Abstract
A Gram-positive, aerobic, nonmotile strain, NM2E3T was identified as Brevibacterium based on the 16S rRNA gene sequence analysis and had the highest similarities to Brevibacterium jeotgali SJ5-8T (97.3 %). This novel bacterium was isolated from root tissue of Prosopis laegivata grown at the edge of a mine tailing in San Luis Potosí, Mexico. Its cells were non-spore-forming rods, showing catalase and oxidase activities and were able to grow in LB medium added with 40 mM Cu2+, 72 mM As5+ and various other toxic elements. Anteiso-C15:0 (41.6 %), anteiso-C17:0 (30 %) and iso-C15:0 (9.5 %) were the major fatty acids. MK-8(H2) (88.4 %) and MK-7(H2) (11.6 %) were the major menaquinones. The DNA G + C content of the strain NM2E3T was 70.8 mol % (Tm). DNA–DNA hybridization showed that the strain NM2E3T had 39.8, 21.7 and 20.3 % relatedness with B. yomogidense JCM 17779T, B. jeotgali JCM 18571T and B. salitolerans TRM 45T, respectively. Based on the phenotypic and genotypic analyses, the strain NM2E3T (=CCBAU 101093T = HAMBI 3627T = LMG 8673T) is reported as a novel species of the genus Brevibacterium, for which the name Brevibacterium metallicus sp. nov., is proposed.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The genus Brevibacterium, a member of the family Brevibacteriaceae of the phylum Actinobacteria, was first proposed by Breed (1953) with Brevibacterium linens as the type species. This species was later emended by Collins et al. (1977). Brevibacterium strains have been isolated from diverse environments such as paintings (Heyman et al. 2004), brown alga (Ivanova et al. 2004), deep sea sediment (Bhadra et al. 2008), sea water (Lee 2008), saline soil (Tang et al. 2008), human skin (Roux and Raoult 2009), moth caterpillars (Kati et al. 2010), indoor wall (Kämpfer et al. 2010), salt lakes (Guan et al. 2010), poultry manure (Tonouchi et al. 2013), river water (Kumar et al. 2013) and activated sludge (Cui et al. 2013; Kim et al. 2013) (LSPN bacterio net; http://www.bacterio.net/). Currently, the genus is comprised of 49 species, including the recently described species Brevibacterium jeotgali (Choi et al. 2013).
In a study about the diversity of endophytic bacteria associated with endemic plants grown in a heavy metal contaminated region in Mexico, a strain NM2E3T was isolated from Prosopis laegivata root tissue. This bacterium is able to grow in Luria–Bertani (LB) medium supplied with high concentration of heavy metals, such as 40 mM Cu2+, 2 mM Zn2+, 12.5 mM Pb2+, 5.8 mM As3+ and 72 mM As5+ (our unpublished data). In the present research, the taxonomic status of the strain NM2E3T is described using a polyphasic approach. Taking into account the results, this strain represents to a novel species within the genus Brevibacterium, and the name Brevibacterium metallicus sp. nov, is proposed. The type strain is NM2E3T (=CCBAU 101093T = HAMBI 3627T = LMG 8673T).
Materials and methods
Bacterial isolation and maintenance
To isolate the endophytic bacteria, 1 g of thin roots (<0.5 mm in diameter) sampled from Prosopis laegivata grown at the edge of a mine tailing at Villa de la Paz in the state of San Luis Potosi (23.7 N, 178.7 W) in Mexico was surface sterilized according to Marquez-Santacruz et al. (2010). Subsequently, 1 g of the sample was macerated in 9 mL of 0.85 % NaCl under aseptic conditions (Barzanti et al. 2007). Aliquots (100 µL) of the tissue extracts (10−1 a 10−3) were plated in triplicate on tryptic soya agar (TSA, Difco) (Gardner et al. 1982). The inoculated medium was incubated at 28 °C for one to 14 days (Sun et al. 2010). Colonies were selected according to their morphology and purified by repeatedly streaking on the same medium. Pure colonies were subcultured and maintained on TSA medium at 4 °C and stored at −70 °C in a liquid medium supplied with 50 % (w/v) of glycerol. To confirm that the disinfection process was successful, aliquots of 100 µL of the sterilized water used in the final rinse during the surface sterilization was inoculated on the same medium and incubated under the same conditions.
Sequencing and phylogenetic analysis of 16S rRNA gene
This analysis was performed for 60 endophytic isolates to scan their taxonomic affinities. Genomic DNA was extracted for each isolate from 5 ml of culture in TS broth incubated for 18 h at 28 °C with shaking (150 rpm), using the protocol of Zhou et al. (1995). Briefly, cells were lysed with 50 mg mL−1 of lysozyme, 70 μL of 10 % (v/w) SDS in 0.1 × TE and 10 mg mL−1 of proteinase K to avoid the difficulty of extraction of DNA from the Gram-positive bacteria. The amplified 16S rRNA gene using the DNA extract as template and the primers fD1 and rD1 (Weisburg et al. 1991) was sequenced according to the protocols described by Sun et al. (2010). The acquired sequences were compared with those in the GenBank database using the program BLAST on the Web site http://blast.ncbi.nlm.nih.gov/Blast.cgi (Altschul et al. 1997). The 16S rRNA gene sequences obtained in this study were aligned using CLUSTAL X (2.0) software (Thompson et al. 1997) together with the closely related sequences extracted by BLAST searching and were manually edited with SEAVIEW software (Galtier et al. 1996). Phylogenetic relationships were estimated by neighbor-joining (NJ) (Saitou and Nei 1987) and the maximum parsimony (MP) algorithms using MEGA 5.2 software (Tamura et al. 2011). In addition, a maximum-likelihood (ML) (Rogers and Swofford 1999) analysis was performed using the PhyML program on the Web site http://www.atgc-montpellier.fr/phyml (Guideon and Gascuel 2003). The MODELTEST 3.06 software (Darriba et al. 2012) was used to select the appropriate model of sequence evolution by the Akaike information criterion (AIC), and the GTR + I + G model (α = 0.5870 for the gamma distribution and p-inv = 0.6480) was chosen. The robustness of the nodes was evaluated with bootstrap analysis using 1000 pseudoreplicates in NJ, MP methods and Shimodaira–Hasegawa (SH)-like test (Guindon et al. 2010). ML tree was visualized with the program MEGA version 5.2 (Tamura et al. 2011). Similarities among sequences were calculated using the MatGAT v.2.01 software (Campanella et al. 2003).
DNA relatedness estimation
Based on the low 16S rRNA similarities with their nearest neighbors, a whitish-yellow pigmented colony coded as NM2E3T was selected for further analyses, including the estimation of DNA relatedness in comparison with the most related type strains Brevibacterium jeotgali JCM 18571T and Brevibacterium yomogidense JCM 17779T (purchased from Japan Collection of Microorganisms), as well as Brevibacterium salitolerans TRM 415T (offered by Dr. Tongwei Guan, Xihua University, Chengdu, China).
The genomic DNA was extracted following the protocol described by Marmur (1961), and the DNA relatedness between NM2E3T and the type strains for related Brevibacterium species was performed according to protocols described by De Ley et al. (1970) using the Spectrophotometer Lambda 35 equipped with PTP-1 temperature control (PerkinElmer Inc, USA), and DNA G + C composition of the strain NM2E3T was determined using a thermal denaturation method (Mandel and Marmur 1968).
Morphological, physiological and biochemical characterization
Cell morphology was observed on well-separated colonies grown on TSA at 28 °C for 3 days. Cell motility was assayed by development of turbidity in test tube containing semisolid TSA medium (0.4 % agar), after 2 days of incubation at 28 °C (Cowan and Steel 1965). Cell dimension and appearance were observed by using a Hitachi S-3400 scanning electron microscope (SEM), and the sample was prepared using the protocol of Jiao et al. (2015). Bacterial growth was tested at 28 °C on LB agar, nutrient agar (NA), International Streptomyces Project (ISP) 1 medium (Pridham and Gottlieb 1948), ISP 4 medium (Shirling and Gottlieb 1966), Czapek’s agar and ISP 5 medium (Pridham and Lyons 1961). Diffusible pigments of strain NM2E3T were analysed on TSA, LB agar, NA, ISP 1 medium, ISP 4 medium and Czapek’s agar, and growth abilities were tested on LB agar and/or ISP 4 plates at different temperatures (4, 10, 20, 28 and 37 °C); at different pH values (5.0–12.0 with 1 pH unit increments and adjusted with 1 mol/L HCl or 1 mol/L NaOH after autoclaving); and with different salinities (NaCl 0, 1, 3, 5, 7, 10, 15 and 20 %). The plates were incubated during 3, 5, 7, 15 and 21 days at 28 °C, except for temperature testing. Antibiotics resistance for the strain NM2E3T and reference species was determined on LB agar medium added with (μg mL−1) tetracycline hydrochloride (5), neomycin sulfate (30), kanamycin sulfate (30), streptomycin sulfate (10), nalidixic acid (30), carbenicillin disodium salt (100), chloramphenicol (30), ampicillin disodium salt (10) and gentamicin sulfate (10). Hydrolysis of starch, Tween 20, Tween 80, and urease, growth on Simmons’s citrate, production of hydrogen sulfide, reduction of nitrate and nitrite, Voges–Proskaüer test, catalase and oxidase activities were tested using the protocols described by Cowan and Steel (1965). Assimilation of various carbon compounds was tested using the Biolog GP2 MicroPlates system according to the manufacturer’s instructions.
Heavy metal resistance assay
The minimum inhibition concentration (MIC), defined as the lowest concentration of heavy metals that completely inhibits the growth of bacteria, was determined in triplicate for the strain NM2E3T by streaking it on LB medium plates supplemented separately with heavy metals Cu2+ (2–40 mM CuSO4), Zn2+ (2–22 mM ZnSO4), As5+ (3–480 mM NaH2AsO4) and As3+ (5.8–28.8 mM NaAsO2). In the case of Pb, the medium proposed by Castañeda-Argullo (1956) was used and the Pb at concentrations of 3.12–20 mM [Pb(NO3)2] was supplied (Luo et al. 2011). Plates were incubated at 28 °C and growth was observed after 4 days of incubation.
Chemotaxonomic analyses
For analysis of fatty acid profile, strain NM2E3T and the related reference strains were cultured on TSA, except of B. jeotgali JCM 18571T which was cultivated on LB agar. All the strains were incubated at 28 °C for 72 h. Preparation and analysis of fatty esters were performed as described by Sasser (2001) using the Microbial Identification System (MIDI Inc.) with the Microbial Identification software package (Sherlock version 6.0; MIDI database: RTSBA6) (Tighe et al. 2000). To identify menaquinones, freezing dried cell mass was prepared from the 3-day culture of strain NM2E3T in TSB at 28 °C. The respiratory quinone was isolated, purified and identified from the strain NM2E3T according to the procedure described by Komagata and Suzuki (1987). The cell-wall peptidoglycan was prepared by using a method described previously (Komagata and Suzuki 1987). Amino acid compositions were determined using an automatic amino acid analyzer (model L-8900; Hitachi) equipped with a Hitachi custom ion exchange resin.
Results and discussion
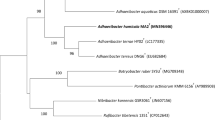
In the phylogenetic analysis of the 16S rRNA gene sequences (c.a. 1480 nucleotides), the 60 isolates were divided into 25 genomic species belonging to Arthrobacter, Bacillus, Brevibacterium, Kocuria, Leucobacter, Microbacterium, Micrococcus, Nocardiopsis, Pseudomonas and Staphylococcus (data not shown). Among these isolates, NM2E3T was clustered within the genus Brevibacterium as shown in the simplified NJ phylogenetic tree of 16S rRNA genes (Fig. 1, complete tree for all the Brevibacterium species available as Supplementary Fig. S1). This strain exhibited similarities of 97.3 % with Brevibacterium jeotgali JCM 18571T, 96.6 % with Brevibacterium yomogidense JCM 17779T and 96.1 % with Brevibacterium salitolerans TRM415T. This phylogenetic relationship was supported by the MV and MP trees (available as Supplementary Figs. S2 and S3), suggesting that the strain NM2E3T represents a possible novel species.
The DNA–DNA relatedness between the strain NM2E3T and B. yomogidense JCM 17779T, B. jeotgali JCM 18571T and B. salitolerans TRM 45T was 39.9, 21.7 and 20.3 %, respectively, which were significantly lower than the threshold (70 %) generally established for the species delineation (Wayne et al. 1987). These results lead to the conclusion that the strain NM2E3T is a novel genomic species within the genus Brevibacterium. The DNA G + C content of the strain NM2E3T was 70.8 mol % (Tm), within the range of Brevibacterium species (62.1–71.7 mol %).
Strain NM2E3T formed colonies on TSA medium after 48 h with whitish-yellow color, smooth surface, circular shape, convex elevation, and entire edge and diameter of 1.5–3.0 mm. Cells are aerobic, Gram-positive, nonmotile and non-spore-forming rods (0.320–0.429 μm in width and 0.794–1.5 μm in length) (see Supplementary Fig. S4). The differential characteristics of strain NM2E3T and the related type strains are given in Table 1. Briefly, this strain can be differentiated from the type strains of the related species based on its utilization of d-arabitol, arbutin, d-cellobiose, d-galactose, l-pyroglutamic acid, and its inability to use l-serine, thymine, as well as its resistance to streptomycin at 10 µg mL−1. The results of both the phylogenetic and DNA relatedness analyses make it evident that NM2E3T represents a novel Brevibacterium species.
The MIC values for strain NM2E3T were 12.5 mM to Pb2+, 40 mM to Cu2+, 72 mM to As5+, 2 mM to Zn2+ and 5.8 mM to As3+. Previously, the resistances to arsenic (1 mM) in Brevibacterium linens AE038-8 (Maizel et al. 2015), to lead, mercury and cadmium in Brevibacterium iodium (De et al. 2008), and to copper (1.57–17.3 mM) in Brevibacterium antarcticum (Tashyreva et al. 2009) have been reported, and the latter bacterium could extract 11–75 % of Cu2+ from the environment depending on cultivation parameters and copper concentration in the medium. Compared with the results in previous studies, strain NM2E3T showed much greater resistance to various heavy metals and to arsenic.
Strain NM2E3T contained MK-8(H2) (88.4 %) and MK-7(H2) (11.6 %) as predominant and minor menaquinones for respiration and 2,6-meso-diaminopimelic acid, l-glutamic acid and l-alanine in its peptidoglycan of the cell wall (Table 1), which matched the genus properties (Cai and Collins 1994; DSMZ 2001; Ivanova et al. 2004; Kim et al. 2013). The fatty acid composition of strain NM2E3T was characterized by predominant amounts of anteiso-C15:0 (41.6 %), anteiso-C17:0 (30 %) and iso-C15:0 (9.5 %). Similar values are also found in the closely related Brevibacterium species (Table 2). The presence of 17:1 ω6c (0.6 %) in strain NM2E3T differentiates it from the closely related species. All of these compounds have been reported as the major fatty acids in the genus Brevibacterium (Kim et al. 2013).
Based on these results, we classify the strain NM2E3T as a novel species within the genus Brevibacterium and name it Brevibacterium metallicus sp. nov.
Description of Brevibacterium metallicus sp. nov
Brevibacterium metallicus (me.tal.li’cus. N. L. masc. adj. metallicus referring to the fact that the bacterium is resistant to various heavy metals).
Cells are Gram-positive, aerobic, nonmotile, non-spore-forming rods with size of 0.32–0.43 μm in width and 0.79–1.5 μm in length. Colonies are whitish-yellow, smooth, circular, convex, with entire borders and 1.5–3.0 mm in diameter after 48 h of incubation on TSA medium at 28 °C. On ISP 4 medium, growth temperature range is 20–28 °C with an optimum of 28 °C and growth salinity range is 0–10 %, with optimum of 1 % (w/v). On LB agar medium, the strain grows well at 10–37 °C with optimum of 28–37 °C, at initial pH range 6–12 with 10–11 as optimum, and in the presence of 0–15 % (w/v) NaCl. NM2E3T grows on LB, TSA, nutritive agar, ISP1, ISP4, ISP 5 and Czapek’s agar and produces a brown diffusible pigment in TSA medium after 2 weeks of incubation at 4 °C. It is resistant to (μg/mL) streptomycin (10), nalidixic acid (30) and tetracycline (5) and is susceptible to carbenicillin (100), chloramphenicol, neomycin and kanamycin (30), ampicillin and gentamycin (10). Oxidase and catalase are positive. Nitrate is reduced to nitrite. Starch and Tween 20 are not hydrolyzed. Urease, assimilation of citrate, H2S production, Vogues–Proskaüer test and nitrite reduction are negative. With the GP2 system, the strain NM2E3T is able to assimilate β-cyclodextrin, Tween 40, Tween 80, l-arabinose, d-arabitol, arbutin, d-cellobiose, d-galactose, ribose, d-xylose, acetic acid, α-hydroxybutyric acid, β-hydroxybutyric acid, α-ketoglutaric acid, α-ketovaleric acid, lactamide, l-lactic acid, l-malic acid, l-pyruvic acid methyl ester, succinic acid monomethyl ester, propionic acid, succinamic acid, l-pyroglutamic acid, l-asparagine, l-alanylglycine, l-alanine, l-alaninamide, pyruvic acid, succinic acid, propionic acid and 2,3-butanodiol, whereas assimilation of gentibiose, α-d-lactose, lactulose, maltose, maltotriose, d-mannitol, d-melezitose, d-melobiose, β-methyl-d-glucose, α-methyl-d-mannoside, palatinose, d-psicose, d-raffinose, l-rhamnose, d-trehalose, ρ-hydroxyphenylacetic acid, putrisine, uridine, uridine-5′monophosphate, α-d-glucose-1-phosphate and d,l-α-glycerolphosphate is negative. In addition, dextrin, glycogen, d-fructose, α-d-glucose, m-inositol, d-mannose, 3-methyl-d-glucose, salicin, d-sorbitol, d-tagatose, xylitol, γ-hydroxybutyric acid, d-lactic acid methyl ester, N-acetyl-l-glutamic acid, d-alanine, glycerol, adenosine, 2´-deoxyadenosine, inosine and adenosine 5´-phosphate are also utilized by the test strain, but the purple color was only developed slightly compared with the compounds mentioned above.
The major fatty methyl esters are anteiso-C15:0 (41.6 %), anteiso-C17:0 (30 %) and iso-C15:0 (9.5 %). The major respiratory quinone is MK-8 (H2) and a minor amount of MK-7 (H2) is also present. Cell-wall diamino acids are 2,6-meso-diaminopimelic acid, l-glutamic acid and l-alanine. The strain NM2E3T presents 70.8 mol % (Tm) of DNA G + C content. The type strain NM2E3T (=CCBAU 101093T = HAMBI 3627T = LMG 8673T) was isolated from the root of Prosopis laegivata grown at the edge of a mine tailing in Villa de la Paz in the state of San Luis Potosi (23.7 N, 178.7 W) in Mexico.
References
Altschul SF, Madden TL, Shäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ (1997) Gapped BLAST and PSI–BLAST: a new generation of protein database search programs. Nucleic Acids Res 25:3389–3402
Barzanti R, Ozino F, Bazzicalupo M, Gabbrielli R, Galardi F, Gonnelli C, Mengon A (2007) Isolation and characterization of endophytic bacteria from the nickel hyperaccumulator plant Alyssum bertolonii. Microb Ecol 53:306–316
Bhadra B, Raghkumar C, Pindi PK, Shivaji S (2008) Brevibacterium oceani sp. nov., isolated from deep-sea sediment of the Chagos Trench, Indian Ocean. Int J Syst Evol Microbiol 58:57–60
Breed RS (1953) The Brevibacteriaceae fam. nov. of order Eubacteriales. Rias Commun VI Congr Int Microbiol Roma 1:13–14
Cai J, Collins MD (1994) Phylogenetic analysis of species of the meso-diaminopimelic acid-containing genera Brevibacterium and Dermabacter. Int J Syst Bacteriol 44:583–585
Campanella JJ, Bitincka L, Smalley J (2003) MatGAT: an application that generates similarity/identity matrices using protein or DNA sequences. BMC Bioinform 4:29
Castañeda-Argullo MS (1956) Studies on biosynthesis of extracellular protease by bacteria. J Gen Physiol 89:369–373
Choi EJ, Lee SH, Jung JY, Jeon CO (2013) Brevibacterium jeotgali sp. nov., isolated from jeotgal a traditional Korean fermented seafood. Int J Syst Evol Microbiol 9:3430–3436
Collins MD, Pirouz T, Goodfellow M, Minnikin DE (1977) Distribution of menaquinones in actinomycetes and corynebacteria. J Gen Microbiol 100:221–230
Cowan ST, Steel KJ (1965) Manual for the identification of medical bacteria. Cambridge University Press, London
Cui Y, Kong MS, Woo SG, Jin L, Kim KK, Park J, Lee M, Lee JT (2013) Brevibacterium daeguense sp. nov., a nitrate-reducing bacterium isolated from 4-chlorophenol enrichment culture. Int J Syst Evol Microbiol 63:152–157
Darriba D, Taboada GL, Doallo R, Posada D (2012) jModelTest 2: more models, new heuristics and parallel computing. Nat Methods 30:772
De Ley J, Cattoir H, Reynaerts A (1970) The quantitative measurement of DNA hybridization from renaturation rates. Eur J Biochem 12:133–142
De J, Ramaiah N, Vardanyan L (2008) Detoxification of toxic heavy metals by marine bacteria highly resistant to mercury. Mar Biotechnol 10:471–477
DSMZ (2001) Catalogue of Strains, 7th edn. Deutsche Sammlung von Mikroorganismen und Zellkulturen, Braunschweig
Galtier N, Gouy M, Gautier C (1996) SEAVIEW and PHYLO_WIN: two graphic tools for sequence alignment and molecular phylogeny. Comput Appl Biosci 12:543–548
Gardner JM, Feldman AW, Zablotowicz RM (1982) Identity and behavior of xylem-residing bacteria in rough lemon roots of Florida citrus trees. Appl Environt Microbiol 43:1335–1342
Guan TW, Zhao K, Xiao J, Liu Y, Xia ZF, Zhang XP, Zhang LL (2010) Brevibacetrium salitolerans sp. nov., an antinobacterium isolated from salt-lake sediment. Int J Syst Evol Microbiol 60:2991–2995
Guideon S, Gascuel O (2003) A simple and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol 52:696–704
Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Hordijk W, Gascuel O (2010) New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of phyml 3.0. Syst Biol 59:307–321
Heyman J, Verbeeren J, Schumann P, Devos J, Swings J, Devos P (2004) Brevibacterium picturae sp. nov., isolated from damaged mural painting at the Saint-Catherine Chapel (Castle Herbersten, Austria). Int J Syst Evol Microbiol 54:1537–1541
Ivanova EP, Christen R, Alexeeva YV, Zhukova NV, Gorshkova NM, Lysenko AM, Mikhailov VV, Nicolau DV (2004) Brevibacterium celere sp. nov., isolated from degraded Thallus of brown alga. Int J Syst Evol Microbiol 54:2107–2111
Jiao YS, Yan H, Ji ZJ, Li YH, Sui XH, Wang ET, Guo BL, Chen WX, Chen WF (2015) Rhizobium sophorae sp. nov. and Rhizobium sophoriradicis sp. nov. nitrogen-fixing rhizobial symbionts of medicinal legume Sophora flavences in China. Int J Syst Ecol Microbiol 65:497–503
Kämpfer P, Schäfer J, Lodders N, Busse HJ (2010) Brevibacterium sandarakium sp. nov., isolated from wall of indoor environment. Int J Syst Evol Microbiol 60:909–1013
Kati H, Ince IA, Demir I, Demirbag Z (2010) Brevibacterium pityocampae sp. nov., isolated from caterpillars of Thaumetopoea pityocampa (Lepidoptera, Thaumetopoeidae). Int J Syst Evol Microbiol 60:312–316
Kim J, Srinivason S, You T, Bang JJ, Park S, Lee SS (2013) Brevibacterium ammoniilyticum sp. nov, an ammonia-degrading bacterium isolated from sludge of a wastewater treatment plant. Int J Syst Evol Microbiol 63:1111–1118
Komagata K, Suzuki KI (1987) Lipid and cell wall analysis in bacteria systematics. Methods Microbiol 19:1–207
Kumar A, Ince I, Kati A, Chakrabarty R (2013) Brevibacterium siliguriense sp. nov., a facultatively oligotrophic bacterium isolated from river water. Int J Syst Evol Microbiol 63:511–515
Lee SD (2008) Brevibacterium marinum sp. nov., isolated from seawater. Int J Syst Evol Microbiol 58:500–504
Luo S, Wan Y, Xiao X, Guo H, Chen L, Xi Q, Zeng G, Liu C, Chen J (2011) Isolation and characterization of endophytic bacterium LRE07 from cadmium hyperaccumulator Solanum nigrum L. and its potential for remediation. Appl Microbiol Biotechnol 89:1637–1644
Maizel D, Utturkar SM, Brown SD, Ferrero MA, Rosen BP (2015) Draft genome sequence of Brevibacterium linens AE038-8, an extremely arsenic resistant bacterium. Genome Announc 3 pii: e00316–15
Mandel M, Marmur J (1968) Use of ultraviolet absorbance temperature profile for determining the guanine plus cytosine content. Methods Enzymol 12B:195–206
Marmur J (1961) A procedure for isolation of deoxyribonucleic acid from microorganisms. J Mol Biol 3:208–218
Marquez-Santacruz HA, Hernández-León R, Orozco-Mosqueda MC, Velázquez-Sepulveda I, Santoyo G (2010) Diversity of bacterial endophytes in roots of Mexican husk tomato plants (Physalis ixocarpa) and their detection in the rhizosphere. Gen Mol Res 9:2372–2380
Pridham TG, Gottlieb D (1948) The utilization of carbon compounds by some actinomycetales as an aid for species determination. J Bacteriol 56:107–114
Pridham TG, Lyons AJ Jr (1961) Streptomyces albus (Rossi-Daria) Waksman et Henria: taxonomic study of strains labeled Streptomyces albus. J Bacteriol 81:431–441
Rogers J, Swofford D (1999) Multiple local maxima for likelihoods of phylogenetic trees: a simulation study. Mol Biol Evol 16:1079–1085
Roux N, Raoult D (2009) Brevibacterium massiliense sp. nov., isolated from a human ankle discharge. Int J Syst Evol Microbiol 59:1960–1964
Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425
Sasser M (2001) Identification of bacteria by gas chromatography of cellular fatty acids. MIDI Tech Note 101:1–6
Shirling EB, Gottlieb D (1966) Methods for characterization of Streptomyces species. Int J Syst Bacteriol 16:313–340
Sun LN, Zhang YF, He LY, Chen ZJ, Wang QY, Quian M, Shen XF (2010) Genetic diversity and characterization of heavy metal-resistant-endophytic bacteria from two copper-tolerant plant species on copper mine wasteland. Biores Technol 101:501–509
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28:2731–2739
Tang SK, Wang Y, Schumann P, Stackebrandt E, Lou K, Jiang CL, Xu LH, Li WJ (2008) Brevibacterium album sp. nov., a novel actinobacterium isolated from a saline soil in China. Int J Syst Evol Microbiol 58:574–577
Tashyreva HO, Iutyns’ka HO, Tashyrev OB (2009) Effect of cultivation parameters of antarctic strains Enterobacter hormaechei and Brevibacterium antarcticum on resistant to copper(II) ions. Mikrobiol Z 71:3–8
Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins D (1997) The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 25:4876–4882
Tighe S, De Lajudie P, Dipietro K, Lindström K, Nick G, Jarvis B (2000) Analysis of cellular fatty acids and phenotypic relationships of Agrobacterium, Bradyrhizobium, Mesorhizobium, Rhizobium and Sinorhizobium species using the Sherlock Microbial Identification System. Int J Syst Evol Microbiol 50:787–801
Tonouchi A, Kimura K, Fujita T (2013) Brevibacterium yomogidense sp. nov., isolated from a soil conditioner made from poultry manure. Int J Syst Evol Microbiol 63:516–520
Wayne LG, Brenner DJ, Colwell RR, Grimont PAD, Kandler O, Krichevsky MI, Moore LH, Moore WEC, Murray RGE, Stackebrant E, Starr HG, Trüper G (1987) International committee on systematic bacteriology. Report of the adhoc committee on reconciliation of approaches to bacterial systematics. Int J Syst Bacteriol 37:463–464
Weisburg WG, Barns SM, Pelletior DA, Lane DJ (1991) 16S ribosomal DNA amplification for phylogenetic study. J Bacteriol 173:697–703
Zhou J, Bruns MA, Tuedje J (1995) DNA recovery from soils of diverse composition. Appl Environ Microbiol 62:316–322
Acknowledgments
This research was funded with Projects SIP-IPN 20130722 and 20130828. B.R.P. received scholarships support from the CONACyT and BEIFI. M.S.V.M., P.E.dl.S. and E.T.W. appreciate the scholarships funded by COFAA and EDI-IPN and SNI-CONACyT. This research was partially supported by China Natural Foundation (No. 31270052) to W. F. C.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare they have no competing interests and they were notified about the content of the manuscript.
Human and animal rights statement
No humans or animals were used in studies for this article.
Additional information
Communicated by Erko Stackebrandt.
The accession number of the 16S rRNA gene for the type strain NM2E3T in GenBank is KM874400.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Román-Ponce, B., Li, Y.H., Vásquez-Murrieta, M.S. et al. Brevibacterium metallicus sp. nov., an endophytic bacterium isolated from roots of Prosopis laegivata grown at the edge of a mine tailing in Mexico. Arch Microbiol 197, 1151–1158 (2015). https://doi.org/10.1007/s00203-015-1156-6
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00203-015-1156-6