analyzed C. pseudointermedia strains rely on permeases responsible for transport of this disaccharide through the plasma membrane and on high-affinity intracellular β-glucosidases, whose expression is highly induced in cellobiose-grown cells and strongly inhibited by glucose. Our data also suggest that cellobiose transport activity might be a limiting step for efficient fermentation of cellobiose in these yeasts. To the best of our knowledge, this is the first study to analyze disaccharide metabolization in C. pseudointermedia strains. We believe it contributes to the fund of knowledge of Brazilian microbiota.
References
Alves SL Jr, Herberts RA, Hollatz C, Miletti LC, Stambuk BU (2007) Maltose and maltotriose active transport and fermentation by Saccharomyces cerevisiae. J Am Soc Brew Chem 65:99–104. https://doi.org/10.1094/ASBCJ-2007-0411-01
Alves SL Jr, Herberts RA, Hollatz C, Trichez D, Miletti LC, de Araujo PS, Stambuk BU (2008) Molecular analysis of maltotriose active transport and fermentation by Saccharomyces cerevisiae reveals a determinant role for the AGT1 permease. Appl Environ Microbiol 74:1494–1501. https://doi.org/10.1128/AEM.02570-07
Alves SL Jr, Thevelein JM, Stambuk BU (2014) Expression of Saccharomyces cerevisiae α-glucoside transporters under different growth conditions. Braz J Chem Eng 31:1–8. https://doi.org/10.1590/S0104-66322014000100001
Alves SL Jr, Müller C, Camargo AF, Bonatto C, Scapini T, Fongaro G, Treichel H (2019) Bioprospecting Enzymes for 2G Ethanol through Metagenomics. In: Treichel H, Alves SL, Fongaro G, Müller C (eds) Ethanol as a green alternative fuel: insight and perspectives, 1st edn. Nova Science Publishers, Hauppauge, NY, pp 179–201
Amoikon TLS, Aké FM, Bakayoko M, Djéni TD, Mougo EBL, Djè MK (2018) Physiological profiles of indigenous yeasts isolated from raffia wine originated of côte d’ivoire. J Glob Biosci 7:5459–5474
Babič MN, Zalar P, Ženko B, Džeroski S, Gunde-Cimerman N (2016) Yeasts and yeast-like fungi in tap water and groundwater, and their transmission to household appliances. Fungal Ecol 20:30–39. https://doi.org/10.1016/j.funeco.2015.10.001
Baffi MA, Tobal T, Henrique J, Lago G, Leite RS, Boscolo M, Gomes E, Da-Silva R (2011) A novel β-glucosidase from Sporidiobolus pararoseus: characterization and application in winemaking. J Food Sci 76:997–1002. https://doi.org/10.1111/j.1750-3841.2011.02293.x
Barbosa AM, Giese EC, Dekker RF, Borsato D, Briones Pérez AI, Ubeda Iranzo JF (2010) Extracellular β-glucosidase production by the yeast Debaryomyces pseudopolymorphus UCLM-NS7A: optimization using response surface methodology. N Biotechnol 27:374–381. https://doi.org/10.1016/j.nbt.2010.05.013
Batista AS, Miletti LC, Stambuk BU (2004) Sucrose fermentation by Saccharomyces cerevisiae lacking hexose transport. J Mol Microbiol Biotechnol 8:26–33. https://doi.org/10.1159/000082078
Bazoti SF, Golunski S, Pereira Siqueira D, Scapini T, Barrilli ÉT, Alex Mayer D, Barros KO, Rosa CA, Stambuk BU, Alves SL Jr, Valério A, de Oliveira D, Treichel H (2017) Second-generation ethanol from non-detoxified sugarcane hydrolysate by a rotting wood isolated yeast strain. Bioresour Technol 244:582–587. https://doi.org/10.1016/j.biortech.2017.08.007
Buchta V, Bolehovská R, Hovorková E, Cornely OA, Seidel D, Žák P (2019) Saprochaete clavata invasive infections: a new threat to hematological-oncological patients. Front Microbiol 10:2196. https://doi.org/10.3389/fmicb.2019.02196
Cadete RM, Rosa CA (2018) The yeasts of the genus Spathaspora: potential candidates for second-generation biofuel production. Yeast 35:191–199. https://doi.org/10.1002/yea.3279
Cadete RM, Santos RO, Melo MA, Mouro A, Gonçalves DL, Stambuk BU, Gomes FC, Lachance MA, Rosa CA (2009) Spathaspora arborariae sp. nov., a d-xylose-fermenting yeast species isolated from rotting wood in Brazil. FEMS Yeast Res 9:1338–1342. https://doi.org/10.1111/j.1567-1364.2009.00582.x
Castresana J (2000) Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol 17:540–552. https://doi.org/10.1093/oxfordjournals.molbev.a026334
Casa-Villegas M, Polaina J, Marín-Navarro J (2018) Cellobiose fermentation by Saccharomyces cerevisiae: comparative analysis of intra versus extracellular sugar hydrolysis. Process Biochem 75:59–67. https://doi.org/10.1016/j.procbio.2018.09.005
Dereeper A, Audic S, Claverie JM, Blanc G (2010) BLAST-EXPLORER helps you building datasets for phylogenetic analysis. BMC Evol Biol 10:8. https://doi.org/10.1186/1471-2148-10-8
Dereeper A, Guignon V, Blanc G, Audic S, Buffet S, Chevenet F, Dufayard JF, Guindon S, Lefort V, Lescot M, Claverie JM, Gascuel O (2008) Phylogeny.fr: robust phylogenetic analysis for the non-specialist. Nucleic Acids Res 36:W465–W469. https://doi.org/10.1093/nar/gkn180
Duval EH, Alves SL Jr, Dunn B, Sherlock G, Stambuk BU (2010) Microarray karyotyping of maltose-fermenting Saccharomyces yeasts with differing maltotriose utilization profiles reveals copy number variation in genes involved in maltose and maltotriose utilization. J Appl Microbiol 109:248–259. https://doi.org/10.1111/j.1365-2672.2009.04656.x
Eliodório KP, Cunha GCGE, Müller C, Lucaroni AC, Giudici R, Walker GM, Alves SL Jr, Basso TO (2019) Advances in yeast alcoholic fermentations for the production of bioethanol, beer and wine. Adv Appl Microbiol 109:61–119. https://doi.org/10.1016/bs.aambs.2019.10.002
Feliciano RJ, Estilo EEC, Nakano H, Gabriel AA (2019) Ultraviolet-C resistance of selected spoilage yeasts in orange juice. Food Microbiol 78:73–81. https://doi.org/10.1016/j.fm.2018.10.003
Gancedo A (1998) Yeast carbon catabolite repression. Microbiol Mol Biol R 62:334–361
Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O (2010) New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol 59:307–321. https://doi.org/10.1093/sysbio/syq010
Guo Z, Duquesne S, Bozonnet S, Cioci G, Nicaud JM, Marty A, O'Donohue MJ (2015) Development of cellobiose-degrading ability in Yarrowia lipolytica strain by overexpression of endogenous genes. Biotechnol Biofuels 8:109. https://doi.org/10.1186/s13068-015-0289-9
Hahn-Hägerdal B, Karhumaa K, Larsson CU, Gorwa-Grauslund M, Görgens J, van Zyl WH (2005) Role of cultivation media in the development of yeast strains for large scale industrial use. Microb Cell Fact 4:31. https://doi.org/10.1186/1475-2859-4-31
Hande A, Mahajan S, Prabhune A (2013) Evaluation of ethanol production by a new isolate of yeast during fermentation in synthetic medium and sugarcane bagasse hemicellulosic hydrolysate. Ann Microbiol 63:63–70. https://doi.org/10.1007/s13213-012-0445-4
Hollatz H, Stambuk BU (2001) Colorimetric determination of activeα-glucoside transport in Saccharomyces cerevisiae. J Microbiol Methods 146:253–259. https://doi.org/10.1016/S0167-7012(01)00281-0
Hoog GS, Smith MTh, Gueho E (1998) Galactomyces redhead and Malloch. In: Kurtzman CP, Fell JW (eds) The yeasts: a taxonomic study, 4th edn. Elsevier, New York, pp 209–213
Iwahara S, Takegawa K, Kawaguchi K, Okamoto G (1993) The presence of trehalose-containing oligosaccharides in yeast extract. Biosci Biotechnol Biochem 57:1220–1221. https://doi.org/10.1271/bbb.57.1220
Kurtzman CP, Fell JW, Boekhout T, Robert V (2011) Methods for isolation phenotypic characterization and maintenance of yeasts. In: Kurtzman CP, Fell JW, Boekhout T (eds) The yeasts: a taxonomic study, 5th edn. Elsevier, NewYork, pp 87–110
Kurtzman CP, Hoog GS (2011) Cephaloascus Hanawa (1920). In: Kurtzman CP, Fell JW, Boekhout T (eds) The yeasts: a taxonomic study, 5th edn. Elsevier, New York, pp 341–344
Kurtzman CP, Robnett CJ (2013) Alloascoidea hylecoeti gen. nov., comb. nov., Alloascoidea africana comb. nov., Ascoidea tarda sp. nov., and Nadsonia starkeyi-henricii comb. nov., new members of the Saccharomycotina (Ascomycota). FEMS Yeast Res 13:423–432. https://doi.org/10.1111/1567-1364.12044
Kurtzman CP, Robnett CJ (1998) Identification and phylogeny of ascomycetous yeasts from analysis of nuclear large subunit (26S) ribosomal DNA partial sequences. Antonie Van Leeuwenhoek 73:331–371. https://doi.org/10.1023/A:1001761008817
Lachance MA, Bowles JM, Starmer WT, Barker JS (1999) Kodamaea kakaduensis and Candida tolerans, two new ascomycetous yeast species from Australian Hibiscus flowers. Can J Microbiol 45:172–177. https://doi.org/10.1139/w98-225
Lagunas R (1993) Sugar transport in Saccharomyces cerevisiae. FEMS Microbiol Rev 10:229–242. https://doi.org/10.1016/0378-1097(93)90598-v
Lefort V, Longueville JE, Gascuel O (2017) SMS: smart model selection in PhyML. Mol Biol Evol 34:2422–2424. https://doi.org/10.1093/molbev/msx149
Limtong S, Kaewwichian R (2015) The diversity of culturable yeasts in the phylloplane of rice in Thailand. Ann Microbiol 65:667–675. https://doi.org/10.1007/s13213-014-0905-0
Lineaweaver H, Burk D (1934) The determination of enzyme dissociation constants. J Am Chem Soc 56:658–666. https://doi.org/10.1021/ja01318a036
Liu D, Zhang R, Yang X, Zhang Z, Song S, Miao Y, Shen Q (2012) Characterization of a thermostable β-glucosidase from Aspergillus fumigatus Z5, and its functional expression in Pichia pastoris X33. Microb Cell Fact 11:25. https://doi.org/10.1186/1475-2859-11-25
Lopes MA, Lara CA, Moura MEF, Uetanabaro APT, Morais PB, Vital MJS, Rosa CA (2018) Characterisation of the diversity and physiology of cellobiose-fermenting yeasts isolated from rotting wood in Brazilian ecosystems. Fungal Biol 122:668–676. https://doi.org/10.1016/j.funbio.2018.03.008
Maciel NO, Piló FB, Freitas LF, Gomes FC, Johann S, Nardi RM, Lachance MA, Rosa CA (2016) The diversity and antifungal susceptibility of the yeasts isolated from coconut water and reconstituted fruit juices in Brazil. Int J Food Microbiol 160:201–205. https://doi.org/10.1016/j.ijfoodmicro.2012.10.012
Marques WL, Raghavendran V, Stambuk BU, Gombert AK (2016) Sucrose and Saccharomyces cerevisiae: a relationship most sweet. FEMS Yeast Res 16:107. https://doi.org/10.1093/femsyr/fov107
Masoud W, Cesar LB, Jespersen L, Jakobsen M (2004) Yeast involved in fermentation of Coffea arabica in East Africa determined by genotyping and by direct denaturating gradient gel electrophoresis. Yeast 21:549–556. https://doi.org/10.1002/yea.1124
Miller MW, Phaff HJ (1998) Metschnikowia Kamienski. In: Kurtzman CP, Fell JW (eds) The yeasts: a taxonomic study, 4th edn. Elsevier, New York, pp 256–267
Molnárová J, Vadkertiová R, Stratilová E (2014) Extracellular enzymatic activities and physiological profiles of yeasts colonizing fruit trees. J Basic Microbiol 54:S74–S84. https://doi.org/10.1002/jobm.201300072
Nakase T, Komagata K, Fukazawa Y (1976) Candida pseudointermedia sp. nov., isolated from "kamaboko", a traditional fish-paste product in Japan. J Gen Appl Microbiol 22:177–182. https://doi.org/10.2323/jgam.22.177
O'Donnell K (1993) Fusarium and its near relatives. In: Reynolds DR, Taylor JW (eds) The fungal holomorph: mitotic, meiotic and pleomorphic speciation in fungal systematic. CAB International, Wallingford, pp 225–233
Parisutham V, Chandran SP, Mukhopadhyay A, Lee SK, Keasling JD (2017) Intracellular cellobiose metabolism and its applications in lignocellulose-based biorefineries. Bioresour Technol 239:496–506. https://doi.org/10.1016/j.biortech.2017.05.001
Peter G, Dlauchy D, Tóbiás A, Fülöp L, Podgoršek M, Čadež N (2017) Brettanomyces acidodurans sp. nov., a new acetic acid producing yeast species from olive oil. Antonie Van Leeuwenhoek 110:657–664. https://doi.org/10.1007/s10482-017-0832-8
Prillinger H, Molnár O, Eliskases-Lechner F, Lopandic K (1999) Phenotypic and genotypic identification of yeasts from cheese. Antonie Van Leeuwenhoek 75:267–283. https://doi.org/10.1023/A:1001889917533
Putignani L, Del Chierico F, Onori M, Mancinelli L, Argentieri M, Bernaschi P, Coltella L, Lucignano B, Pansani L, Ranno S, Russo C, Urbani A, Federici G, Menichella D (2011) MALDI-TOF mass spectrometry proteomic phenotyping of clinically relevant fungi. Mol Biosyst 7:620–629. https://doi.org/10.1039/c0mb00138d
Reis AL, de Fátima R, de Souza R, Baptista Torres RR, Leite FC, Paiva PM, Vidal EE, de Morais MA (2014) Oxygen-limited cellobiose fermentation and the characterization of the cellobiase of an industrial Dekkera/Brettanomyces bruxellensis strain. Springerplus 3:38. https://doi.org/10.1186/2193-1801-3-38
Riley R, Haridas S, Wolfe KH, Lopes MR, Hittinger CT, Göker M, Salamov AA, Wisecaver JH, Long TM, Calvey CH, Aerts AL, Barry KW, Choi C, Clum A, Coughlan AY, Deshpande S, Douglass AP, Hanson SJ, Klenk HP, LaButti KM, Lapidus A, Lindquist EA, Lipzen AM, Meier-Kolthoff JP, Ohm RA, Otillar RP, Pangilinan JL, Peng Y, Rokas A, Rosa CA, Scheuner C, Sibirny AA, Slot JC, Stielow JB, Sun H, Kurtzman CP, Blackwell M, Grigoriev IV, Jeffries TW (2016) Comparative genomics of biotechnologically important yeasts. Proc Natl Acad Sci USA 113:9882–9887. https://doi.org/10.1073/pnas.1603941113
Ríos-Fránquez FJ, González-Bautista E, Ponce-Noyola T, Ramos-Valdivia AC, Poggi-Varaldo HM, García-Mena J, Martinez A (2017) Expression of a codon-optimized β-glucosidase from Cellulomonas flavigena PR-22 in Saccharomyces cerevisiae for bioethanol production from cellobiose. Arch Microbiol 199:605–611. https://doi.org/10.1007/s00203-016-1333-2
Rosa CA, Morais PB, Lachance MA, Santos RO, Melo WG, Viana RH, Bragança MA, Pimenta RS (2009) Wickerhamomyces queroliae sp. nov. and Candida jalapaonensis sp. nov., two yeast species isolated from Cerrado ecosystem in North Brazil. Int J Syst Evol Microbiol 59:1232–1236. https://doi.org/10.1099/ijs.0.006411-0
Rosa CA, Resende MA, Barbosa FAR, Morais PB, Franzot SP (1995) Yeast diversity in a mesotrophic lake on the karstic plateau of Lagoa Santa, MG-Brazil. Hydrobiologia 308:103–108. https://doi.org/10.1007/BF00007394
Saha BC, Bothast RJ (1996) Production, purification, and characterization of a highly glucose tolerant novel β-glucosidase from Candida peltata. Appl Environ Microbiol 62:3165–3170
Santangelo GM (2006) Glucose signaling in Saccharomyces cerevisiae. Microbiol Mol Biol Rev 70:253–282. https://doi.org/10.1128/MMBR.70.1.253-282.2006
Santos RO, Cadete RM, Badotti F, Mouro A, Wallheim DO, Gomes FC, Stambuk BU, Lachance MA, Rosa CA (2011) Candida queiroziae sp. nov., a cellobiose-fermenting yeast species isolated from rotting wood in Atlantic Rain Forest. Antonie Van Leeuwenhoek 99:635–642. https://doi.org/10.1007/s10482-010-9536-z
Shen XX, Opulente DA, Kominek J, Zhou X, Steenwyk JL, Buh KV, Haase MAB, Wisecaver JH, Wang M, Doering DT, Boudouris JT, Schneider RM, Langdon QK, Ohkuma M, Endoh R, Takashima M, Manabe RI, Čadež N, Libkind D, Rosa CA, DeVirgilio J, Hulfachor AB, Groenewald M, Kurtzman CP, Hittinger CT, Rokas A (2018) Tempo and mode of genome evolution in the budding yeast subphylum. Cell 175:1533–1545. https://doi.org/10.1016/j.cell.2018.10.023
Silveira MCF, Carvajal E, Bon EPS (1996) Assay for in vivo yeast invertase activity using NaF. Anal Biochem 238:26–28. https://doi.org/10.1006/abio.1996.0244
Srivastava N, Srivastava M, Mishra PK, Gupta VK, Molina G, Rodriguez-Couto S, Manikanta A, Ramteke PW (2018) Applications of fungal cellulases in biofuel production: advances and limitations. Renew Sust Energ Rev 82:2379–2386. https://doi.org/10.1016/j.rser.2017.08.074
Stambuk BU (1999) A simple experiment illustrating metabolic regulation: induction versus repression of yeast α-glucosidase. Biochem Educ 27:177–180. https://doi.org/10.1016/S0307-4412(98)00302-1
Suh SO, Blackwell M (2006) Three new asexual arthroconidial yeasts, Geotrichum carabidarum sp. nov., Geotrichum histeridarum sp. nov., and Geotrichum cucujoidarum sp. nov., isolated from the gut of insects. Mycol Res 110:220–228. https://doi.org/10.1016/j.mycres.2005.08.008
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 30:2725–2729. https://doi.org/10.1093/molbev/mst197
Villena MA, Iranzo JFU, Gundllapalli SB, Otero RRC, Pérez AIB (2006) Characterization of an exocellular β-glucosidase from Debaryomyces pseudopolymorphus. Enzyme Microb Technol 39:229–234. https://doi.org/10.1016/j.enzmictec.2005.10.018
Wallecha A, Mishra S (2003) Purification and characterization of two β-glucosidases from a thermo-tolerant yeast Pichia etchellsii. Biochim Biophys Acta 1649:74–84. https://doi.org/10.1016/S1570-9639(03)00163-8
Wang X, Liu ZL, Weber SA, Zhang X (2016) Two New Native β-Glucosidases from Clavispora NRRL Y-50464 Confer its dual function as cellobiose fermenting ethanologenic yeast. PLoS ONE 11:e0151293. https://doi.org/10.1371/journal.pone.0151293
Wang Y, Ren YC, Li Y, Hui FL (2015) Molecular phylogeny and taxonomy of Yamadazyma dushanensis f.a., sp. nov., a cellobiose-fermenting yeast species from China. Curr Microbiol 71:268–273. https://doi.org/10.1007/s00284-015-0847-1
White TJ, Bruns T, Lee S, Taylor J (1990) Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfand DH, Sninsky JJ, White TJ (eds) PCR protocols: a guide to methods and applications. Academic Press, San Diego, pp 315–322
Acknowledgements
The authors thank the National Council for Scientific and Technological Development (CNPq, Grant Numbers 454215/2014-2, 308627/2015-6 and 308389/2019-0) and Research and Innovation Support Foundation of Santa Catarina State (FAPESC, Grant Number 749/2016; T.O. 2016TR2188). ETB and VT are grateful to the Support Program for Scientific and Technological Initiation from Federal University of Fronteira Sul (PRO-ICT/UFFS), and CM to the Coordination for the Improvement of Higher Education Personnel (CAPES), for scholarships. The authors also thank R. Philip Lindeman, MD, PhD (Liberty Medical Communications, LLC) for language editing.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflict of interest.
Additional information
Communicated by Erko Stackebrandt.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Barrilli, É.T., Tadioto, V., Milani, L.M. et al. Biochemical analysis of cellobiose catabolism in Candida pseudointermedia strains isolated from rotten wood. Arch Microbiol 202, 1729–1739 (2020). https://doi.org/10.1007/s00203-020-01884-1
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00203-020-01884-1