Abstract
A novel aerobic bacterium, strain HT23T, able to grow on 500 mM sodium arsenate was isolated from a hot-spring sediment sample collected from Athamallik, Orissa, India. Cells of this isolate were Gram negative. Heterotrophic growth was observed at pH 6.0–11.0 and 20–45 °C. Optimum growth was observed at 37 °C and pH 7.0–10.0. The major polar lipids are diphosphatidyl glycerol, phosphatidyl glycerol, phosphatidyl ethanolamine, phosphatidyl choline and phosphatidyl monomethyl ethanolamine. The major isoprenoid quinone was Q-10. 16S rRNA gene sequence analysis indicated that the bacterium clustered with the genus Pannonibacter and showed 98.9 % similarity with Pannonibacter phragmitetus C6-19T (DSM 14782T) and 98 % with the P. phragmitetus group B and P. phragmitetus group E strains. Levels of DNA–DNA relatedness between the strain HT23T and P. phragmitetus C6-19T (DSM 14782T) and other strains of P. phragmitetus group B and group E strains were below 55 %. On the basis of phenotypic and chemotaxonomic characteristics, 16S rRNA gene sequence analysis and DNA–DNA hybridization data, strain HT23T is considered to represent a novel species of the genus Pannonibacter, for which the name Pannonibacter indica sp. nov. is proposed. The type strain is HT23T (=JCM 16851T = DSM 23407T = LMG 25769T).
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Exploration of bacterial species in hot springs is of importance owing to the intriguing biogeochemistry and extreme conditions of these environments. Over the past years, microbiological studies based on molecular and cultural approaches revealed a high phylogenetic and physiological diversity of microorganisms within terrestrial thermal springs (Hall et al. 2008). The vast majority of bacterial species belonging to the most recent lines of descent, such as the Proteobacteria, are mesophilic, although slightly, moderately and even extremely thermophilic species have been described. Besides many mesophilic species, Alphaproteobacteria includes a few slightly and moderately thermophilic species, namely Rubritepida flocculans (Alarico et al. 2002), Phenylobacterium lituiforme (Kanso and Patel 2004), Acidicaldus organivorus (Johnson et al. 2006), Nesiotobacter exalbescens (Donachie et al. 2006) and Rubellimicrobium thermophilum (Denner et al. 2006).
The genus Pannonibacter was proposed by Borsodi et al. (2003), and at the time of writing, the genus comprised only one recognized species, P. phragmitetus, of which the type strain is C6-19T (Borsodi et al. 2003). Subsequent studies showed that the type strain P. phragmitetus C6-19T is phenotypically atypical due to its inability to produce acid from sucrose and salicin as opposed to other P. phragmitetus strains (Holmes et al. 2006). Detailed studies on 16S rRNA gene sequence analysis, G+C content, DNA–DNA homology, fatty acid composition and biochemical characterization revealed that the Achromobacter groups B, Achromobacter groups E and P. phragmitetus C6-19T belong to the Alphaproteobacteria (Borsodi et al. 2003; Holmes et al. 2006). The description of the genus Pannonibacter was emended by Biebl et al. (2007) with respect to its lipid pattern and fatty acid profile.
In this study, we describe a novel bacterium (strain HT23T) isolated from a tropical hot spring at Athamallik, Orissa, India, which is able to grow in the presence of 500 mM sodium arsenate. It has a phenotypic, chemotaxonomic and phylogenetic characteristic that allows its assignment to a novel species within the genus Pannonibacter for which the name Pannonibacter indica sp. nov. is proposed.
Materials and methods
Isolation and phenotypic characterization of bacteria
Samples were collected from the hot spring at Athamallik, Orissa, India. The surface temperature of the sediment sample was 43 °C and the pH was 7.4. Sediment samples were transported to the laboratory without temperature control, and subsequently, 5.0 g (wet weight) sediment sample was transferred into a 250-ml conical flask containing 50 ml nutrient broth pH 7.2 (Difco) and incubated on a shaker (ISF-I–V; Adolf Kuhner AG) at 200 rpm and 43 °C. After incubation overnight, the suspension was serially diluted and plated onto nutrient agar medium pH 7.2 (Difco) and incubated at 43 °C for 3 days. Several colonies that developed at 43 °C were picked and purified by repeated streaking on the same medium. A colony with a cream colour as being representative of several similar colonies and having the ability to grow on 100 mM sodium arsenate was selected for further analysis; this strain was designated HT23T. For short-term preservation, the culture was streaked on nutrient agar and stored at 4o C. For long-term preservation, the culture was stored at −80 °C in 15 % glycerol. Strain HT23T and closely related strains of P. phragmitetus C6-19T (DSM14728T), LMG5410, LMG5411, LMG5412, LMG5421, LMG5430, and LMG5431 were characterized under comparable conditions.
Cell morphology was examined by transmission electron microscopy (model FEI Morgagni 268D). The presence of flagella was demonstrated by using cells placed on a carbon-coated grid and negatively stained with 2 % phosphotungstic acid (Jyoti et al. 2010). Gram staining was carried out using a commercial kit (Hi-Media, Mumbai, India) according to manufacturer’s instructions. Oxidase activity was assayed using discs impregnated with dimethyl-p-phenylenediamine (Hi-Media). Catalase activity was assayed by mixing a pellet of a freshly centrifuged culture with a drop of hydrogen peroxide (30 % v/v). Anaerobic growth was tested using the BD GasPak EZ system (Becton, Dickinson and Company, Maryland, USA). Acid production using different carbon sources was tested in ammonium salt medium supplemented with d-adonitol, l-arabinose, d-xylose, d-galactose, d-fructose, glycerol, l-rhamnose, dulcitol, d-raffinose, d-cellobiose, inositol, d-maltose, ethanol, d-mannitol, salicin, d-sorbitol, sucrose, d-trehalose or d-lactose. Glucose oxidation and fermentation were tested according to Hugh and Leifson (1953). Biochemical tests were carried out using API ZYM, API 50CH test kits (bioMe’rieux) and Biolog GN2 (Biolog, USA) according to the manufacturer’s instructions. Hydrolysis of starch, gelatin, hippurate, casein, Tween 80 and phosphatase activity were examined following the methods of Smibert and Krieg (1994). Other biochemical tests were performed as described by Cruickshank et al. (1975) and Hi25 TM (Hi-Media, Mumbai, India) identification kits as recommended by the manufacturers. The pH range for growth was examined in nutrient broth prepared in buffered solutions in the range 3.0–11.0 in steps of one pH unit at 37 °C for 4 days. Similarly, temperature in the range of 15–50 °C was tested for growth in nutrient broth at pH 7.2 for 4 days. Yeast extract-peptone broth containing 0–5.0 % (w/v) NaCl was inoculated and incubated at 37 °C for 4 days to test for salt tolerance. Antibiotic resistance of strain HT23T was checked on nutrient agar containing different antibiotics at the concentration of ampicillin (50 μg/ml), streptomycin (50 μg/ml), kanamycin (50 μg/ml), neomycin (20 μg/ml), chloramphenicol (20 μg/ml), tetracycline (15 μg/ml), nalidixic acid (20 μg/ml) or rifampicin (20 μg/ml).
16S rRNA gene sequence analysis
For 16S rRNA gene sequence analysis, DNA was isolated according to Sambrook and Russell (2001). The 16S rRNA gene of the strain HT23T was amplified by the method described earlier (Panday et al. 2011). Primers used for the amplification of 16S rRNA were 5′-GAG TTT GAT CCT GGC TCA G-3′ (forward primer) and 5′-AGA AAG GAG GTG ATC CAG CC-3′ (reverse primer). 16S rRNA amplification was performed with a Thermal Cycler, Model PCT-200 (M.J. Research, Waltham, MA, USA), with the following temperature conditions: initial denaturation step at 94 °C for 4 min, and then 30 cycles of denaturation at 94 °C for 1 min, renaturation at 62 °C for 1 min and extension at 72 °C for 1.5 min. This is followed by a final extension period of 7 min at 72 °C. After amplification, the reaction mixture was subjected to electrophoresis on a 0.7 % agarose gel using a horizontal electrophoresis apparatus (Bio-Rad, Hercules, CA, USA). The PCR products were purified using the QIAQuick Gel Extraction Kit (Qiagen, Hilden, Germany) and sequenced using a CEQ Dye terminator cycle sequencing kit in an automated DNA sequencer (Model CEQ 8000; Beckman Coulter, Fullerton, CA, USA). The nucleotide sequences obtained were assembled using the sequence alignment editor program Bioedit (http://www.mbio.ncsu.edu/BioEdit/bioedit.html). The 16S rRNA gene sequence obtained was compared with those in GenBank after BLAST searches (Altschul et al. 1997) and using the EzTaxon server (Chun et al. 2007). The 16S rRNA gene sequence-based phylogenetic tree was constructed according to the Kimura 2 parameter model (Kimura 1980) using the MEGA 4.1 (Kumar et al. 2004) software package (The Biodesign Institute, Arizona, USA). A phylogenetic tree was constructed using the neighbour-joining method of Saitou and Nei (1987) and with the maximum likelihood algorithms.
DNA–DNA homology study
The DNA–DNA re-association study was performed with the strain HT23T and other closely related species showing 16S rRNA sequence similarities ≥97 %. DNA from Escherichia coli strain HB101 was taken as an unrelated negative control. DNA (1 μg) of each strain was transferred on to a positively charged nylon membrane (Hybond-N+; Amersham) by using a dot-blot apparatus (Bio-Rad). The membrane was air-dried and cross-linked and the DNA probe for each strain was labelled with [α-32P]-CTP (BRIT) by using NEBlot-kit (New England Biolabs). Hybridization was performed overnight at 65 °C following the method of Ezaki et al. (1989) and Bhadra et al. (2008). Prior to autoradiography, the blot was washed twice, each time with 2 × SSC (standard saline citrate) containing 0.1 % sodium dodecyl sulphate (SDS) for 20 min, after which an identical treatment with 0.2 × SSC and 0.1 % SDS was performed at 65 °C.
Determination of resistance to arsenate
Resistance of strain HT23T and P. phragmitetus C6-19T (DSM 14782T) for As(V) was determined by growing in modified low-phosphate liquid medium (Oden et al. 1994) supplemented with increasing concentrations of As(V) (from 100, 200, 300, 400, 500 to 600 mM). Modified liquid medium contained 80 mM NaCl, 20 mM KCl, 20 mM NH4Cl, 3 mM (NH4)2SO4, 1 mM MgCl2 and 0.12 mM Tris base, supplemented with 0.5 % glucose, 2 mg/ml of thiamine, 1 % peptone, 0.1 mM CaCl2, pH 7.0. The cells were initially grown in low-phosphate liquid medium without arsenate and were used as inocula. Cultures were grown at 37 °C, and the growth was monitored by measuring the optical density at 600 nm in a Cary100 spectrophotometer (Varian, USA) at different time intervals.
Analytical methods
For determination of the DNA G+C content, DNA was degraded enzymatically into nucleosides as described by Mesbah et al. (1989). The obtained nucleoside mixture was then separated by HPLC (Shimadzu) using an analytical column (Vydac 201 SP54, C18, 5 mm; 250 × 4.6 mm) equipped with a guard column (201 GD54H; Vydac). The operating conditions were as follows: 45 °C, 10 μl sample, 0.3 M (NH4)H2PO4/acetonitrile, 40:1 (v/v), pH 4.4, as solvent and flow rate of 1.3 ml/min, as described by Tomaoka and Komagata (1984). Lambda phage DNA (Sigma) and three bacterial DNAs (Bacillus subtilis DSM 402, Xanthomonas campestris pv. campestris DSM 3586T and Streptomyces violaceoruber DSM 40783) with published genome sequences were used as calibration standards. For the analysis of lipids, quinone and diaminopimelic acid (DAP), cells were grown in liquid medium (Yamada and Komagata 1970) to mid-log phase in a rotary shaker at 37 °C. DAP was detected by the method of Rhuland et al. (1955) with the modification that TLC on cellulose sheets (Merck, cat no. 1.05577, Germany) was used instead of paper chromatography. Isoprenoid quinones analysis of strain HT23T was performed by HPLC as described by Groth et al. (1996). Polar lipids were determined by 2D-TLC and application of specific spray reagents according to the method described by Minnikin et al. (1979). Cellular fatty acids were extracted from cells grown on rich medium (Yamada and Komagata 1972) agar plates at 37 °C for 3 days. Cells were saponified and transmethylated as described by Kuykendall et al. (1988). The fatty acid methyl ester mixtures were separated using the Sherlock Microbial Identification System (MIDI; Microbial ID), which consisted of an Agilent model 6890N GC.
SDS–PAGE of cellular proteins
SDS–PAGE was performed by using the method of Laemmli (1970) with minor modifications. Strain HT23T and the reference strains used in this study were grown in nutrient broth (Difco) at 37 °C overnight. Protein samples were prepared by heating 50 mg (wet weight) of cells at 100 °C for 2.0 min in 1.0 ml sample treatment buffer (Laemmli 1970). The resulting extracts were centrifuged at 11,000g for 15 min. Electropherograms were developed in a Protean II xi vertical electrophoresis cell (Bio-Rad) at 8 mA and 10 °C for 12 h. The protein banding pattern was visualized by staining with 0.25 % Coomassie brilliant blue R-250 in 50 % (v/v) methanol and 10 % acetic acid. Protein patterns were grouped by using the Fingerprinting II software package v. 3.0 (Bio-Rad) developed by Applied Maths, under normalized densitometry. Levels of similarity between the strains were calculated by using the Dice coefficient, and a UPGMA dendrogram was constructed as described by Vauterin and Vauterin (1992).
Results and discussion
Morphological and physiological characterization
On nutrient agar medium, colonies appeared were round, smooth, cream coloured and about 1–1.5 mm in diameter. Single colonies appeared within 48 h at 37 °C. Cells were rod shaped, 0.6–0.8 μm wide and 1.8–2.0 μm long and motile by a single polar flagellum (Fig. 1). Strain HT23T is a Gram-negative, aerobic rod. Both oxidase and catalase activities were positive. Growth occurred at 20–45 °C and pH 6.0–11.0. Optimum growth was observed at 37 °C and pH 7.0–10.0. This bacterium grew in yeast extract-peptone broth containing 0–4.0 % (w/v) NaCl, but not with 5.0 % NaCl. Differential traits between the strain HT23T and the reference strains are given in Table 1. Detailed phenotypic properties of strain HT23T are given in the species description and in supplementary Table S1.
DNA base composition
The G+C content of the genomic DNA of strain HT23T was 63.3 mol%, a value close to that reported for the genus Pannonibacter (Borsodi et al. 2003; Holmes et al. 2006).
Chemotaxonomic properties
The peptidoglycan of strain HT23T contained meso-DAP as also reported for P. phragmitetus C6-19T (DSM 14782T). The major isoprenoid quinone detected was Q-10. Minor amounts of Q-9 occurred additionally. The quinone pattern of strain HT23T differed from that of the type strain P. phragmitetus C6-19T by the absence of Q-7 (Borsodi et al. 2003). Polar lipids of strain HT23T included diphosphatidyl glycerol (DPG), phosphatidyl glycerol (PG), phosphatidyl ethanolamine (PE), phosphatidyl choline (PC), phosphatidyl monomethyl ethanolamine (PMME) and two unknown amino lipids (AL1-2) (Supplementary figure 1). However, strain HT23T could be differentiated from P. phragmitetus C6-19T (DSM 14782T) by the presence of PE and the occurrence of an additional amino lipid (Biebl et al. 2007). The major cellular fatty acid composition of strain HT23T was C14:0 3OH (3.41 %), C16:0 (7.22 %), C17:0 (3.42 %), C18:0 (8.65 %), C18:0 3OH (2.16 %), C18:1 w7c (62.89 %), C18:1 w7c 11-methyl (7.35 %) and C20:1 w7c (1.27 %). As seen from supplementary Table 2, complete fatty acid compositions were almost identical, although strain HT23T differed slightly by having a higher proportion of C18:0 (8.65 %) and C16:0 (7.22 %).
Phylogenetic analysis of the 16S rRNA gene sequence
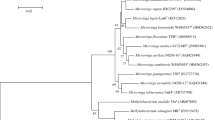
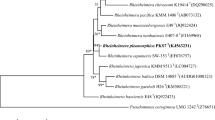
Altogether, 1,405 nucleotides of 16S rRNA were sequenced from strain HT23T and compared with those of closely related strains retrieved from the GenBank database. Strain HT23T showed 98.9 % 16S rRNA gene sequence similarity with P. phragmitetus C6-19T (DSM 14782T), 98 % with the P. phragmitetus group B and P. phragmitetus group E strains, 95.7 % similarity with Polymorphum gilvum SL003B-26A1T, 95.4 % similarity with Labrenzia marina mano18T and Labrenzia alba CECT 5094T, 94.5 % similarity with Labrenzia aggregate IAM 12614T, 94.2 % with Labrenzia alexandrii DFL-11T, 94.2 % similarity with Stappia indica B106T and 92.5 % similarity with Pseudovibrio denitrificans DN34T. Phylogenetically, strain HT23T clustered with P. phragmitetus strains (Fig. 2). The phylogenetic tree presented is similar in its topology to the tree generated by the maximum likelihood algorithm. The statistical significance of branch points was calculated by 1,000 bootstrap re-samplings of the data (Felsenstein 1985). Similar tree topology was obtained using the Jukes and Cantor (1969) parameter model.
DNA–DNA re-association
DNA–DNA hybridization is considered as a standard method for bacterial species delineation (Wayne et al. 1987). Hence, DNA–DNA hybridization experiments were carried out between strain HT23T and the reference strains showing 16S rRNA gene sequence similarities higher than 97 %. Using a labelled probe of strain HT23T, the mean DNA–DNA re-association value was 55 % with P. phragmitetus C6-19T. The mean re-association values obtained with other strains of P. phragmitetus group B and group E strains were within values ranging from 34 to 51 %. However, no hybridization signal was observed with the E. coli DNA used as a negative control. Therefore, given the recommended DNA–DNA relatedness cut-off point for species delineation of 70 % (Wayne et al. 1987), strain HT23T should be regarded as a novel species of the genus Pannonibacter.
Numerical analysis by electrophoretic protein patterns
SDS–PAGE of whole-cell protein extracts is used widely in bacterial taxonomy and is suitable for grouping strains at the species level (Diouf et al. 2000; Tan et al. 1997). The protein patterns of strain HT23T were different from those of its closest related reference strains (Supplementary figure S2a). Based on this dendrogram, the level of protein similarity between strain HT23T, P. phragmitetus C6-19T, group B and group E strains of P. phragmitetus was less than 70 % (Supplementary figure S2b). Thus, SDS–PAGE analysis also suggested that strain HT23T should be regarded as representing a novel species of the genus Pannonibacter.
Growth on arsenate
Time course of growth of the strain HT23T and P. phragmitetus C6-19T (DSM 14782T) showed resistance toward As(V). Growth of HT23T was observed in the presence of 100–500 mM, but not in 600 mM (Fig. 3a), and that of P. phragmitetus C6-19T was observed in the presence of 100–400 mM, but not in 500 mM As(V) (Fig. 3b).
Effect of sodium arsenate on the growth response of a Pannonibacter indica strain HT23T and b Pannonibacter phragmatitus C6-19T in low-phosphate medium. Symbols: filled circle without sodium arsenate; filled triangle 100 mM sodium arsenate; filled inverted triangle 200 mM sodium arsenate; filled diamond 300 mM sodium arsenate; empty circle 400 mM sodium arsenate; filled square 500 mM sodium arsenate; empty triangle 600 mM sodium arsenate
Description of Pannonibacter indica sp. nov
Pannonibacter indica (in’ di. ca. L. fem. adj. indica of India, the geographical origin of the type strain). Cells are motile rods and occur singly. Strictly aerobic. Growth at 20–45 °C and pH 6.0–11.0. Optimum growth was observed at 37 °C and pH 7.0–10.0. Gram negative. Catalase and oxidase positive. Colonies on nutrient agar are smooth, round and cream coloured. Positive for urease, phosphatase, growth on MacConkey’s agar, growth on King’s A and King’s B medium, lysine decarboxylase, aesculin hydrolysis, β-galactosidase production, citrate utilization, casein digestion and arginine hydrolysis. Nitrate and nitrite reduction, Methyl Red and Voges–Proskauer test, indole production, phenylalanine deamination, malonate utilization, ornithine decarboxylase, pigment production, DNase production, 3-ketolase production, H2S production, growth on cetrimide agar, hydrolysis of tyrosine, brown pigment production on tyrosine agar, KCN tolerance, growth at 5 °C, hydrolysis of starch, cellulose, hippurate, gelatin and lecithinase production tests were negative. In Biolog GN2 microplates, the test proved positive for oxidation of Tween 80, d-arabitol, d-cellobiose, d-fructose, l-fucose, α-d-glucose, d-mannitol, d-mannose, d-melibiose, acetic acid, citric acid, formic acid, β-hydroxy butyric acid, propionic acid, l-asparagine, l-glutamic acid, glycyl-l-aspartic acid, urocanic acid, uridine and thymidine. The test proved negative for α-cyclodextrin, dextrin, glycogen, Tween 40, N-acetyl d-galactosamine, N-acetyl d-glucosamine, adonitol, l-arabinose, d-erythritol, d-galactose, gentiobiose, m-inositol, α-d-lactose, lactulose, maltose, β-methyl d-glucoside, d-psicose, d-raffinose, l-rhamnose, sucrose, d-trehalose, turanose, d-sorbitol, xylitol, pyruvic acid methyl ester, succinic acid mono-methyl ester, cis-aconitic acid, d-galactonic acid lactone, d-galacturonic acid, d-gluconic acid, d-glucosaminic acid, d-glucuronic acid, α-hydroxybutyric acid, γ-hydroxy butyric acid, p-hydroxy phenylacetic acid, itaconic acid, α-ketobutyric acid, α- ketoglutaric acid, α-ketovaleric acid, d,l-lactic acid, malonic acid, quinic acid, d-saccharic acid, sebacic acid, succinic acid, bromo succinic acid, succinamic acid, glucuronamide, l-alaninamide, d-alanine, l-alanine, l-alanyl-glycine l-aspartic acid, glycyl-l-glutamic acid, l-histidine, hydroxy-l-proline, l-leucine, l-ornithine, l-phenylalanine, l-proline, l-pyroglutamic acid, d-serine, l-serine, l-threonine, d,l-camitine, γ-aminobutyric acid, inosine, phenylethylamine, putrescine, 2-aminoethanol, 2,3-butendiol, glycerol, d,l,α-glycerol phosphate, α-d-glucose-1-phosphate and d-glucose-6-phosphate. In API 50CH strips and in ammonium salt sugar medium, strain HT23T was tested positive for acid production from l-arabinose, d-xylose, d-galactose, d-glucose, d-fucose, l-fucose, d-arabinose, d-ribose, l-rhamnose, d-cellobiose and d-melibiose. Strain HT23T was negative for both acid and gas production from glucose in peptone water medium. The strain was tested negative for production of acid from d-lactose, d-fructose, glycerol, erythritol, ethanol, l-xylose, d-adonitol, methyl-β-d-xylopyranoside, d-mannose, l-sorbose, dulcitol, inositol, d-mannitol, d-sorbitol, methyl-α-d-mannopyranoside, methyl-α-d-glucopyranoside, N-acetylglucosamine, amygdalin, arbutin, salicin, d-maltose, d-saccharose (sucrose), d-trehalose, inulin, d-melezitose, d-raffinose, amidon (starch), glycogen, xylitol, gentiobiose, d-turanose, d-lyxose, d-tagatose, d-arabitol, l-arabitol, potassium gluconate, potassium 2-ketogluconate and potassium 5-ketogluconate. Enzymatic activities determined using API ZYM strip were positive for alkaline phosphatase, leucine arylamidase, trypsin, acid phosphatase, naphthol-AS-BI-phosphohydrolase, α-galactosidase and β-galactosidase, whereas strain HT23T was negative for esterase, lipase, valine arylamidase, cystine arylamidase, α-chymotrypsin, β-glucuronidase, α-glucosidase, β-glucosidase, N-acetyl-β-glucosaminidase, α-mannosidase and α-fucosidase. Strain HT23T was susceptible to ampicillin (50 μg/ml), streptomycin (50 μg/ml), kanamycin (50 μg/ml), neomycin (20 μg/ml), chloramphenicol (20 μg/ml), tetracycline (15 μg/ml), nalidixic acid (20 μg/ml) or rifampicin (20 μg/ml).
The cellular fatty acid composition of strain HT23T was C10:0 (0.04 %), C11:0 (0.06 %), C12:0 (0.09 %), C14:0 iso (0.02 %), C14:0 (0.12 %), C14:0 3OH (3.41 %), C15:0 3OH (0.28 %), C15:0 iso 3OH (0.03 %), C15:0 iso (0.02 %), C15:1 w8c (0.02 %), C16:0 (7.22 %), C16:0 iso (0.04 %), C16:0 3OH (0.11 %), C16:1 w7c/16:1 w6c (0.47 %), C17:0 (3.42 %), C17:1 w6c (0.60 %), C17:1 w8c (0.28 %), C17:0 3OH (0.11 %), C17:0 iso 3OH (0.14 %), C18:0 (8.65 %), C18:1 2OH (0.04 %), C18:0 3OH (2.16 %), C18:1 w7c (62.89 %), C18:1 w7c 11-methyl (7.35 %), C18:1 w5c (0.21 %), C19:0 (0.18 %), C19:0 iso (0.03 %), C19:0 iso I (0.01 %), C19:0 cyclo w10c/19w6 (0.67 %), C20:0 (0.06 %) and C20:1 w7c (1.27 %). Polar lipids of strain HT23T included diphosphatidyl glycerol (DPG), phosphatidyl glycerol (PG), phosphatidyl ethanolamine (PE), phosphatidyl choline (PC), phosphatidyl monomethyl ethanolamine (PMME) and two unknown amino lipids (AL1-2).
The DNA G+C content of the type strain is 63.3 mol%. The type strain is HT23T (=JCM 16851T = DSM 23407T = LMG 25769T). GenBank/EMBL accession number for the 16S rRNA gene of P. indica sp. nov. HT23T is EF608175. This bacterium was isolated from a hot spring.
References
Alarico S, Rainey FA, Empadinhas N, Schumann P, Nobre MF, da Costa MS (2002) Rubritepida flocculans gen. nov., sp. nov., a new slightly thermophilic member of the alpha-1 subclass of the Proteobacteria. Syst Appl Microbiol 25:198–206
Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25:3389–3402
Bhadra B, Raghukumar C, Pindi PK, Shivaji S (2008) Brevibacterium oceani sp. nov., isolated from deep-sea sediment of the Chagos Trench, Indian Ocean. Int J Syst Evol Microbiol 58:57–60
Biebl H, Pukall R, Lunsdorf H, Schulz S, Allgaier M, Tindall BJ, Wagner-Dobler I (2007) Description of Labrenzia alexandrii gen. nov., sp. nov., a novel alphaproteobacterium containing bacteriochlorophyll a, and a proposal for reclassification of Stappia aggregata as Labrenzia aggregata comb. nov., of Stappia marina as Labrenzia marina comb. nov. and of Stappia alba as Labrenzia alba comb. nov., and emended descriptions of the genera Pannonibacter, Stappia and Roseibium, and of the species Roseibium denhamense and Roseibium hamelinense. Int J Syst Evol Microbiol 57:1095–1107
Borsodi AK, Micsinai A, Kovács G, Tóth E, Schumann P, Kovács AL, Böddi B, Márialigeti K (2003) Pannonibacter phragmitetus gen. nov., sp. nov., a novel alkalitolerant bacterium isolated from decomposing reed rhizomes in a Hungarian soda lake. Int J Syst Evol Microbiol 53:555–561
Chun J, Lee J-H, Jung Y, Kim M, Kim S, Kim BK, Lim YW (2007) EzTaxon: a web-based tool for the identification of prokaryotes based on 16S ribosomal RNA gene sequences. Int J Syst Evol Microbiol 57:2259–2261
Cruickshank R, Duguid JP, Marmion BP, Swain RHA (1975) Medical microbiology. The practice of medical microbiology, vol 2, 12th edn. Churchill Livingstone, Edinburgh, pp 301–310
Denner EB, Kolari M, Hoornstra D, Tsitko I, Kampfer P, Busse HJ, Salkinoja-Salonen M (2006) Rubellimicrobium thermophilum gen. nov., sp. nov., a red-pigmented, moderately thermophilic bacterium isolated from coloured slime deposits in paper machines. Int J Syst Evol Microbiol 56:1355–1362
Diouf A, de Lajudie P, Neyra M, Kersters K, Gillis M, Martinez-Romero E, Gueye M (2000) Polyphasic characterization of rhizobia that nodulate Phaseolus vulgaris in West Africa (Senegal and Gambia). Int J Syst Evol Microbiol 50:159–170
Donachie SP, Bowman JP, Alam M (2006) Nesiotobacter exalbescens gen. nov., sp. nov., a moderately thermophilic alphaproteobacterium from an Hawaiian hypersaline lake. Int J Syst Evol Microbiol 56:563–567
Ezaki T, Hashimoto Y, Yabuuchi E (1989) Fluorometric deoxyribonucleic acid deoxyribonucleic acid hybridization in microdilution wells as an alternative to membrane filter hybridization in which radioisotopes are used to determine genetic relatedness among bacterial strains. Int J Syst Bacteriol 39:224–229
Felsenstein J (1985) Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:783–791
Groth I, Schumann P, Weiss N, Martin K, Rainey FA (1996) Agrococcus jenensis gen nov., sp. nov., a new genus of actinomycetes with diaminobutyric acid in the cell wall. Int J Syst Evol Microbiol 46:234–239
Hall JR, Mitchell KR, Jackson-Weaver O, Kooser AS, Corn BR, Crossey IJ, Takacs-Vesbach CD (2008) Molecular characterization of the diversity and distribution of a thermal spring microbial community by using rRNA and metabolic genes. Appl Environ Microbiol 74:4910–4922
Holmes B, Segers P, Coenye T, Vancanneyt M, Vandamme P (2006) Pannonibacter phragmitetus described from a Hungarian soda lake in 2003, had been recognized several decades earlier from human blood cultures as Achromobacter groups B and E. Int J Syst Evol Microbiol 56:2945–2948
Hugh R, Leifson E (1953) The taxonomic significance of fermentative versus oxidative metabolism of carbohydrates by Gram negative bacteria. J Bacteriol 66:24–26
Johnson DB, Stallwood B, Kimura S, Hallberg KB (2006) Isolation and characterization of Acidicaldus organivorus, gen. nov., sp. nov.: a novel sulfur-oxidizing, ferric iron-reducing thermo-acidophilic heterotrophic Proteobacterium. Arch Microbiol 185:212–221
Jukes TH, Cantor CR (1969) Evolution of protein molecules. In: Munro HN (ed) Mammalian Protein Metabolism. Academic Press, New York, pp 21–132
Jyoti V, Narayan KD, Das SK (2010) Gulbenkiania indica sp. nov., isolated from a sulfur spring. Int J Syst Evol Microbiol 60:1052–1055
Kanso S, Patel BK (2004) Phenylobacterium lituiforme sp. nov., a moderately thermophilic bacterium from a subsurface aquifer, and emended description of the genus Phenylobacterium. Int J Syst Evol Microbiol 54:2141–2146
Kimura M (1980) A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol 16:111–120
Kumar S, Tamura K, Nei M (2004) MEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief Bioinform 5:150–163
Kuykendall LD, Roy MD, O’Neill JJ, Devine TE (1988) Fatty acids, antibiotic resistance and deoxyribonucleic acid homology groups of Bradyrhizobium japonicum. Int J Syst Bacteriol 38:358–361
Laemmli UK (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680–685
Mesbah M, Premachandran U, Whitman WB (1989) Precise measurement of the G+C content of deoxyribonucleic acid by high-performance liquid chromatography. Int J Syst Bacteriol 39:159–167
Minnikin DE, Collins MD, Goodfellow M (1979) Fatty acid and polar lipid composition in the classification of Cellulomonas, Oerskovia and related taxa. J Appl Bacteriol 47:87–95
Oden KL, Gladysheva TB, Rosen BP (1994) Arsenate reduction mediated by the plasmid-encoded ArsC protein is coupled to glutathione. Mol Microbiol 12:301–306
Panday D, Schumann P, Das SK (2011) Rhizobium pusense sp. nov., isolated from the rhizosphere of chickpea (Cicer arietinum L.). Int J Syst Evol Microbiol 61:2632–2639
Rhuland LE, Work E, Denman RF, Hoare DS (1955) The behaviour of the isomers of α, ε-diaminopimelic acid on paper chromatograms. J Am Chem Soc 77:4844–4846
Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425
Sambrook J, Russell DW (2001) Molecular cloning: a laboratory manual, 3rd edn. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY
Smibert RM, Krieg NR (1994) Phenotypic characterization. In: Gerhardt P, Murray RGE, Wood WA, Krieg NR (eds) Methods for general and molecular bacteriology. American Society for Microbiology, Washington, DC, pp 603–711
Tan ZY, Xu XD, Wang ET, Gao JL, Martinez-Romero E, Chen WX (1997) Phylogenetic and genetic relationships of Mesorhizobium tianshanense and related rhizobia. Int J Syst Bacteriol 47:874–879
Tomaoka J, Komagata K (1984) Determination of DNA base composition by reversed phase high performance liquid chromatography. FEMS Microbiol Lett 25:125–128
Vauterin L, Vauterin P (1992) Computer aided objective comparison of electrophoretic patterns for grouping and identification of microorganisms. Eur Microbiol 1:37–41
Wayne LG, Brenner DJ, Colwell RR, Grimont PAD, Kandler O, Krichevsky MI, Moore LH, Moore WEC, Murray RGE et al (1987) International Committee on Systematic Bacteriology. Report of the ad hoc committee on reconciliation of approaches to bacterial systematics. Int J Syst Bacteriol 37:463–464
Yamada K, Komagata K (1970) Taxonomic studies on coryneform bacteria. II. Principal amino acids in the cell wall and their taxonomic significance. J Gen Appl Microbiol 16:103–113
Yamada K, Komagata K (1972) Taxonomic studies on coryneform bacteria. IV. Morphological, cultural, biochemical and physiological characteristics. J Gen Appl Microbiol 18:103–113
Acknowledgments
We are grateful to the All India Institute of Medical Sciences, New Delhi, for electron microscopic facility. Pannonibacter phragmitetus C6-19T (DSM 14782T) and the representative of Pannonibacter phragmitetus group B strains (LMG 5410, LMG 5411, LMG 5412 and LMG 5421) and Pannonibacter phragmitetus group E strains (LMG 5430 and LMG 5431) used in this study were provided by the DSMZ, Braunschweig, Germany and the BCCM/LMG Bacteria collection, Universiteit Gent, Belgium. This work was supported by the Department of Biotechnology, Ministry of Science and Technology, Govt. of India. The author, S. Bandyopadhyay, acknowledges the Council of Scientific and Industrial Research (CSIR), Government of India, New Delhi, for providing the research fellowship.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by Erko Stackebrandt.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Bandyopadhyay, S., Schumann, P. & Das, S.K. Pannonibacter indica sp. nov., a highly arsenate-tolerant bacterium isolated from a hot spring in India. Arch Microbiol 195, 1–8 (2013). https://doi.org/10.1007/s00203-012-0840-z
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00203-012-0840-z