Abstract
Genes encoding dextranolytic enzymes were isolated from Paenibacillus strains Dex40-8 and Dex50-2. Single, similar but non-identical dex1 genes were isolated from each strain, and a more divergent dex2 gene was isolated from strain Dex50-2. The protein deduced from the Dex40-8 dex1 gene sequence had 716 amino acids, with a predicted Mr of 80.8 kDa. The proteins deduced from the Dex50-2 dex1 and dex2 gene sequences had 905 and 596 amino acids, with predicted Mr of 100.1 kDa and 68.3 kDa, respectively. The deduced amino acid sequences of all three dextranolytic proteins had similarity to family 66 glycosyl hydrolases and were predicted to possess cleavable N-terminal signal peptides. Homology searches suggest that the Dex40-8 and Dex50-2 Dex1 proteins have one and two copies, respectively, of a carbohydrate-binding module similar to CBM_4_9 (pfam02018.11). The Dex50-2 Dex2 deduced amino acid sequence had highest sequence similarity to thermotolerant dextranases from thermophilic Paenibacillus strains, while the Dex40-8 and Dex50-2 Dex1 deduced protein sequences formed a distinct sequence clade among the family 66 proteins. Examination of seven Paenibacillus strains, using a polymerase chain reaction-based assay, indicated that multiple family 66 genes are common within this genus. The three recombinant proteins expressed in Escherichia coli possessed dextranolytic activity and were able to convert ethanol-insoluble blue dextran into an ethanol-soluble product, indicating they are endodextranases (EC 3.2.1.11). The reaction catalysed by each enzyme had a distinct temperature and pH dependence.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Dextrans are polydisperse, long-chain polymers of glucose linked predominantly through α-1,6-glycosidic bonds. A variety of other linkages found in natural dextrans introduce branching into the polymer (Walker 1978). Dextrans are produced by members of the Lactobacillaceae and are biotechnologically useful as chromatography supports (Porath 1997), blood expanders (Tollofsrud et al. 2001) and potential drug delivery vehicles (Davis and Anseth 2002). Despite their usefulness, naturally occurring dextrans are detrimental as a component of dental plaque (Loesche 1986) and as a contaminant of deteriorated sugarcane and beets (Tilbury and French 1974; de Bruijn 2000).
Dextranases (EC 3.2.1.11) cleave the α-1,6-glycosidic linkages in the interior of dextrans, producing linear oligosaccharides. Based on protein sequence comparisons, the characterised dextranases fall into two distinct groups of glycosyl hydrolases. The dextranases from five Streptococcus species (Wanda and Curtiss 1994; Igarashi et al. 1995, 2001, 2004; Ohnishi et al. 1995) belong to glycosyl hydrolase sequence family 66. The cycloisomaltooligosaccharide glucanotransferases (citases) from Bacillus circulans also attack dextran endolytically and have high sequence similarity to the family 66 hydrolases, but catalyse a translocation reaction that produces cyclic oligosaccharides (Oguma et al. 1994). Unlike the enzymes from the bacilli, the dextranases from strains of the actinobacterium Arthrobacter globiformis belong to glycosyl hydrolase family 49 (Oguma et al. 1999). This family also includes the dextranase sequences from Penicillium minioluteum and P. funiculosum (Coutinho and Henrissat 1999).
During a survey to discover potential biotechnologically useful endodextranases, ten dextranolytic Paenibacillus strains showing a wide range of optimal growth temperatures and containing multiple dextranolytic proteins were isolated from garden compost and sugar-mill samples (Finnegan et al. 2004a). The gene sequences of two approximately 70-kDa thermostable dextranases from different thermotolerant Paenibacillus strains were determined and found to encode glycosyl hydrolase family 66 proteins (Finnegan et al. 2004b). Both recombinant enzymes had optimal activity at about 60°C and pH 5.5. The sequence and biochemical diversity of dextranolytic enzymes produced by the non-thermophilic Paenibacillus strains Dex50-2 and Dex40-8 are explored here. Dex50-2 and Dex40-8 have the most diverse 16S rRNA gene sequences of the non-thermophilic Paenibacillus strains examined (Finnegan et al. 2004a). These strains also possess the most diverse sets of monomeric dextranolytic enzymes. Dex50-2 produces three enzymes, of 70, 90 and 105 kDa, while Dex40-8 produces up to five enzymes, of 68, 70, 85, 105 and 120 kDa (Finnegan et al. 2004a).
Materials and methods
Bacterial strains and growth
Dextranolytic Paenibacillus strains Dex40-8 and Dex50-2, isolated from Australian sugarcane-mill samples, were grown to confluence at their optimal growth temperature of 45°C on indicator plates containing dextran coupled with Cibacron Blue (Blue Dextran, Amersham Biosciences, Sydney, Australia), as described (Finnegan et al. 2004a). Escherichia coli TOP10F′ cells (One-Shot Competent Cells, Invitrogen, Mount Waverley, VIC, Australia) were used for all DNA cloning and protein-expression experiments.
DNA manipulation and screening of genomic libraries
Unless otherwise stated, DNA was manipulated using established methods (Sambrook et al. 1989). Libraries were constructed from genomic DNA purified (Morris et al. 1995) from freshly grown Paenibacillus strains Dex40-8 and Dex50-2. Approximately 10 μg genomic DNA was partially digested with Sau3AI and twice size-selected for 4- to 8-kb fragments on agarose gels stained with methylene blue (Flores et al. 1992). Size-selected DNA fragments were ligated into pBluescript II (Stratagene) previously digested with BamHI and treated with shrimp alkaline phosphatase (Amersham Biosciences) as instructed by the supplier prior to transformation into E. coli. The libraries were screened for dextranolytic activity as described (Finnegan et al. 2004b), except dextran degradation was performed at 37°C. Dextranolytic E. coli were recovered by coring the colonies from plates and growing in liquid Luria–Bertani medium before isolating the plasmid DNAs carrying putative dextranase genes. The recognition sites of selected restriction endonucleases within each plasmid insert were mapped.
DNA sequencing and sequence analysis
Plasmid DNA sequencing templates were purified from small-scale SDS-alkaline lysis preparations, using a commercial kit (Concert Rapid PCR Purification System, Invitrogen), and sequencing reactions were performed using fluorescent dideoxy terminator chemistry (BigDye, Applied Biosystems, Scoresby, VIC, Australia), according to the manufacturers’ instructions. The sequencing products were separated (DNA Sequencing Facility, Macquarie University, Sydney, Australia), analysed (MacVector Sequence Analysis Software, Accelrys, Sydney, NSW, Australia) and compared to public database sequences, using the BLAST and rpsblast algorithms available at the National Center for Biotechnology Information Web site (http://www.ncbi.nlm.nih.gov/BLAST/). The GenBank accession numbers for the genomic DNA fragments encoding the identified Paenibacillus dextranase genes are AY326309 (Dex40-8 dex1), AY326310 (Dex50-2 dex1) and AY326311 (Dex50-2 dex2). Deduced amino acid sequences were analysed using SignalP, version 3.0 (Bendtsen et al. 2004), to detect possible N-terminal signal sequences and predict cleavage sites.
Assay of recombinant dextranases
E. coli cells transformed with plasmids carrying genes encoding dextranolytic enzymes were lysed by resuspending in B-Per II detergent solution (Pierce, Rockford, Ill., USA). Dextranolytic activity was determined as previously described (Finnegan et al. 2004b), either by monitoring the release of Cibacron Blue dye from Blue dextran (Wynter et al. 1995) or by monitoring the release of reducing sugars from Dextran T-2000 (Amersham Biosciences) using p-hydroxybenzoic acid hydrazide (Lever 1973). Lysates of non-transformed E. coli lacked detectable dextranolytic activity.
Oligonucleotide primer design
BLOCKMAKER software, available at the Baylor College of Medicine Web site (http://www.bcm.edu), was used to identify blocks of amino acid sequence that were highly conserved across family 66 protein sequences. The aligned sequence blocks corresponding to Val143–Tyr153 and Phe306–Glu316 of the Paenibacillus strain Dex40-8 Dex1 protein were used to design a convergent pair of degenerate oligonucleotide primers, using the CODEHOP strategy (Rose et al. 1998; http://www.bioinformatics.weizmann.ac.il/blocks/codehop.html). The primer sequences were 5′-GTTTCAACTGATTGGACCAAATWYCCNCGNTAYG-3′, which corresponds to the sense sequence encoding Val143–Tyr153, and 5′-TCTCCAATTGTATCACCATGCMWNCCRTCAA-3′, which corresponds to the antisense sequence of the region encoding Phe306–Glu316.
PCR assays
Each 20-μl reaction contained 10 mM Tris-HCl (pH 8.3), 50 mM KCl, 1.5 mM MgCl2, 0.2 mM each dNTP, 0.2 μM each primer and 0.8 U thermostable DNA polymerase (MasterTaq, Eppendorf, Sydney, NSW, Australia). The templates were added by touching fresh bacterial colonies with a sterile plastic pipette tip and transferring the smallest visible amount of biomass to the reaction tube. Reactions were cycled using a touchdown protocol: (95°C, 10 min) × 1 cycle; (95°C, 30 s; 65°C, 30 s; 72°C, 1 min) × 15 cycles, with the annealing temperature (initially 65°C) decreasing 1°C per cycle; (95°C, 30 s; 50°C, 30 s; 72°C, 1 min) × 30 cycles; and (72°C, 10 min) × 1 cycle. The products were separated on agarose gels and the approximately 525-bp product excised, purified (Concert Rapid PCR Purification System, Invitrogen) and ligated into a T-tailed vector (TA Cloning Kit, Invitrogen), using commercial kits as directed by the manufacturers. Recombinant plasmids were obtained, purified and sequenced.
Results
Isolation of genes encoding dextranolytic enzymes
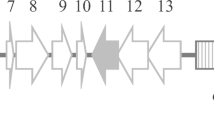
Plasmid-based genomic DNA libraries were constructed for the dextran-degrading Paenibacillus strains Dex40-8 and Dex50-2. Library screening showed that the Dex40-8 and Dex50-2 libraries contained three and five plasmids, respectively, that were able to confer dextranolytic activity onto E. coli. Mapping of restriction sites within each plasmid insert showed that the three Dex40-8 library plasmids contained the same 5.3-kb insert (Fig. 1). The five Dex50-2 library plasmids covered two distinct genomic regions, of 4.2 kb and 5.4 kb (Fig. 1).
Localisation of dextranolytic enzyme genes on Paenibacillus genomic DNA fragments. The locations of open reading frames (ORFs) (boxes) found within each genomic DNA fragment are shown. The BLAST algorithm at the National Center for Biotechnology Information Web site (http://www.ncbi.nlm.nih.gov) was used to determine the most likely function of each ORF (I, II, III and V permeases of ABC sugar transport systems, IV oligo-1,6-glucosidase, VI and VII two component sensor histidine kinase/methyl accepting chemotaxis proteins). ORFs I, IV, V and VII are incomplete. The recognition sites for BamHI (B), BstXI (X), EcoRI (E), EcoRV (V), HindIII (H), KpnI (K), SacI (C), SalI (S) and SmaI (M) are shown
The DNA sequence was determined from both strands of each isolated genomic DNA fragment. One open reading frame (ORF) on each fragment encoded a protein with significant sequence similarity to the Streptococcus mutans family 66 dextranase (Fig. 2) and other family 66 glycosyl hydrolases (Fig. 3). Of the remaining ORFs, only incomplete ORF IV on the 4.2-kb Dex50-2.1 genomic fragment (Fig. 1) encoded a protein with homology to a known glycosylase. The protein encoded by incomplete ORF IV had greatest identity (59%) to the Bacillus cereus ATCC7064 oligo-1,6-glucosidases, a family 13-glycosyl hydrolase. The dextranolytic activity of E. coli transformed with the Dex50-2.1 fragment was probably not due to oligo-1,6-glucosidase activity encoded by ORF IV, because, by analogy, a B. cereus oligo-1,6-glucosidase with an N-terminal truncation similar to that of the ORF IV-encoded protein was inactive (Watanabe et al. 2001).
An alignment of Paenibacillus family 66 amino acid sequences. The amino acid sequences deduced from the Paenibacillus Dex40-8 dex1 (40-8.1, accession AY326309) and Paenibacillus Dex50-2 dex1 (50-2.1, accession AY326310) and dex2 (50-2.2, accession AY326311) genes were aligned, along with the Streptococcus mutans dextranase sequence (SmDex, accession BAA08409), using the Clustal W algorithm (Baylor College of Medicine Search Launcher, http://searchlauncher.bcm.tmc.edu/). Gaps were introduced to maximise the alignment. Numbers on the left indicate residue positions within each sequence. Amino acid identities (black) or similarities (grey) present in three or more of the sequences are boxed. The boundaries of the N-terminal and C-terminal variable regions with the conserved region (Igarashi et al. 2002) are marked (σ). Putative signal peptide cleavage sites ( [ ) are indicated by vertical arrows. The aspartate residue that is absolutely conserved among family 66 dextranases and has been implicated in catalysis is indicated (*), as is the site of the approximately 120-amino acid residue insertion present in the Bacillus circulans citase sequences (θ, accessions P94286 and P70873). The boundaries of the repeated sequences found in Dex50-2 dex1 protein are indicated by the double-headed arrows above the alignment. The residues within the repeat sequences that are identical to the consensus sequence derived from 49 proteins containing the CBM_4_9 (pfam02018.11) carbohydrate-binding domain are underlined
A phylogram of the family 66 glycosylases. Included in the comparison are amino acid sequences of the Paenibacillus Dex40-8 dex1, and Paenibacillus Dex50-2 dex1 and dex2 dextranases described here, the Paenibacillus Dex70-1B (accession AY326312) and Paenibacillus Dex70-34 (accession AY326313) dextranases, the Streptococcus salivarius (accession Q59979), S. mutans (accession BAA08409) and Streptococcus sobrinus (accession T30291) dextranases and the B. circulans Cit1 (accession P94286) and Cit2 (accession P70873) citases. The phylogram was constructed using a Clustal W alignment package (European Bioinformatics Institute, http://www.ebi.ac.uk). Branch lengths and associated numerals indicate the fraction of positions that differ between sequences
The Dex40-8 family 66 ORF was designated dex1. The family 66 ORF located on the Dex50-2.1 genomic DNA fragment (Fig. 1) was also designated dex1, because the deduced protein sequence was more similar to the protein sequence deduced from the Dex40-8 dex1 gene (78% identical) than to the protein sequence deduced from the family 66 ORF on the Dex50-2.2 genomic fragment (32% identical, Figs. 2, 3). The family 66 ORF on Dex50-2.2 was designated dex2.
Characteristics of the deduced family 66 proteins
The deduced Dex40-8 dex1 protein had 715 amino acid residues and a predicted Mr of 80.8 kDa. The amino acid sequence was similar to other family 66 glycosylases over its entire length, with 25% sequence identity with the analogous portion of the S. mutans dextranase (Fig. 2). The deduced Dex50-2 dex1 protein had 905 amino acid residues and a predicted Mr of 100.1 kDa. This protein sequence was 78% and 24% identical to the analogous portions of the Dex40-8 dex1 protein and the S. mutans dextranase, respectively. The Dex50-2 dex1 protein also had a 200 amino acid residue C-terminal extension relative to the Dex40-8 dex1 protein (Fig. 2). The deduced Dex50-2 dex2 protein had 596 amino acid residues with a predicted Mr of 68.3 kDa, making it the smallest family 66 glycosylase so far characterised. The Dex50-2 dex2 protein had sequence similarity across its entire length to all family 66 glycosylases, including 32% identity with both the Dex40-8 and Dex50-2 dex1 proteins and 25% identity with the S. mutans dextranase (Fig. 2). The Dex50-2 dex2 protein sequence was 56% and 57% identical to the sequences deduced from dex1 genes encoding thermoactive family 66 glycosylases in Paenibacillus strains Dex70-34 and Dex70-1B, respectively (Finnegan et al. 2004b). The Dex40-8 and Dex50-2 dex1 protein sequences had 32–36% identity to the Dex70-1B and Dex70-34 protein sequences.
Structural relationships among Paenibacillus family 66 hydrolases
All three Paenibacillus family 66 proteins described here had the aspartate residue that is absolutely conserved in all characterised family 66 glycosylases (Asp312 in Dex40-8 Dex1, Fig. 2) and is necessary for catalytic function of the S. mutans dextranase (Igarashi et al. 2002). Like the streptococcal dextranases, all three Paenibacillus sequences lack the insertion of approximately 120 residues found in the citases of family 66 (Fig. 2). The Paenibacillus dextranases have a truncated N-terminal variable region compared to the S. mutans and other streptococcal dextranase sequences (Igarashi et al. 2002). Despite the shorter N-terminal variable regions, the SignalP algorithm predicted that all three Paenibacillus family 66 proteins possess cleavable N-terminal signal peptides (Fig. 2). After processing, the mature Paenibacillus proteins would be 76.7 kDa (Dex40-8 dex1), 97.7 (Dex50-2 dex1) and 64.5 (Dex50-2 dex2) kDa. The N termini produced by the predicted cleavage of the Dex40-8 and Dex50-2 dex1 proteins (Fig. 2) would coincide well with the N-terminal sequences directly determined for other bacterial dextranases. A strain identified as P. illinoisensis produces three dextranolytic proteins, each with the N-terminal sequence ASTGL (Khalikova et al. 2003), while the N terminus of a dextranase from Thermoanaerobacter strain Rt364 has the sequence **TGNLIQRVYTDCARYNPGDLVTIAANLI, where the first two residues (designated *) were not determined (Wynter et al. 1997).
The Paenibacillus family 66 proteins differ from one another in length largely due to variation in their C-terminal variable regions (Fig. 2), as defined by Igarashi et al. (2002) from sequence comparisons among the streptococcal family 66 proteins. In alignments of deduced amino acid sequences (Fig. 2), the C terminus of the Dex50-2 dex2 protein is almost precisely at the designated boundary between the central conserved core region and the C-terminal variable region of the S. mutans family 66 dextranase (Igarashi et al. 2002). Unlike the Dex50-2 dex2 protein, the Dex40-8 and Dex50-2 dex1 proteins have C-terminal variable regions, of 98 and 298 amino acid residues, respectively. While the C-terminal regions have high sequence similarity (69% sequence identity) over the 98 residues of overlap (Fig. 2), they have only limited similarity to the corresponding region of the S. mutans family 66 dextranase. The 98 residue overlap between C-terminal variable regions of the Dex40-8 and Dex50-2 dex1 proteins covers nearly one complete iteration of a 134 amino acid imperfect direct repeat sequence located within the C-terminal variable region of Dex50-2 dex1 (Fig. 2). A rpsblast search of the Conserved Domain Database (Marchler-Bauer et al. 2003) through the National Center for Biotechnology Information Web site revealed that the N-terminal 75 amino acid residues of each repeat motif had limited sequence similarity with the N-terminal half of the CBM_4_9 (pfam02018.11) carbohydrate-binding domain (Fig. 2). The CBM_4_9 motif is found in a number of glycosylases, where it has been shown to bind xylan and a variety of β-glucans.
Multiple family 66 genes among the Paenibacillus
A PCR-based assay was used to investigate whether other dextranolytic Paenibacillus strains harbour more than one family 66 gene. Degenerate primers were designed to anneal to the Paenibacillus family 66 gene sequences that encode highly conserved amino acid sequences. The primers were used to amplify a 521-bp DNA fragment corresponding to Val143–Glu316 of the Dex40-8 dex1 protein from strains Dex40-4, Dex40-5, Dex40-6, Dex40-8 and Dex50-2. Paenibacillus strains Dex70-1B and Dex70-34 (Finnegan et al. 2004a) were also included in this analysis to determine if these thermotolerant strains contained additional family 66 genes to those previously isolated by library screening (Finnegan et al. 2004b). The amplified products were ligated into a plasmid vector and individual inserts sequenced. Fragments from multiple family 66-like genes were obtained for strains Dex40-4, Dex40-5 and Dex50-2, while fragments from single genes were found for Dex40-6, Dex40-8, Dex70-1B and Dex70-34. The two Dex50-2 sequences were identical to the corresponding dex1 and dex2 gene sequences. Sequences homologous to both dex1 and dex2 were obtained from Dex40-4 and Dex40-5, but Dex40-5 also yielded a third divergent family 66 gene sequence. The Dex40-4 dex1 and dex2 partial sequences were identical to the homologous sequences from the corresponding Dex50-2 genes, supporting the conclusion that Dex40-4 and Dex50-2 are independent isolates of the same species (Finnegan et al. 2004a). The partial Dex40-5 dex1 sequence was identical to the corresponding region of Dex40-8 dex1, but the two other Dex40-5 family 66 partial gene sequences that were obtained were unique. While fragments from single dex1-like genes were isolated from Dex40-6, Dex40-8, Dex70-1B and Dex70-34, the sequencing of more recombinant plasmids is needed to verify that there are no other genes. The family 66 gene fragments isolated for Dex70-1B and Dex70-34 were identical in sequence to the respective dex1 genes isolated by library screening (Finnegan et al. 2004b).
Biochemical characteristics of the family 66 glycosylases
Each Paenibacillus family 66 enzyme was expressed in E. coli and assayed for dextranolytic activity. Incubation of the recombinant enzymes with blue dextran resulted in the time-dependent conversion of the dextran-bound Cibacron Blue dye from an ethanol-insoluble to an ethanol-soluble form. The solubilisation of Cibacron Blue went to completion if adequate time was allowed. These observations suggested the family 66 enzymes were endo acting (Wynter et al. 1997). When Dextran T-2000 was used as the substrate, reducing sugars were released, indicating these family 66 enzymes catalysed a hydrolysis reaction. Thus, the Paenibacillus enzymes appear to be dextranases (EC 3.2.1.11) and not citases.
The Dex40-8 dex1 and Dex50-2 dex2 enzymes had similar thermoactivity profiles (Fig. 4a), with maximal activity between 35°C and 40°C, under the assay condition used here. Both enzymes declined in activity at higher incubation temperatures and were largely inactive above 50°C. The enzyme from the Dex50-2 dex1 gene was more thermotolerant, having maximal activity at about 45°C with the assay conditions used.
Temperature and pH dependence of Paenibacillus dextranases. The activity of recombinant Paenibacillus Dex40-8 dex1 (black circles) and Paenibacillus Dex50-2 dex1 (black squares) and dex2 (black triangles) dextranases expressed in Escherichia coli was measured by determining the amount of reducing sugar equivalents released from Dextran T-2000 during a 30-min incubation. For a given enzyme, the activities are expressed relative to the maximal activity obtained in each experiment. a Cleared lysates were assayed at the indicated temperatures in reaction mixtures at pH 7.5. b Cleared lysates were assayed at the indicated pH, at the temperature found in a to give maximal activity. Each point represents the average of triplicate determinations of the reducing sugar released in triplicate reactions
The reaction catalysed by each enzyme had a unique pH dependence (Fig. 4b). The Dex40-8 dex1 enzyme had >80% maximal activity from pH 6.5 – 8, with highest activity between pH 7.0–7.5. The Dex50-2 dex1 enzyme had a similarly broad, but somewhat more acidic, pH dependence profile, with maximal activity at pH 7.0. The Dex50-2 dex2 protein had a substantially narrower pH dependence profile, due to being largely inactive within one pH unit below its pH optimum of 6.0–6.5.
Discussion
ORFs encoding family 66 glycosyl hydrolases from Paenibacillus strains Dex40-8 or Dex50-2 conferred dextranolytic activity on transformed E. coli. This conclusion is based upon the presence of ORFs encoding family 66 hydrolases on all three genomic DNA fragments characterised here, the absence of any other set of homologous ORFs common to all three DNA fragments and the previous demonstration that family 66 enzymes from other Paenibacillus strains (Finnegan et al. 2004b), Streptococcus spp. (Wanda and Curtiss 1994; Igarashi et al. 1995) and B. circulans (Oguma et al. 1994) are dextranolytic. Indeed, all three Paenibacillus family 66 proteins characterised here are clearly true dextranases (EC 3.2.1.11), as each enzyme completely converts Blue Dextran into an ethanol-soluble form in plate and solution assays. Moreover, the degradation of both Blue Dextran and Dextran T-2000 in solution assays produced reducing sugars, demonstrating that the dextranolytic activity was due to hydrolysis of glycosidic bonds rather than a citase-like translocation, which does not produce reducing ends (Oguma et al. 1994).
Of the non-family 66 proteins encoded by the characterised genomic fragments, only the partial oligo-α-1,6-glucosidase encoded by the N-terminally truncated ORF IV (Fig. 1) has sequence similarity to enzymes known to hydrolyse dextran. The truncated Dex50-2 oligo-α-1,6-glucosidase is unlikely to be active, because the enzyme lacks residues essential for substrate binding and catalysis (Watanabe et al. 2001). In addition, the activity of an oligo-α-1,6-glucosidase would remove successive glucose residues from the non-reducing end of dextran and stop at non-α-1,6 branch points. Therefore, only a true dextranase, not encoded by ORF IV, can account for the fact that lysates of the Dex50-2.1 clone are able to fully convert Blue Dextran into ethanol-soluble products.
The three Paenibacillus dextranase protein sequences characterised here are fairly typical of family 66 proteins (Igarashi et al. 2004), having the conserved aspartate residue that has been implicated in catalysis (Igarashi et al. 2002) and a central core region whose sequence is well conserved across the family. Like the five characterised streptococcal proteins (Igarashi et al. 2001, 2004), the Paenibacillus proteins were predicted to have cleavable N-terminal signal peptides, consistent with the presence of dextranase activity in Paenibacillus culture supernatants (Finnegan et al. 2004a). In sequence comparisons (Fig. 3), the deduced Dex50-2 dex2 protein sequence clustered with the thermoactive enzymes from Dex70-1B and Dex70-34 (Finnegan et al. 2004b), despite being one of the least thermoactive of the Paenibacillus dextranases examined so far, while the sequences inferred from the Dex40-8 and Dex50-2 dex1 genes formed a separate clade within the previously characterised family 66 proteins.
The clustering of the Dex40-8 and Dex50-2 dex1 protein sequences was in part due to the proteins having homologous C-terminal variable regions. These regions are absent from some family 66 proteins, including the Dex50-2 dex2 protein (Fig. 2). The extensions in the Dex40-8 and Dex50-2 dex1 proteins shared little or no similarity with the C-terminal variable regions defined for the streptococcal family 66 proteins (Igarashi et al. 2001) and contained one and two copies, respectively, of a sequence with limited similarity to the CBM_4_9 carbohydrate-binding module. The CBM_4_9 motif has been shown to bind xylan and a number of β-glucans, and it is counterintuitive that the Dex40-8 and Dex50-2 dex1 proteins, which hydrolyse the α-1,6 linkages in dextran, would have the ability to bind to xylans or β-glucans. It is more likely, given the low sequence identity between the Paenibacillus motifs and CBM_4_9 (Fig. 2), that the Paenibacillus domains represent a novel dextran-binding module. It is also likely, by analogy to other family 66 proteins, that all three Paenibacillus family 66 proteins possess at least one dextran-binding module within their core conserved regions. As shown by Morisaki et al. (2002), N-terminal deletions of the family 66 enzyme of S. mutans abolished catalytic activity, but not dextran binding. Enzyme with all but 20 residues of the C-terminal extension deleted also retained dextran-binding activity, but the affinity (and activity) was lost if 117 residues were deleted from the C-terminus of the core region. The nature of the dextran-binding domain within the conserved core region is unclear, but the conservation across the Paenibacillus, Bacillus and Streptococcus family 66 proteins of invariant amino acid residues within the C-terminal 117 residues of the conserved core (Fig. 2, Finnegan et al. 2004b; Igarashi et al. 2004) will give guidance for further study.
Paenibacillus strains Dex40-8 and Dex50-2 possess five and three dextranolytic proteins, respectively (Finnegan et al. 2004a). However, library screening and PCR using degenerate primers only revealed two family 66 genes for Dex50-2 and a single family 66 gene for Dex40-8. This could mean that there are no other dextranase genes in these species, and that the multiple dextranolytic proteins seen on activity gels are the result of post-translational modifications such as glycosylation or proteolysis. However, we cannot rule out the existence of other dextranase genes, which might have escaped detection because they failed to produce active enzymes in the library screening and did not hybridise with the degenerate primers used for PCR amplification.
The gene sequence diversity seen here among three Paenibacillus family 66 proteins, coupled with the size diversity of dextranolytic proteins among Paenibacillus isolates revealed by electrophoretic profiling (Finnegan et al. 2004a), indicates that dextranase sequence diversity may be considerably wider than illustrated by the five Paenibacillus family 66 gene sequences currently available. The genus Paenibacillus, then, is an important and readily accessible repository for diverse dextranase genes and potentially novel carbohydrate binding domains.
References
Bendtsen JD, Nielsen H, von Heijne G, Brunak S (2004) Improved prediction of signal peptides: SignalP 3.0. J Mol Biol 340:783–795
Bruijn JM de (2000) Processing of frost-damaged beets at CSM and the use of dextranases. Zuckerindustrie 125:898–902
Coutinho PM, Henrissat B (1999) Carbohydrate-active enzymes: an integrated database approach. In: Gilbert HJ, Davies G, Henrissat B, Svensson B (eds) Recent advances in carbohydrate bioengineering. The Royal Society of Chemistry, Cambridge, pp 3–12
Davis KA, Anseth KS (2002) Controlled release from crosslinked degradable networks. Crit Rev in Ther Drug Carrier Syst 19:385–423
Finnegan PM, Brumbley SM, O’Shea MG, Nevalainen KMH, Bergquist PL (2004a) Paenibacillus isolates possess diverse dextran-degrading enzymes. J Appl Microbiol 97:477–485. DOI 10.1111/j.1365-2672.2004.02325.x
Finnegan PM, Brumbley SM, O’Shea MG, Nevalainen KMH, Bergquist PL (2004b) Isolation and characterisation of genes encoding thermoactive and thermostable dextranases from two thermotolerant soil bacteria. Curr Microbiol 49:327–333. DOI 10.1007/s00284-004-4308-5
Flores N, Valle F, Bolivar F, Merino E (1992) Recovery of DNA from agarose gels stained with methylene blue. Biotechniques 13:203–205
Igarashi T, Yamamoto A, Goto N (1995) Sequence analysis of the Streptococcus mutans Ingbritt dexA gene encoding extracellular dextranase. Microbiol Immunol 39:853–860
Igarashi T, Yamamoto A, Goto N (2001) Nucleotide sequence and molecular characterization of a dextranase gene from Streptococcus downei. Microbiol Immunol 45:341–348
Igarashi T, Morisaki H, Yamamoto Y, Goto N (2002) An essential amino acid residue for catalytic activity of the dextranase of Streptococcus mutans. Oral Microbiol Immunol 17:193–196
Igarashi T, Morisaki H, Goto N (2004) Molecular characterization of dextranse from Streptococcus rattus. Microbiol Immunol 48:155–162
Khalikova E, Susi P, Usanov N, Korpela T (2003) Purification and properties of extracellular dextranase from a Bacillus sp. J Chromatogr B 796:315–326
Lever M (1973) Colorimetric and fluorometric carbohydrate determination with p-hydroxybenzoic acid hydrazide. Biochem Med 7:274–281
Loesche WJ (1986) Role of Streptococcus mutans in human dental decay. Microbiol Rev 50:353–380
Marchler-Bauer A, Anderson JB, DeWeese-Scott C, Fedorova ND, Geer LY, He S, Hurwitz DI, Jackson JD, Jacobs AR, Lanczycki CJ, Liebert CA, Liu C, Madej T, Marchler GH, Mazumder R, Nikolskaya AN, Panchenko AR, Rao BS, Shoemaker BA, Simonyan V, Song JS, Thiessen PA, Vasudevan S, Wang Y, Yamashita RA, Yin JJ, Bryant SH (2003) CDD: a curated Entrez database of conserved domain alignments. Nucleic Acids Res 31:383–387
Morisaki H, Igarashi T, Yamamoto A, Goto N (2002) Analysis of a dextran-binding domain of the dextranase of Streptococcus mutans. Lett Appl Microbiol 35:223–227
Morris DD, Reeves RA, Gibbs MD, Saul DJ, Bergquist PL (1995) Correction of the β-mannase domain of the celC pseudogene from Caldicellulosiruptor saccharolyticus and activity of the gene product on kraft pulp. Appl Environ Microbiol 61:2262–2269
Oguma T, Kouichiro T, Kobayashi M (1994) Purification and properties of a novel enzyme from Bacillus spp T-3040, which catalyzes the conversion of dextran to cyclic isomaltooligosaccharides. FEBS Lett 345:135–138
Oguma T, Kurokawa T, Tobe K, Kitao S, Kobayashi M (1999) Cloning and sequence analysis of the gene for glucodextranase from Arthrobacter globiformis T-3044 and expression in Escherichia coli cells. Biosci Biotechnol Biochem 63:2174–2182
Ohnishi Y, Kubo S, Ono Y, Nozaki M, Gonda Y, Okano H, Matsuya T, Matsushiro A, Morita T (1995) Cloning and sequencing of the gene coding for dextranase from Streptococcus salivarius. Gene 156:93–96
Porath J (1997) From gel filtration to adsorptive size exclusion. J Prot Chem 16:463–468
Rose TM, Schultz ER, Henikoff JG, Pietrokovski S, McCallum CM, Henikoff S (1998) Consensus-degenerate hybrid oligonucleotide primers for amplification of distantly related sequences. Nucleic Acids Res 26:1628–1635
Sambrook J, Fritsch EF, Maniatis T (1989) Molecular cloning: a laboratory manual, 2nd edn. Cold Spring Harbor Laboratory Press, Cold Spring Harbor
Tilbury RH, French SM (1974) Further studies on enzymic hydrolysis of dextrans in mill juice by dextranases and amylases. In: Proceedings of the 15th congress of the international society of sugar cane technology, pp 1277–1287
Tollofsrud S, Elgjo GI, Prough DS, Williams CA, Traber DL, Kramer GC (2001) The dynamics of vascular volume and fluid shifts of lactated Ringer’s solution and hypertonic-saline-dextran solutions infused in normovolemic sheep. Anesth Analg 93:823–831
Walker GJ (1978) Dextrans. In: Manners DJ (ed) Biochemistry of carbohydrates, vol 16. University Park, Baltimore, pp 75–126
Wanda S-Y, Curtiss R III (1994) Purification and characterization of Streptococcus sobrinus dextranase produced in recombinant Escherichia coli and sequence analysis of the dextranase gene. J Bacteriol 176:3839–3850
Watanabe K, Miyake K, Suzuki Y (2001) Identification of catalytic and substrate-binding site residues in Bacillus cereus ATCC7064 oligo-1,6-glucosidase. Biosci Biotechnol Biochem 65:2058–2064
Wynter CVA, Galea CF, Cox LM, Dawson MW, Pate BK, Hamilton S, De Jersey J, Inkerman PA (1995) Thermostable dextranases: screening, detection and preliminary characterization. J Appl Bacteriol 79:203–212
Wynter CVA, Chang M, De Jersey J, Patel B, Inkerman PA, Hamilton S (1997) Isolation and characterization of a thermostable dextranase. Enzyme Microb Technol 20:242–247
Acknowledgements
This work was funded by grants from BSES Limited, Indooroopilly, Queensland, Australia, the External Collaborative Research Grant scheme of Macquarie University, Sydney, Australia and the SPIRT scheme of the Australian Research Council.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Finnegan, P.M., Brumbley, S.M., O’Shea, M.G. et al. Diverse dextranase genes from Paenibacillus species. Arch Microbiol 183, 140–147 (2005). https://doi.org/10.1007/s00203-004-0756-3
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00203-004-0756-3