Abstract
Rhizobium leguminosarum biovar trifolii strain TA1 polysaccharide synthesis (pss) mutants in the pssD, pssP, pssT and pssO genes and altered in exopolysaccharide (EPS) synthesis were investigated. EPS-deficient mutants were also changed in lipopolysaccharide structure. All mutants exhibited varied sensitivities to detergents, ethanol and antibiotics, thus indicating changes in bacterial membrane integrity. Using pss mutants marked with the gusA gene, EPS-deficient mutants were found to have abnormalities in nodule development and to provoke severe plant defence reactions. The pss mutants that produced altered quantities of EPS with a changed degree of polymerisation generally occupied the younger developmental zones of the nodules and elicited moderate plant defence reactions.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Gram-negative bacteria collectively termed rhizobia are capable of establishing nitrogen-fixing symbioses with leguminous plants. Rhizobia living in the rhizosphere attach to the plant roots, invade plant tissues via structures called infection threads and colonise nodule cortical cells, where they differentiate into bacteroids which provide fixed nitrogen for the plant in exchange for carbon (for a review, see Perret et al. 2000).
Exopolysaccharide (EPS) produced by Rhizobium leguminosarum bv. trifolii is crucial for establishing a nitrogen-fixing symbiosis with clover (Trifolium sp.). Mutants that are defective in EPS production usually induce non-nitrogen-fixing nodules (Skorupska et al. 1995; Rolfe et al. 1996) or fail to nodulate their host plant at all (for a review, see Becker and Pühler 1998).
EPS produced by R. leguminosarum is a species-specific, high molecular weight (HMW) polymer of an octasaccharide repeating unit composed of glucose, glucuronic acid and galactose, modified by O-acetyl or organic acid residues. Several polysaccharide synthesis (pss) genes have been identified in R. leguminosarum as involved in EPS biosynthesis, processing, transport and regulation (Ivashina et al. 1994; van Workum et al. 1997; Mazur et al. 2003).
The rhizobia invading a plant host are usually recognised as non-pathogenic bacteria and only moderate-scale plant defence mechanisms are activated. EPSs are considered as a barrier that suppresses plant defence responses and protects the bacteria against detrimental environmental factors (Niehaus and Becker 1998). We isolated and characterised several pss mutants of R. leguminosarum bv. trifolii strain TA1 (RtTA1). They can be divided into two groups: (i) mutants entirely deficient in EPS production, affected in the genes essential for EPS biosynthesis that induce non-nitrogen-fixing nodules on clover and (ii) mutants that produced altered amounts of EPS in comparison with the wild-type strain and form effective or ineffective nodules on clover. Both the amount of EPS produced and the distribution of HMW and low molecular weight (LMW) fractions of EPS in these mutants were changed when compared with the wild-type strain RtTA1.
In the present study, the pss mutants of RtTA1 were further characterised with respect to a variety of free-living and symbiotic phenotypes. Our results revealed that deficiency in EPS production was also accompanied by changes in lipopolysaccharide (LPS) structure. Generally the pss mutations resulted in altered tolerance to detergents, ethanol and several classes of antibiotics. Moreover, we demonstrated that the plant defence reactions were markedly enhanced in response to an invasion by the pss mutants, especially in the case of those with EPS production deficiency.
Materials and methods
Bacterial strains and growth conditions
The bacterial strains used in this study are listed in Table 1. R. leguminosarum strains were grown at 28°C in 79CA complete medium with 1% mannitol (Vincent 1970). Escherichia coli strains were grown at 37°C in LB medium. The pJBA21Tc plasmid carrying the gusA gene (Wielbo and Skorupska 2001) was transferred from E. coli DH5α into R. leguminosarum bv. trifolii wild-type strain and pss mutants by conjugation.
Sensitivity assays
The sensitivity of RtTA1 strains to sodium deoxycholate (DOC), sodium dodecyl sulfate (SDS) and ethanol was studied and the particular agent’s minimal inhibitory concentration was determined. Rhizobium strains (10 μl of liquid culture, with an optical density at 550 nm of 0.2) were plated on 79CA agar medium containing a defined concentration of DOC (1.5–6 mM), SDS (0.0125–0.05% w/v) or ethanol (1.5–4.25% v/v) and incubated for 3 days. For Triton X-100 experiments, two-layer plates were prepared as follows: 100 μl of culture was added to 3 ml of 79CA soft agar and poured onto 79CA plates (20 ml). After 30 min, paper disks (6 mm diam.) were applied on the agar surface and 5 μl of the appropriate Triton X-100 solution (5–60% v/v) was dropped onto them. The plates were incubated for 3 days and then the diameter of the zone of bacterial growth inhibition was measured. Antibiotic sensitivity assays were performed using commercially available filter disks with the appropriate antibiotic: erythromycin (15 μg), chloramphenicol (30 μg), tetracycline (30 μg; Mast Diagnostics, Merseyside, UK), amikacin (30 μg) and cefadroxil (30 μg; Bristol-Myers International, N.Y., USA). Filter disks were placed on the surface of 79CA where 100 μl of liquid rhizobial cultures was previously spread. The diameter of the growth inhibition zone was measured after 3 days of incubation.
LPS analysis
LPS from R. leguminosarum strains was isolated using a microextraction method (Apicella et al. 1994). Samples were separated by 12.5% SDS-PAGE and visualised by silver staining (Tsai and Frasch 1982).
Plant growth condition and β-glucuronidase histochemistry
Clover (T. pratense cv. Ulka) seeds were germinated, inoculated and grown on nitrogen-free Fåhraeus medium (Vincent 1970), as described by Skorupska et al. (1995). Nodules were harvested weekly (starting from 1 week after inoculation) and stained histochemically for β-glucuronidase (GUS) activity according to Wilson et al. (1992). Starch staining was conducted in 20% (v/v) water/Lugol solution. Lipid-accumulating nodule cells were stained for 30 min at room temperature with 0.2% Sudan III solution (in 70% ethanol). Stained nodule sections were analysed by light and fluorescence microscopy (Nikon Optiphot 2, using Nomarsky optics). For the detection of phenolic compounds in plant nodule cells, autofluorescence of nodule sections was studied with an extinction filter (510 nm).
Results and discussion
Previously, we described several RtTA1 pss mutants affected in EPS synthesis. In the Rt133 mutant, Tn5 insertion in the pssD gene coding for glycosyltransferase resulted in an EPS-deficient phenotype (Król et al. 1998). This mutant induced non-nitrogen-fixing nodules on clover. Three other mutants, designated RtP20, RtP22 and RtP23, were affected in the pssP gene. The PssP protein belongs to the membrane-periplasmic auxiliary protein family, whose members are involved in the synthesis and polymerisation of HMW EPS (Mazur et al. 2002). The RtP22 mutant deleted for the entire pssP gene is EPS-deficient and does not fix nitrogen in symbiosis with clover. RtP20, with the C-terminal domain of the PssP protein disrupted, produces reduced amounts of EPS (43% of RtTA1 EPS) with a slightly changed HMW/LMW distribution of EPS fractions when compared with the RtTA1 wild-type strain. The RtP23 mutant strain with the PssP N-terminal domain disrupted also synthesises reduced amounts of EPS but the ratio of HMW:LMW EPS is markedly different (15:85) from that observed in the RtTA1 strain (50:50). Both pssP mutants (RtP20, RtP23) are able to elicit nitrogen-fixing nodules on clover (Mazur et al. 2002). The RtO12 strain is merozygotic in the pssO gene (unpublished data) and produces highly reduced amounts of EPS (45% of RtTA1 EPS) with a slightly changed HMW/LMW EPS ratio. The PssO protein function in EPS biosynthesis is unknown, but the protein’s secondary structure indicates outer-membrane localisation. The RtO12 mutant induces non-nitrogen-fixing nodules on clover roots. The RtAH1 mutant carries a disrupted pssT gene. The PssT protein is an inner membrane transport protein with 12 membrane-spanning domains. It belongs to the polysaccharide-specific transport family of proteins, whose members constitute a component of the type I polysaccharide transport system (Mazur et al. 2003). The RtAH1 mutant produces a significantly increased amount of EPS (132% of RtTA1 EPS) with an altered distribution of HMW/LMW forms (57:43), and it elicits an increased number of nitrogen-fixing nodules on clover.
Sensitivity assays of pss mutants to SDS, Triton X100, DOC and ethanol
Taking into consideration the putative localisation of the studied proteins in both the outer and inner membranes, the question arises whether the altered phenotype and symbiotic behaviour of EPS mutants results directly from the changes in EPS quality and/or structure, or rather from the pleiotropic effects of cell envelope distortion (both inner and outer membranes) caused by the presence of misfolded protein(s) inserted into the membrane(s), or the lack of important protein(s), e.g. those involved in transport of EPS, leading to instability or disorganisation of translocation complexes. Indirect evidence for membrane alterations could be a change in sensitivity to such compounds as detergents, ethanol and antibiotics.
The ionic detergent SDS sensitivity test revealed moderate differences in the growth of the mutants on plates supplemented with different concentrations of this detergent. The RtAH1 mutant was more resistant than the other strains tested (Table 2). The Rt133, RtP22, RtP20 and RtP23 mutants showed significant DOC sensitivity. Of the remaining mutants, RtO12 and RtAH1 especially exhibited increased resistance to DOC in comparison with the wild-type strain (Table 2). Mutants Rt133, RtP22 and RtP20 grew poorly on ethanol in contrast to the other mutants and the RtTA1 strain (Table 2). The wild-type strain and the Rt133, RtP22, RtO12, RtAH1 mutants showed a comparable level of resistance to the non-ionic detergent Triton X-100. However, an essential increase in sensitivity to Triton X-100 was detected in the case of RtP20 and RtP23, both mutated in the pssP gene (data not shown). These results demonstrate that the quantity or degree of EPS polymerisation does not seem to correlate with mutant susceptibility to the tested compounds and that EPS is not the sole barrier protecting rhizobia against deleterious compounds. Moreover, these data indicate that mutations affecting the PssP, PssT and PssO proteins could affect membrane integrity, resulting in an altered sensitivity to the tested compounds.
LPS analysis
An increased sensitivity to detergents (especially DOC) and ethanol is a common characteristic of mutants altered in LPS biosynthesis (Campbell et al. 2002; Nikaido 2003). The SDS-PAGE LPS analysis revealed significant changes in banding pattern in the case of the Rt133 and RtP22 mutants (Fig. 1, lanes 1, 7, respectively). Their LPS displayed an apparently diminished smooth LPS species with an upward shift in both rough and smooth LPS in comparison with the wild-type LPS (Fig. 1, lane 2). They can be attributed either to an alteration in the O-antigen chain length or to a significant reduction of this moiety. The parental strain RtTA1 and mutants altered in the level of EPS production or polymerisation manifested both rough and smooth LPS. These results prove that EPS deficiency in the Rt133 and RtP22 mutants is accompanied by changes in LPS. It also explains the increased sensitivity of these mutants to DOC and ethanol, previously observed for the Sinorhizobium meliloti LPS mutants (Le Vier and Walker 2001). The link between LPS and EPS synthesis is well established and mutations of particular genes whose products are involved in these pathways might have pleiotropic effects (Becker and Pühler 1998; Guerreiro et al. 2000). Campbell et al. (2003) showed that mutation in genes not recognised to play a role in LPS synthesis could affect LPS structure, resulting in the complexity of phenotypes observed by PAGE analysis, including additional rough LPS bands and alterations in the molecular weight distribution of the smooth LPS.
Sensitivity to antibiotics
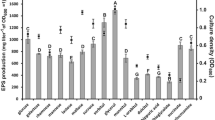
To further characterize the EPS mutants of R. leguminosarum bv. trifolii, they were tested for sensitivity to both hydrophilic antibiotics, such as chloramphenicol, amikacin (aminoglycoside) and cefadroxil (β-lactam), and to hydrophobic ones, such as erythromycin (macrolide) and tetracycline (Fig. 2). The most remarkable increase in resistance to chloramphenicol was observed in the case of the RtP20, RtP23 and RtO12 mutants. The same mutants were also more sensitive to cationic aminoglycoside amikacin and hydrophobic erythromycin (RtP20, RtO12). Unexpectedly, the RtAH1 mutant displayed an increased sensitivity to chloramphenicol, amikacin and erythromycin. Both EPS-deficient mutants (Rt133, RtP22) differently responded to the tested antibiotics. The Rt133 mutant was nearly 2.5-fold more resistant to erythromycin than the wild-type strain (Fig. 2).
Relative resistance of R. leguminosarum bv. trifolii pss mutants to antibiotics, determined by measuring the diameter of growth-inhibition zones. The values for the wild-type strain RtTA1 were as follows: chloramphenicol 16.2±1.5 mm, cefadroxil 34.2±3.2 mm, amikacin 20.6±2.2 mm, erythromycin 29.6±0.9 mm, tetracycline 57.3±1.5 mm
The PssT, PssP and PssO proteins engaged in EPS synthesis and transport possibly form a protein complex embedded in the outer and inner membranes. It might be expected that mutations in these proteins could affect the permeability and, consequently, susceptibility of bacterial cells to antibiotics (Hancock 1997). In fact, the increase in resistance to chloramphenicol of RtP20, RtP22, RtP23 and RtO12 suggests that the outer membrane becomes less permeable to this antibiotic. It is probable that a mutation in the pssP or pssO genes leads to an alteration in non-specific porin channels in the outer membrane, which are used by chloramphenicol (Nikaido 2003).
Nodule colonisation by R. leguminosarum bv. trifolii pss mutants
Using R. leguminosarum bv. trifoli EPS mutants constitutively expressing GUS from a stably maintained vector, we revealed differences in the capabilities of the mutated rhizobia for clover root nodule invasion. This work confirmed earlier observations that the pattern of clover root nodule invasion was closely dependent on EPS production (Chakravorty et al. 1982; Skorupska et al. 1995).
RtP22 and Rt133 EPS-deficient and LPS-altered mutants induced nodules on clover roots with an approximately weekly delay, as compared with the RtTA1 wild-type strain. The nodules were white and small, and they manifested several morphological abnormalities. Until 3–4 weeks after inoculation, the bacteria were not detected in the nodule cells by the GUS staining assay. In nodules at 4–5 weeks, only a few plant cells were invaded by the rhizobia and the infected cells were grouped or dispersed in the nodule (Fig. 3d–f). In addition, several plant defence reactions elicited by the Rt133 mutant were observed in the nodules, including: (i) emergence of local necrosis, seen as a cluster of brown-coloured cells (Fig. 3d), (ii) accumulation of phenolic compounds revealing bright autofluorescence in the thickened plant cell walls (Fig. 3f) and (iii) deposition of red-coloured (after staining with Sudan III) lipid substances (probably suberine) in the nodule cell walls (Fig. 3e). In the case of the EPS-deficient RtP22 mutant with the pssP gene deleted, the nodule occupancy was even weaker than that of Rt133 (Fig. 3g,h). The symptoms of plant defence reactions were not seen up to 5–6 weeks after inoculation and were not as evident, probably because of the very low number of infected cells. Slight necrotic changes (Fig. 3h) and a moderate accumulation of phenolic compounds were detected in 7-week-old nodules (Fig. 3i).
Light microscopy of T. pratense nodules (in section) inoculated with R. leguminosarum bv. trifolii mutants harbouring plasmid pJBA21Tc with the gus reporter gene. a–c RtTA1 wild-type nodules at 5, 10, 35 days post-infection (d.p.i.), respectively, d–f Rt133 nodules at 35 d.p.i., g–i RtP22 nodules at 28, 49 d.p.i., j–m RtP20 nodules at 14, 21, 35 d.p.i., n,o RtO12 nodules at 14, 35 d.p.i., respectively, p,q RtP23 nodules at 35 d.p.i., r RtAH1 nodules at 35 d.p.i. All nodules were stained for GUS activity. f,i,m Fluorescence microscopy of nodules in section, e Rt133 nodules additionally stained for the presence of lipids, k RtP20 nodules stained for the presence of starch. fl Autofluorescence, lp lipids, nec necrosis, st starch. Bars 0.5 mm
The RtP20 mutant elicited two different types of nodules on clover roots. Most of them were large and longitudinal, similar to wild-type nodules (Fig. 3k,l). They emerged without delay and were infected by rhizobia (Fig. 3j). However, in well developed nodules, the bacteria occupied the younger zones (Fig. 3k,l). In uninfected zones of these nodules, a massive starch accumulation was seen, probably as a result of a less efficient nitrogen fixation (Fig. 2k). No plant defensive response was detected in these nodules.
The other type of nodules induced by the RtP20 mutant was small and round, with no distinguishable developmental zones. The rhizobia occupied only a limited number of plant cells and an accumulation of phenolic compounds in the plant cell walls was observed (Fig. 3m).
In the case of the RtO12 mutant, nodule appearance and morphology were similar to the wild-type nodules, but the pattern of their colonisation was different. In young nodules, bacterial colonisation was less efficient (Fig. 3n) and the younger zones were occupied in well developed nodules (Fig. 3o). Symptoms of plant defensive reactions (i.e. small groups of necrotic cells) were rarely observed (Fig. 3o).
The RtP23 mutant (Fig. 3p,q) that produced most of its EPS in LMW form was able to colonise younger zones of the nodules and was capable of nitrogen fixation. This confirms the significant role of LMW EPS in establishing an effective symbiosis (Wang et al. 1999) and shows that even a significant decrease in HMW EPS does not deleteriously affect the symbiosis of R. leguminosarum bv. trifolii with clover. Interestingly, the EPS-overproducing RtAH1 mutant (which was more resistant to ionic detergents and ethanol and more sensitive to several antibiotics in comparison with the wild-type strain) induced infected nodules with bacteroids that were capable of nitrogen fixation (Fig. 3r). The alteration of PssT protein influenced the permeability of the outer and inner membranes but nevertheless did not affect the efficiency of symbiosis. We did not detect any plant defensive reactions in these nodules. In summary, these findings present evidence that the EPS amount and rate of polymerisation affect nodule colonisation and plant defence responses to a lesser extent than a deficiency in EPS accompanied by LPS alteration.
References
Apicella M, Griffiss J, Schneider H (1994) Isolation and characterization of lipopolysaccharides, lipooligosaccharides and lipid A. Methods Enzymol 235:242–252
Becker A, Pühler A (1998) Production of exopolysaccharides. In: Spaink HP, Kondorosi A, Hooykaas PJJ (eds) The Rhizobiaceae. Molecular biology of model plant-associated bacteria. Kluwer, Dordrecht, pp 97–118
Campbell GRO, Reuhs BL, Walker GC (2002) Chronic intracellular infection of alfalfa nodules by Sinorhizobium meliloti requires correct lipopolysaccharide core. Proc Natl Acad Sci USA 99:3938–3943
Campbell GRO, Sharypova LA, Scheidle H, Jones KM, Niehaus K, Becker A, Walker GC (2003) Striking complexity of lipopolysaccharide defects in a collection of Sinorhizobium meliloti mutants. J Bacteriol 185:3853–3862
Chakravorty AK, Zurkowski W, Shine J, Rolfe BG (1982) Symbiotic nitrogen fixation: molecular cloning of Rhizobium genes involved in exopolysaccharide synthesis and effective nodulation. J Mol Appl Genet 1:585–596
Guerreiro N, Ksenzenko VN, Djordjevic MA, Ivashina TV, Rolfe BG (2000) Elevated levels of synthesis of over 20 proteins results after mutation of the Rhizobium leguminosarum exopolysaccharide synthesis gene pssA. J Bacteriol 182:4521–4532
Hancock REW (1997) The bacterial outer membrane as a drug barrier. Trends Microbiol 5:37–42
Ivashina TV, Khmelnitsky MI, Shlyapnikov MG, Kanapin AA, Ksenzenko VN (1994) The pss4 gene from Rhizobium leguminosarum bv. viciae VF39: cloning, sequence and the possible role in polysaccharide production and nodule formation. Gene 150:111–116
Król J, Wielbo J, Mazur A, Kopcińska J, Łotocka B, Golinowski W, Skorupska A (1998) Molecular characterisation and symbiotic importance of pssD gene of Rhizobium leguminosarum bv. trifolii strain TA1: pssD mutant is affected in exopolysaccharide synthesis and endocytosis of bacteria. Mol Plant Microbe Interact 11:1142–1148
Le Vier K, Walker GC (2001) Genetic analysis of the Sinorhizobium meliloti BacA protein: differential effects of mutations on phenotypes. J Bacteriol 183:6444–6453
Mazur A, Król J, Wielbo J, Urbanik-Sypniewska T, Skorupska A (2002) Rhizobium leguminosarum bv. trifolii PssP protein is required for exopolysaccharide biosynthesis and polymerization. Mol Plant Microbe Interact 15:388–397
Mazur A, Król J, Marczak M, Skorupska A (2003) Membrane topology of PssT, the transmembrane protein component of the type I exopolysaccharide transport system in Rhizobium leguminosarum bv. trifolii strain TA1. J Bacteriol 185:2503–2511
Niehaus K, Becker A (1998) The role of microbial surface polysaccharides in the Rhizobium–legume interaction. Subcell Biochem 29:73–116
Nikaido H (2003) Molecular basis of bacterial outer membrane permeability revisited. Microbiol Mol Biol Rev 67:593–656
Perret X, Staehelin C, Broughton W (2000) Molecular basis of symbiotic promiscuity. Microbiol Mol Biol Rev 64:180–201
Rolfe BG, Carlson RW, Ridge RW, Dazzo FB, Mateos PF, Pankhurst CE (1996) Defective infection and nodulation of clovers by exopolysaccharide mutants of Rhizobium leguminosarum bv. trifolii. Aust J Plant Physiol 23:285–303
Skorupska A, Białek U, Urbanik-Sypniewska T, Lammern A van (1995) Two types of nodules induced on Trifolium pratense by mutants of Rhizobium leguminosarum bv. trifolii deficient in exopolysaccharide production. J Plant Physiol 147:93–100
Tsai CM, Frasch CE (1982) A sensitive silver stain for detecting lipopolysaccharides in polyacrylamide gels. Anal Biochem 119:115–119
Vincent JM (1970) A manual for the practical study of the root-nodule bacteria. Blackwell, Oxford
Wang LX, Wang Y, Pellock B, Walker GC (1999) Structural characterization of the symbiotically important low-molecular-weight succinoglycan of Sinorhizobium meliloti. J Bacteriol 181:6788–6796
Wielbo J, Skorupska A (2001) Construction of improved vectors and cassettes containing gusA and antibiotic resistance genes for studies of transcriptional activity and bacterial localization. J Microbiol Methods 45:197–205
Wilson KJ, Hughes SG, Jefferson RA (1992) The Escherichia coli gus operon, induction and expression of the gus operon in E. coli and the occurrence and use of GUS in other bacteria. In: Gallagher S (ed) GUS protocols, using the GUS gene as a reporter of gene expression. Academic, New York, pp 7–23
Workum WAT van, Canter Cremers HCJ, Wijfjes AHM, Kolk C van der, Wijffelman CA, Kijne JW (1997) Cloning and characterization of four genes of Rhizobium leguminosarum bv. trifolii involved in exopolysaccharide production and nodulation. Mol Plant Microbe Interact 10:290–301
Acknowledgements
This work was supported by the Polish Committee for Scientific Research (no. 2 P04A 034 26). A.M. is the recipient of a fellowship from The Foundation for Polish Science.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Wielbo, J., Mazur, A., Król, J. et al. Complexity of phenotypes and symbiotic behaviour of Rhizobium leguminosarum biovar trifolii exopolysaccharide mutants. Arch Microbiol 182, 331–336 (2004). https://doi.org/10.1007/s00203-004-0723-z
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00203-004-0723-z