Abstract
Vegetative wild-type and DNA repair-deficient (homologous recombination, recA and nucleotide excision repair, uvrB) Bacillus subtilis cells were exposed to UV-C radiation. Colony formation, DNA bipyrimidine photoproducts and gene expression were measured during cell recovery. Gene expression was measured after 60 min cell recovery where 50% (wild-type), 30% (recA) and 8% (uvrB), respectively, of the UV-C induced DNA photoproducts were repaired. We examined changes in the gene expression following UV exposure in wild-type and both repair-deficient strains. A set of known and unknown genes were found to be significantly up-regulated in wild-type B. subtilis cells, whereas no or lower gene induction was determined for both mutant strains. In addition, the possible roles of newly identified UV-responsive genes are discussed with respect to cellular recovery following exposure to UV irradiation.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
UV-C radiation exposure to microorganisms gives rise to DNA lesions such as bipyrimidine photoproducts that are able at least to transiently block essential biochemical processes such as replication and transcription (Friedberg et al. 1995). Cells respond to DNA damaging stresses by up-regulating the expression of several genes that promote repair of DNA lesions and restore replication (Goranov et al. 2006). Gene expression in response to DNA damage produced by UV-C radiation and a few other environmental agents has been collectively termed the SOS response (Courcelle et al. 2001; Au et al. 2005). A variety of DNA repair mechanisms have been identified in both pro and eukaryotic cells. These include direct damage reversal, base excision repair, nucleotide excision repair (NER) (Cadet et al. 2005), mismatch correction and recombination repair. Among these repair pathways, NER is a versatile process that is implicated in the removal of a variety of bulky DNA lesions such as the UV-induced cyclobutane pyrimidine dimers (CPDs) and pyrimidine (6-4) pyrimidone photoproducts (6-4 PPs) (Koehler et al. 1996). NER is a complex process that involves successively recognition of the DNA damage, unwinding of the double helix at the DNA lesion site, single strand incision on both sides of the modification, excision of the lesion-containing single stranded DNA fragment, DNA repair synthesis to replace the gap and ligation of the remaining nick (Friedberg et al. 1995). Thus, CPDs and 6-4 PPs formed in cellular DNA upon UV-C irradiation are removed as oligonucleotides and the gap thus created is filled up upon insertion of normal nucleotides during repair synthesis. Certain genes of B. subtilis that have been shown to be involved in recombination DNA repair (Fernandez et al. 1998; Zahradka et al. 2002). The latter DNA repair process is closely connected to the cellular replication system. DNA repair including general homologous recombination plays a critical role in maintaining gene diversification and genome stability (Pastink et al. 2001). It may be noted that RecA coordinates the initiation of repair of DNA damage. RecA binds to ssDNA exposed during DNA damage and catalyzes recombination processes inside the cell. RecA has also been shown to mediate replication fork reversal (a mechanism used to repair a stalled replication fork on the leading strand) and to alter the accessibility of the 3′ ends of DNA to DNA polymerase (Au et al. 2005).
In this communication we analyzed the recovery of wild-type and DNA repair-deficient Bacillus subtilis following UV-C irradiation by colony formation ability, assessment of the nature and levels of UV photoproducts in DNA at various post-irradiation times and gene expression after 60 min recovery. Further on, unknown up-regulated genes were aligned on their genomic information and functional characterization according to the world-wide-web database SubtiList-server http://www.genolist.pasteur.fr/SubtiList (Moszer et al. 2002).
Material and methods
Biological material
Bacillus subtilis trpC2 strain 168 (DSM 402) was obtained from the DSMZ—German Collection of Microorganisms and Cell Cultures, Braunschweig, Germany. While strain 168 possess wild-type DNA repair capability, strain WN463 is defective in the major recombinogenic protein RecA (obtained from W. L. Nicholson) while strain TKJ6312, kindly provided by N. Munakata, is deficient in NER-repair pathway (uvrB) (Moeller et al. 2007). Both strains are isogenic with B. subtilis 168 wild-type strain. Cells were cultivated under vigorous aeration at 37°C in nutrient broth (NB) medium (Difco Detroit, MI, USA). The exponential growth phase was measured by optical density readings (OD 0.6–0.8) at 600 nm. Cells were harvested by centrifugation at 4,000×g for 15 min at 4°C, washed twice with 0.1 M phosphate-buffered saline (PBS) (pH 7.5), resuspended in the chemical-defined Spizizen minimal medium with a supplement of 0.5% glucose, which has been previously used for cell recovery experiments (Billen et al. 1971; Hadden 1979; Hanlin et al. 1985).
UV-C irradiation and cell recovery
Hundred milliliters of the cell suspension (107/ml) in Petri dishes of 200 mm in diameter were exposed to a fluence of 25 J/m2 UV-C radiation emitted by a mercury low-pressure lamp (NN 8/15, Heraeus, Berlin, Germany) with a major emission line at 253.65 nm. The spectral irradiance was determined spectrophotometrically (Bentham 150 double monochromator) and the fluence rate was 90.5 W/m2 as measured with a UV-X radiometer (UVP Ultra-Violet Products, Cambridge, UK) (Moeller et al. 2005; Pogoda de le Vega et al. 2005). UV-C irradiations were carried out at 4°C for preventing the onset of DNA repair during irradiation. The suspension was stirred continuously to ensure homogeneous exposure. Untreated cells were handled similarly except they were not UV-C irradiated. Immediately after irradiation, samples of 100 μl were taken from the irradiated cell suspension for survival analysis. To measure cell recovery ability the irradiated cells in suspension were incubated under vigorous stirring at 37°C up to 4 h. Every 60 min samples were taken for determination of colony forming units (CFU) (100 μl), DNA photoproducts (10 ml) and gene expression analyses (20 ml at 60 min cell recovery). CFUs were determined from the samples after appropriate dilution in 0.1 M PBS, plating on NB agar and incubation at 37°C overnight. Previous experiments have shown that there were no significant differences in the gene expression and the photoproduct repair by a temperature shift directly after the irradiation (4°C) and 60 min cell recovery at 37°C (Moeller et al., unpublished data).
Quantification of DNA bipyrimidine photoproducts
High molecular weight DNA was isolated from the irradiated and subsequently incubated cells using the Wizard Genomic DNA Purification Kit (Promega GmbH, Mannheim, Germany). Isolated DNA samples from irradiated and germinating spores were enzymatically digested by nuclease P1 and phosphodiesterase I as described (Douki and Cadet 2001; Douki et al. 2005; Moeller et al. 2007). The digested samples were analyzed by high-performance liquid chromatography coupled with tandem mass spectrometry (HPLC-MS/MS) for dimeric photoproducts involving adjacent pyrimidines that were released as modified dinucleoside monophosphates (Douki et al. 2005; Pogoda de la Vega et al. 2005). The following photoproducts were quantified as dinucleoside monophosphates using analytical methods described previously (Douki et al. 2005): cyclobutane pyrimidine dimers (CPD) of thymine-thymine (CPD TT), of thymine–cytosine (CPD TC), of cytosine–thymine (CPD CT) and of cytosine–cytosine (CPD CC) as well as the pyrimidine (6-4) pyrimidone photoproducts (6-4 PP) of thymine–thymine (6-4 TT), and of thymine–cytosine (6-4 TC). Quantitation of the dimeric pyrimidine photoproducts was performed in the multiple reaction-monitoring (MRM) mode using transitions reported previously (Douki et al. 2005; Moeller et al. 2007). Additional experiments were carried out in the product ion scan mode. For this purpose, the full fragmentation spectra (mass range 200–450) of pseudomolecular ions at 545, 530 and 531 m/z were recorded during the HPLC analysis.
RNA isolation, cDNA labeling and microarray hybridization
Total RNA was isolated from B. subtilis cells by the acid phenol method described by Moeller et al. (2006). Isolated RNA samples were photometrically quantified using the NanoDrop spectrophotometer (Kisker, Steinfurt, Germany) and checked for RNA integrity with the 2100 BioAnalyzer (Agilent, Böblingen, Germany). Only RNA samples with a RIN (RNA integrity number) above 9.0 were used. In order to generate cDNA probes 15 μg of RNA was taken for each labeling reaction as described in the LabelStar protocol (Qiagen GmbH, Hilden, Germany). For the RT-PCR reaction B. subtilis cDNA labeling primers (Sigma-Genosys, Haverhill, UK) were used for priming, whereas the labeling was achieved using either the Cy3-dUTP or Cy5-dUTP fluorophore (Amersham Pharmacia Biotech, Freiburg, Germany). The labeling efficiency was checked by using a NanoDrop spectrophotometer (Kisker, Steinfurt, Germany). Each total RNA preparation was labeled with both Cy3-dUTP and Cy5-dUTP. To hybridize a single glass slide, the Cy3-dUTP-labeled probe from one condition was mixed with the Cy5-dUTP-labeled probe using the so-called color swap procedure and vice versa. Each experiment was repeated four times, requiring 8 slides. Equal amounts of either Cy3-dUTP or Cy5-dUTP-labeled probe were applied to each slide. Each DNA microarray comprised all of the B. subtilis genes found in the genome data release R16.1 on the SubtiList website (Moszer et al. 2002) as capture probes. The hybridization was carried out at 42°C for 18 h with the microarray slide hybridization buffer containing formamide (Ambion, Huntingdon, UK). Post hybridization microarray procedures, i.e. slide washing and drying were performed as described in detail by Au et al. (2005) and Goranov et al. (2006).
DNA microarray data analysis
Scanning of microarrays was performed with 10 μm resolution using a DNA microarray laser scanner (DNA Microarray Scanner BA, Agilent Technologies, Agilent, Böblingen, Germany). Features were extracted from the raw image data using the Agilent Technologies image analysis software [G2567AA Feature Extraction (FE) Software, Version A7.5] and default settings for non-Agilent microarrays. The local background subtraction method was used and the background was globally adjusted to zero. Dye normalization was done by selecting normalization features with the rank consistency method and applying a global approach with linear scaling together with local weighted regression normalization (LOWESS) (Draghici 2002). The ratio between the channels, the log ratio error and the P value as measures of differential expression were calculated using default settings (Eichenberger et al. 2003). The user manual and detailed information describing the FE software can be retrieved on the Agilent Technologies Inc. (Santa Clara, CA, USA) website (https://www.chem.agilent.com). Tab-delimited files of the individual hybridizations were used for signal intensity analyses. All microarray experiments were done with six independent replicates and evaluated by using significance analysis of microarrays (Tusher et al. 2001). Two-fold cutoff was applied for all experiments, i.e., only effects that were ≥2.0-fold change were considered significant and are reported.
Numerical and statistical analysis
Survival after UV-C irradiation and recovery incubation was determined from CFUs as N/N 0 where N gives the CFUs after irradiation and x min of incubation, and N 0 gives the CFUs of non-irradiated samples after 0 min of incubation. The cell recovery curve was obtained by plotting the survival versus recovery time. Similarly, the photoproduct repair curve was obtained by plotting the number of dimeric pyrimidine lesions per 104 bases versus cell recovery time. All experiments were repeated at least four times, the data shown are mean values with standard deviation. The significance of differences in the bipyrimidine photoproduct repair rates and colony formation was assessed by variance analysis (ANOVA and Student’s t test) by using Analyse-it software, version 1.62 (Analyse-It Software, Ltd, Leeds, UK). Values were analyzed in multigroup pairwise combinations, and differences with P values of ≤0.05 were considered statistically significant (Moeller et al. 2005; Pogoda de la Vega et al. 2005).
Results
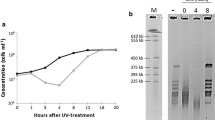
Exposure of B. subtilis 168 exponential growth phase cells to UV-C radiation at a fluence of 25 J/m2 led to a 90.2 ± 4.1% (wild-type), 99.3 ± 0.5% (recA) and 99.8 ± 0.04% (uvrB) cell inactivation (Fig. 1a). During the subsequent cell recovery period, the survival increased exponentially with incubation time and reached almost the same level as without irradiation after a 4-h period. The cell recovery rate was estimated to be 3331 ± 267 (wild-type), 1916 ± 109 (recA) and 198 ± 30 (uvrB) repaired cells/min. As expected, the non-irradiated control cells of all strains did not exhibit a significant increase in colony forming ability during the incubation period (total increase of less than >17 ± 3%). In order to correlate the recovery of cell proliferation capacities with the extent of DNA repair, the level of bipyrimidine photoproducts (PP) was assessed over the same period of recovery (Fig. 1b). The total number of bipyrimidine photoproducts decreased exponentially with the recovery time at a rate of 32 ± 5 (wild-type), 23 ± 3 (recA) and 8 ± 2 (uvrB) PP removed per min. The distribution of UV-induced photoproducts for all three strains determined immediately after irradiation (Fig. 2a) does not indicate genotype specific differences. The distribution pattern is similar to that found previously upon UV-C irradiation of isolated DNA (Ravanat et al. 2001; Cadet et al. 2005). CPDs at TT and TC sequences on the one hand and 6-4 TC on the other hand were found to be main bipyrimidine dimers formed by UV-C radiation in the DNA of the cells, whereas CPDs at CC and CT sites together with 6-4 TT lesions were produced in lower yields. In addition small amounts of dewar photoproducts at TT and TC sites were found to be generated although in a poor yield (<5%). The ratio between (6-4) photoproducts and cyclobutane pyrimidine dimers was found to depend on the nature of the bipyrimidine sites. For B. subtilis the ratio was close to 2 at TC sites and to 0.1 at TT sites. Time-course study of the removal of photoproducts revealed that CPDs and 6-4 PPs were repaired with a similar efficiency (Fig. 2). Approximately, 50% the total initial lesions were found to be removed in the wild-type cells within the first 60 min after irradiation, whereas only 30 and 8%, respectively, for recA and uvrB were removed (Fig. 2b). Interestingly, for all types of induced bipyrimidine photoproducts similar repair rates were measured in wild-type cells and with lower repair rates in both repair-deficient strains (Table 1).
Cell recovery curve (a) and photoproduct repair curve (b) after UV-C irradiation at a fluence of 25 J/m2 and subsequent recovery incubation of wild-type (solid circles), recA mutant (solid triangles) and uvrB mutant (solid squares) B. subtilis 168 cells; non-irradiated cells (open circles). Data are expressed as averages and standard deviations (n = 5)
Gene activation during cell recovery
In order to determine the transcriptional regulation during cell recovery of the UV-irradiated cells, total RNA was extracted from the treated cells after 60 min of recovery as well as from the non-treated cells. The process was followed by performing several measures including reverse transcription, cDNA labeling, hybridization using DNA microarrays and analyses of the fluorescent intensity differences (ratio of UV-C irradiated to non-irradiated cells). The gene expression of the B. subtilis 168 genome (Kunst et al. 1997) was analyzed in response to UV-C irradiation and the ensuing DNA repair by transcriptional profiling of six independent experiments. Prior to quantification of the array data, the quality and reproducibility of the array experiment were estimated by comparing the normalized spot intensities in the scattered diagram for wild-type cells as example (Fig. 3). Array data from hybridizations of independent samples representing the same cultivation condition always yielded high Pearson correlation coefficient with r ≥ 0.900 (Blencke et al. 2003), measured for all three strains. Using a two-fold balanced differential expression as the appropriate threshold level, 278 of the 4,107 known and unknown genes were found to be up-regulated in the wild-type strain (Fig. 3), whereas only 195 (recA) and 154 (uvrB) genes were induced after UV-C irradiation. Table 2 lists the 50 most highly up-regulated (activated) genes after 60 min of recovery for the UV-irradiated cells of all three strains, as inferred from the determination of the fluorescent intensity ratio ± standard deviation of six microarray experiments. Further on, for all 50 genes their putative LexA-binding site according to Au et al. (2005) and their response to further DNA damaging agents, e.g. mitomycin C were determined by bibliographic search (for detailed information see footnotes in Table 2). Among the 50 highly up-regulated transcripts many genes have described functions that may be directly involved in DNA repair processes. These include typical DNA repair genes (Au et al. 2005) such as uvrA and uvrB genes were highly up-regulated (Table 2). They are definitely directly responsible for the decrease in the level of UV induced DNA photoproducts: in this case the uvrAB-operon, coding for the exonuclease subunits A and B that are essential for the nucleotide excision repair pathway (Smith et al. 2002). Deficiencies in these genes lead to dramatic decrease in the DNA PP removal after UV-irradiation as can see in Table 1 and Fig. 1. Four differnet DNA polymerases, dnaX (DNA polymerase III), yorL (DNA polymerase III located in the prophage SPβ region), yqjH (a putitative DinB induced DNA polymerase IV) and yshC (DNA polymerase family X) also significantly were up-regulated. Several rec-genes like recA, recF, recG and recO that are likely to be also implicated in SOS-induced DNA repair processes (homologous recombination) were also significantly induced by UV-C irradiation. Further on, two DNA damage-inducible genes dinB and former dinC (tagC) showed increased fluorescent values. YefB, a putative site-specific recombinase, with high sequence similarity to Bacillus sp. NRRL B-14911 (Moszer et al. 2002), was significantly up-regulated in all three strains, while for recA and uvrB the wild-type induction was the two–three-fold higher than in the mutants.
Scatter diagram of normalized spot intensities. Gene intensities of UV-C irradiated are plotted versus non-irradiated in wild-type B. subtilis samples after 60 min cell recovery. The significance threshold of two-fold induction or repression is symbolized by the two dashed lines. Data are expressed as mean values (n = 6). r = Pearson correlation coefficient
We noted that in addition to genes known to be involved in DNA repair of UV-induced damage, a large set of highly up-regulated genes with unknown function was detected after 60 min cell recovery (Table 2). In order to annotate their physiological role, we mainly used the Smith-Waterman-reports that are available on the SubtiList-database. YneB, a coding sequence for a site-specific resolvase and yjcD, a likely ATP-dependent DNA helicase were found to exhibit the highest values for the group of unknown, up-regulated genes. In particular, yneB shows significant sequence similarity to a B. licheniformis (ATCC 14580) gene that encodes for a site-specific recombinase/resolvase (91% similarity). In addition, a large number of genes with uncertain function, such as ydaG, yraA and ytxG, which code for several general stress proteins were identified. From the Smith–Waterman-report it may be suggested that yraA displays either protease or peptidase functions as an adaptive response to atypical conditions. YdaG is a sigma-B-dependent gene, which codes for a general stress response protein. YtxG, another sigma-B controlled general stress gene, is induced by the addition of salt during logarithmical growth and it was observed to be under the influence of sigma-B regulated bacterial promoters (Petersohn et al. 2001). After validation of the DNA microarray it was found that not only sigB (RNA polymerase general stress sigma factor) was up-regulated (2.2) but also sigV, which exhibited with a ratio of 9.6 ± 2.1 the highest intensity of all sig-genes. Further on, YwaC encoding a putative GTP-pyrophosphokinase was found to be up-regulated during DNA repair and recently described as a member of the sigV regulon (Zellmeier et al. 2005)
Discussion
It was the overall aim of the present work to determine the DNA damage extent and gene expression profile in cells of B. subtilis in response to UV-C irradiation Therefore, the kinetics of DNA photoproduct repair, colony forming ability and gene activation after a 60 min recovery period were assessed under identical conditions for wild-type, HR- and NER-deficient B. subtilis cells. The formation of DNA bipyrimidine dimers was the major biochemical and physiological consequence of the UV-C radiation-induced stress. We observed that CPD TT, CPD TC and 6-4 TC were the major UV-C induced DNA lesions. It may be pointed out that a similar pattern of DNA damage was shown to be induced upon UV-C irradiation of other microorganisms such as Escherichia coli and the highly radiation resistant bacterium Deinococcus radiodurans (Koehler et al. 1996; Pogoda de la Vega et al. 2005). During subsequent recovery period, an increase in cell survival occurred concomitantly with a decrease in the level of the UV-C induced DNA photoproducts. The latter result further underlines the major role played by DNA photoproducts in UV-C induced cell lethality (Cadet et al. 2005). It is interesting to note that in B. subtilis DNA the 6-4 PPs and CPDs photoproducts were removed at a similar rate, whereas in UV-C irradiated E. coli the repair kinetics of CPDs and 6-4 PPs removal differ about two-fold shortly after 40 J/m2 (Koehler et al. 1996). This result points to strong differences in the unspecific PP dimer removal in the DNA repair of B. subtilis (as can see in Table 1) and to the necessity of a better assessment of genes involved in such processes. In conclusion, our work shows that the DNA PP repair efficiency in vegetative cells of B. subtilis corresponds to their genetic makeup and mutations in HR and NER leading to dramatic lowering of the PP repair.
The use of DNA microarray technology constitutes a relevant approach to determine gene expression profiles after UV-C induced stress. In the present study emphasis was placed on the application of the DNA microarray assay to study expression of genes involved in DNA repair during cell recovery. The majority of the up-regulated wild-type genes are known to be directly involved in DNA biochemical processes comprising mostly DNA repair mechanisms such as nucleotide and base excision repair, homologous recombination. In this respect recA exhibits the highest ratio of gene activation upon UV-C irradiation and damage recovery. As expected, a large group of dedicated repair genes including adaB and alkA (both DNA glycosylases involved in the adaptative response to DNA alkylation), dinB and tagC (DNA damage inducible genes), ligA and ligB (DNA ligases), dps (encoding a DNA-protecting protein; stress-induced gene), mutS (DNA mismatch repair recognition protein), ruvAB (Holliday junction DNA helicases, involved in HR) were found to be up-regulated after UV irradiation. The uvrAB operon encodes essential two excinucleases for the NER mechanism in order to cope with the elimination of DNA lesions such as CPDs and 6-4 PPs.
From the 278 wild-type highly up-regulated genes 106 genes (38.1%) were characterized as yet unknown whereas some were functionally classified according to their sequence homology with other microbial transcripts (data not shown). Thus hypothetical functions are proposed for these so called “y”-genes on the basis of either observed of sequence similarity with the highest match of protein databanks or on experimental evidence that did not lead to a change in the gene name (Sonenshein et al. 2002). In fact descriptions of the latter genes in databases such as SubtiList-database start with the words “putative”, “probable” or “possible” (Kunst et al. 1997; Moszer et al. 2002). In the present experiments, we used this information as a working hypothesis for the data analysis of the highly up-regulated unknown “y”-genes. Twenty-five, highly up-regulated “y”-genes are listed in Table 2 according to their group affiliation and their fluorescent intensity. This set of the unknown up-regulated genes could be separated in three groups, namely: direct DNA repair processes (e.g. potential DNA repair; yhaZ, yjhB, yqjH, ywqL, ywjD), indirect DNA process (e.g. ensuring DNA integrity: yefB, yisT, yjcD, yneB, yorL, yqfR, yshC) and other functions (e.g. detoxification processes and radiation stress response: ydaG, yerA, yhjE, yneA, yppQ, ytxG).
Our data suggest that two “unknown” genes are closely involved in the DNA repair processes of UV-C irradiated cells of B. subtilis during cell recovery, namely yneB and yfeB putative site-specific recombinases/resolvases, which are likely to be involved in recombination DNA repair (yneAB operon a recently described a SOS-inducible inhibitor of cell division (Au et al. 2005)), and yjcD, a ATP-dependent DNA helicase, expected to play an important role in genomic integrity and stability. YjcD shows interesting features according to the Smith-Waterman-report with significant sequence homology to a Bacillus clausii KSM-K16 (E value 1e-169) gene that encodes for an ATP-dependent DNA helicase, UvrD-like and controlled by the lexA. Recently, reports on new functional characterized DNA enzymes, e.g. a RuvABC resolvase and PcrA (Petit et al. 1998; Zahradka et al. 2002), an essential B. subtilis DNA helicase show that there is a need of a close interaction between DNA mechanisms, e.g. NER and cell recovery to ensure cell survival. This suggestion is mainly based on the information inferred from microarray analysis. Further detailed characterizations of the highly up-regulated “y”-genes are required (work in progress). Some of these finding, e.g. yneB (a site-specific resolvase) and yqjH (a potential DinB-induced DNA polymerase IV, involved in the translesion synthesis) obtained from B. subtilis and in homologue in the E. coli genome are in good agreement with previous reports on general and radiation stress-induced global transcriptional analysis by Au et al. (2005), Courcelle et al. (2001) and Goranov et al. (2006). We identified and therefore suggest that in addition to the recognized genes other “yet unknown” genes are involved and play an important role in DNA repair of UV-C irradiated cells of B. subtilis during radiation stress response and cell recovery. This is likely the case for sigV, a possible UV irradiation response sigma factor for which the delineation of its exact role is awaiting further experiments, complementary to previous studies performed with sigB-depending stress genes (Haldenwang et al. 1995; Petersohn et al. 2001). Our results show that the precise details of DNA repair after UV-C irradiation of B. subtilis are yet not completely clarified and that the use of modern techniques like DNA microarray technology allows for the first time simultaneous monitoring of genome-wide analysis for known and unknown genes. It is essential to identify the transcriptional response of DNA damage induction and repair with regard to UV-C induced bipyrimidine lesions. Several “y”-genes such as yneA, yneB, yhaZ and ytxG were recently, found to be directly involved in DNA repair and/or stress response (Au et al. 2005; Petersohn et al. 2001). Follow-on experiments based on the comparison of the microarray-based gene expression analysis after UV-A irradiation ≥320 nm to 254-nm data and the verification of selected candidate genes (e.g. yefB) via RT-PCR are in progress (Moeller et al., in preparation). Herewith, further information on the genetic level of microbial UV photobiology and DNA repair strategies (according to Zahradka et al. 2006) will be obtained.
References
Au N, Kuester-Schoeck E, Mandava V, Bothwell LE, Canny SP, Chachu K, Colavito SA, Fuller SN, Groban ES, Hensley LA, O’Brien TC, Shah A, Tierney JT, Tomm LL, O’Gara TM, Goranov AI, Grossman AD, Lovett SM (2005) Genetic composition of the Bacillus subtilis SOS system. J Bacteriol 187:7655–7666
Billen D, Carreira LB, Hadden CT, Silverstein SJ (1971) Evidence suggestive of compartmentalization of deoxyribonucleic acid-synthesizing systems in freeze-treated Bacillus subtilis. J Bacteriol 108:1250–1256
Blencke HM, Homuth G, Ludwig H, Mader U, Hecker M, Stulke J (2003) Transcriptional profiling of gene expression in response to glucose in Bacillus subtilis: regulation of the central metabolic pathways. Metab Eng 5:133–149
Cadet J, Sage E, Douki T (2005) Ultraviolet radiation-induced damage to cellular DNA. Mutat Res 571:3–17
Courcelle J, Khodursky A, Peter B, Brown PO, Hanawalt PC (2001) Comparative gene expression profiles following UV exposure in wild-type and SOS-deficient Escherichia coli. Genetics 158:41–64
Douki T, Cadet J (2001) Individual determination of the yield of the main UV-induced dimeric pyrimidine photoproducts in DNA suggests a high mutagenicity of CC photolesions. Biochemistry 40:2495–2501
Douki T, Setlow B, Setlow P (2005) Effects of the binding of alpha/beta-type small, acid-soluble spore proteins on the photochemistry of DNA in spores of Bacillus subtilis and in vitro. Photochem Photobiol 81:163–169
Draghici S (2002) Statistical intelligence: effective analysis of high-density microarray data. Drug Discov Today 7:55–63
Eichenberger P, Jensen ST, Conlon EM, van Ooij C, Silvaggi J, Gonzalez-Pastor JE, Fujita M, Ben-Yehuda S, Stragier P, Liu JS, Losick R (2003) The sigmaE regulon and the identification of additional sporulation genes in Bacillus subtilis. J Mol Biol 327:945–972
Fernandez S, Sorokin A, Alsonso JC (1998) Genetic recombination in Bacillus subtilis 168: effects of recU and recS mutations on DNA repair and homologous recombination. J Bacteriol 180:3405–3409
Friedberg EC, Walker GC, Siede W (1995) DNA repair and mutagenesis. ASM Press, Washington pp 698–735
Goranov AI, Kuester-Schoeck E, Wang JD, Grossman AD (2006) Characterization of the global transcriptional responses to different types of DNA damage and disruption of replication in Bacillus subtilis. J Bacteriol 188:5595–5605
Hadden CT (1979) Pyrimidine dimer excision in a Bacillus subtilis Uvr-mutant. J Bacteriol 139:247–255
Haldenwang WG (1995) The sigma factors of Bacillus subtilis. Microbiol Rev 59:1–3
Hanlin JH, Lombardi SJ, Slepecky RA (1985) Heat and UV light resistance of vegetative cells and spores of Bacillus subtilis rec-mutants. J Bacteriol 163:774–777
Helmann JD, Wu MF, Gaballa A, Kobel PA, Morshedi MM, Fawcett P, Paddon C (2003) The global transcriptional response of Bacillus subtilis to peroxide stress is coordinated by three transcription factors. J Bacteriol 85:243–253
Helmann JD, Wu MF, Kobel PA, Gamo FJ, Wilson M, Morshedi MM, Navre M, Paddon C (2001) Global transcriptional response of Bacillus subtilis to heat shock. J Bacteriol 183:7318–7328
Koehler DR, Courcelle J, Hanawalt PC (1996) Kinetics of pyrimidine(6–4)pyrimidone photoproduct repair in Escherichia coli. J Bacteriol 178:1347–1350
Kunst F, Ogasawara N, Moszer I, Albertini AM, Yoshikawa H, Danchin A et al (1997) The complete genome sequence of the gram-positive bacterium Bacillus subtilis. Nature 390:249–256
Moeller R, Douki T, Cadet J, Stackebrandt E, Nicholson WL, Rettberg P, Reitz G, Horneck G (2007) UV radiation induced formation of DNA bipyrimidine photoproducts in Bacillus subtilis endospores and their repair during germination. Int Microbiol 10:39–46
Moeller R, Horneck G, Facius R, Stackebrandt E (2005) Role of pigmentation in protecting Bacillus sp. endospores against environmental UV radiation. FEMS Microbiol Ecol 51:231–236
Moeller R, Horneck G, Rettberg P, Mollenkopf HJ, Stackebrandt E, Nicholson WL (2006) A method for extracting RNA from dormant and germinating Bacillus subtilis strain 168 endospores. Curr Microbiol 53:227–231
Moszer I, Jones LM, Moreira S, Fabry C, Danchin A (2002) SubtiList: the reference database for the Bacillus subtilis genome. Nucleic Acids Res 30:62–65
Pastink A, Eeken JCJ, Lohman PHM (2001) Genomic integrity and the repair of double-strand DNA breaks. Mutat Res 480:37–50
Petersohn A, Brigulla M, Haas S, Hoheisel JD, Voelker U, Hecker M (2001) Global analysis of the general stress response of Bacillus subtilis. J Bacteriol 183:5617–5631
Petit MA, Dervyn E, Rose M, Entian KD, McGovern S, Ehrlich SD, Bruand C (1998) DNA repair: bacteria PcrA is an essential DNA helicase of Bacillus subtilis fulfilling functions both in repair and rolling-circle replication. Mol Microbiol 29:261–273
Pogoda de la Vega U, Rettberg P, Douki T, Cadet J, Horneck G (2005) Sensitivity to polychromatic UV-radiation of strains of Deinococcus radiodurans differing in their DNA repair capacity. Int J Radiat Biol 81:601–611
Ravanat J-L, Douki T, Cadet J (2001) Direct and indirect effects of UV radiation on DNA and its components. J Photochem Photobiol Biol B 63:88–102
Smith BT, Grossman AD, Walker GC (2002) Localization of UvrA and effect of DNA damage on the chromosome of Bacillus subtilis. J Bacteriol 184:488–493
Sonenshein AL, Hoch JA, Losick R (2002) Appendix: functional classification of the Bacillus subtilis protein-encoding genes. In: Sonenshein AL, Hoch JA, Losick R (eds) Bacillus subtilis and its closest relatives: from genes to cells, Appendix 2. ASM Press, Washington, pp 570–615
Steil L, Hoffmann T, Budde I, Volker U, Bremer E (2003) Genome-wide transcriptional profiling analysis of adaptation of Bacillus subtilis to high salinity. J Bacteriol 185:6358–6570
Tusher VG, Tibshirani R, Chu G (2001) Significance analysis of microarrays applied to ionizing radiation response. Proc Natl Acad Sci USA 98:5116–5121
Zahradka K, Slade D, Bailone A, Sommer S, Averbeck D, Petranovic M, Lindner AB, Radman M (2006) Reassembly of shattered chromosomes in Deinococcus radiodurans. Nature 443:569–573
Zahradka D, Zahradka K, Petranovic M, Dermic D, Brcic-Kostic K (2002) The RuvABC resolvase is indispensable for recombinational repair in sbcB15 mutants of Escherichia coli. J Bacteriol 184:4141–4147
Zellmeier S, Hofmann C, Thomas S, Wiegert T, Schumann W (2005) Identification of sigma(V)-dependent genes of Bacillus subtilis. FEMS Microbiol Lett 253:221–229
Acknowledgments
The authors thank Joerg Angermann and Ina Wagner for their excellent technical assistance during the DNA microarray experiments. These results will be included in the PhD thesis of Ralf Moeller.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by Jorge Membrillo-Hernández.
Rights and permissions
About this article
Cite this article
Moeller, R., Stackebrandt, E., Douki, T. et al. DNA bipyrimidine photoproduct repair and transcriptional response of UV-C irradiated Bacillus subtilis . Arch Microbiol 188, 421–431 (2007). https://doi.org/10.1007/s00203-007-0263-4
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00203-007-0263-4