Abstract
It is generally challenging to quantify the fidelity of surrogate models without additional system evaluations. Standard error measures, such as the mean squared error and cross-validation error, often do not adequately capture the fidelity of the model trained using all available sample points. This paper introduces a new model-independent approach to quantify surrogate model fidelity, called Predictive Estimation of Model Fidelity (PEMF). In PEMF, intermediate surrogates are iteratively constructed over heuristic subsets of sample points. The median and the maximum errors estimated over the remaining points are used to determine the respective error distributions at each iteration. The estimated modes of the error distributions are represented as functions of the density of intermediate training points through nonlinear regression, assuming a smooth decreasing trend of errors with increasing sample density. These regression functions are then used to predict the expected median and maximum errors in the final surrogate model (trained using all available sample points). A Monotonic Trend criterion is defined to statistically test if the regression function is reasonably reliable in predicting the model fidelity, failing which a stable implementation of k-fold cross-validation (based on modal error) is used to predict the final surrogate error. To compare the accuracy and robustness of PEMF with that of the popular leave-one-out cross-validation, numerical experiments are performed using Kriging, RBF, and E-RBF models. It is observed that the model fidelities estimated by PEMF is up to two orders of magnitude more accurate and statistically more stable compared to those based on cross-validation.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
1.1 Approximation models
Mathematical approximation models are commonly used to provide a tractable and inexpensive approximation of the actual system behavior in many routine engineering analysis and design activities, e.g., domain exploration, sensitivity analysis, development of empirical models, and optimization. One of the most popular classes of approximation models are surrogate models or metamodels (Kleijnen 1975), which are purely mathematical models, and are not directly derived from the physics of the system being modeled. Major surrogate modeling methods include Polynomial Response Surfaces (Jin et al. 2000), Kriging (Simpson et al. 2001; Forrester and Keane 2009), Radial Basis Functions (RBF) (Hardy 1971), Neural Networks (Yegnanarayana 2004), Support Vector Regression (Gunn 1998), and hybrid surrogate models (Queipo et al. 2005; Zhang et al. 2012). Constructing a surrogate model typically involves the following steps:
-
1.
Performing a design of experiments (DOE);
-
2.
Executing the sample experiments (e.g., high fidelity simulations or physical experiments) to generate the training data;
-
3.
Selecting and training the appropriate surrogate model using the training data; and
-
4.
Testing the accuracy of the surrogate model.
In the fourth step, specialized error measures can be used to assess the accuracy of the surrogate model in representing the actual system behavior. The knowledge of the regional and global accuracy of a surrogate is crucial (i) for model selection, (ii) for further improvement of the surrogate, e.g., using adaptive sampling (Lehmensiek et al. 2002) or active learning (Sugiyama 2006), (iii) for iterative surrogate-based optimization (Jones et al. 1998; Mehmani et al. 2012), and (iv) for quantifying the uncertainty associated with the surrogate. Other possible applications include the construction of hybrid surrogate models and conservative surrogate models. The state-of-the-art measures of surrogate model error are subject to one or more of the following limitations:
-
They are model-dependent (and hence may not be suitable for global model comparison, and effective model selection); or
-
They require additional system evaluations (and hence are expensive); or
-
They quantify errors in intermediate surrogates (e.g., PRESS), which are often not representative of the error in the actual surrogate model that will be used for design and analysis; and
-
They generally do not provide an understanding of the uncertainty introduced by a surrogate model in a design process (i.e., instead provides deterministic error measures).
In this paper, we conceive and develop a new error quantification method for surrogate models that seek to address the above limitations. In the following subsection, the existing major methods for assessing the accuracy of surrogates are briefly reviewed.
1.2 Measures of surrogate model fidelity
Methods for quantifying the fidelity of or (inversely) the error in statistical models can be broadly classified into: (i) methods that require additional data, and (ii) methods that use existing data (Meckesheimer et al. 2002). The former can be significantly expensive and is thus not a practical option in a majority of surrogate modeling applications. Error quantification methods can also be classified into global and local error estimation methods (Goel et al. 2007). The performance of the surrogate over the entire domain is evaluated by global error measures, while local or point-wise error measures provide the surrogate accuracy in different locations of the design domain. Table 1 provides a list of popular surrogate modeling methods, and the error estimation techniques commonly used for these surrogates.
Popular approaches to quantifying model independent global error measures include (Queipo et al. 2005): split sample, cross-validation, bootstrapping, and Akaike’s information criterion (AIC) method. In a split sample strategy, the sample data is divided into training and test data. The former is used to construct a surrogate; and the latter is used to test the performance of the surrogate. Cross-validation is a popular technique to estimate the error of a surrogate without investing any additional system evaluations. In q-fold cross-validation approach, the data set is split randomly into q (approximately) equal subsets. The surrogate is constructed q times, each time leaving out one of the subsets from the training set. The omitted subset, at each iteration, is used to evaluate the cross-validation error (Viana et al. 2009). A k-fold cross-validation approach is a variation of q-fold approach, in which all the possible subsets of size k are used to evaluate cross-validation error. In leave-one-out cross-validation approach (k=1), at each iteration the training set is created by using all sample points except one, and the left out point is used for estimating the error between the surrogate prediction and the actual value. The bootstrapping approach generates m sub-samples from the sample points. Each sub-sample is a combination of all samples with replacement. Different variants of the bootstrapping approach can be used for model identification and determining confidence intervals for surrogates (Queipo et al. 2005). In Akaike’s original AIC, the performance of the surrogate was predicted based on a penalized likelihood. AIC is equal to the sum of a negative log likelihood and a penalty term, as given by
In this equation, \(L(\hat {\theta })\) is the maximized likelihood function, and k is the number of free parameters in the model, which is a measure of complexity or the compensation for the bias in the lack of fit when the maximum likelihood estimators are used (Bozdogan 2000).
Standard local error measures include: (i) the mean squared errors for Kriging (Jones et al. 1998), and (ii) the linear reference model (LRM) (Nguyen et al. 2011). In stochastic surrogate models like Kriging, the errors at two different points of the design domain are not independent; and the correlation between the points is related to the distance between them. When the distance between the two points is smaller, the correlation tends to one, and when the distance is larger, the correlation tends to zero. According to this correlation strategy, if the point x ∗ is close to sample points, the prediction confidence at that point is higher than that when the point is far away from all the sample points. This concept is reflected in the local error measurement method for Kriging predictor at the special point x ∗. This error is equal to zero at the sample points and is equal to the approximation error variance in the stochastic process. The LRM is a model independent method for quantifying the local performance of a surrogate. The LRM considers the region with oscillations as a high-error location. This method categorizes errors of a surrogate in the design domain based on the deviation of the surrogate from the local linear interpolation (Nguyen et al. 2011).
Another popular cross-disciplinary error measure is the mean squared error (MSE) or root mean square error (RMSE), which provides a measure of global error over the entire design domain. The RMSE evaluated at a set of test points (N T e s t ) is given by:
where y i and \(\hat {y}_{i}\) are the actual and the predicted values at the i th test point, respectively. The RMSE thus provides information about the accuracy of the actual surrogate. However RMSE requires additional system evaluations at test points in the case of interpolating surrogates. The maximum absolute error (MAE) and relative absolute error (RAE) are indicative of local deviations:
The prediction sum of square (PRESS) is based on the leave-one-out cross-validation error:
where \(\hat {y}_{i}\) and \(\hat {y}_{i}^{(-i)}\) are the surrogate estimations at the i th training point respectively predicted by the surrogate constructed using all sample points and the surrogate constructed using all sample points except the i th point (Queipo et al. 2005). Goel et al. (2009, 2009) compared different error measures including PRESS and MSE for Kriging, and found that PRESS provided better performance compared to the other methods, as a fidelity prediction and as a surrogate model selection criterion.
Meckesheimer et al. (2002) used the root mean square error of k-fold cross-validation (RMSE C V ), i.e., root mean square of PRESS (PRESS R M S ), to measure the global accuracy of the surrogate over the entire design domain:
where k is the number of omitted sample points. The variation of k from 1 to 10 was studied; for each k value, the average of the error measured on all possible combinations of k points were used to provide a measure of the global accuracy of the surrogate. They also compared this error measure with the actual error estimated on additional test points to show the practicality of the RMSE C V as a fidelity characterizing method without using additional system evaluations.
Viana et al. (2009) applied the RMSE C V approach as a criterion in surrogate model selection and in constructing a weighted average surrogate. They also showed that better results can be achieved using the leave-one-out approach. Later, Viana et al. (2010) used RMSE C V to estimate the safety margin for a conservative surrogate model. Zhang et al. (2014) applied relative absolute error of cross-validation (RAE C V ) for characterizing the uncertainty in surrogate models, where the normalized RAE C V at the i th training point is defined based on the leave-one-out approach. This characterization was used in conjunction with support vector machines to segregate the design space into sub-spaces based on the level of model errors.
1.3 Model refinement using surrogate error measures
Another important area of application of surrogate model error measures is “model refinement”. Over the last two decades, different statistical strategies have been developed to improve the accuracy and robustness of surrogate models by adding infill points where additional evaluations of the high fidelity model / experiment are desired to be performed. Infill points can be added in a fully sequential manner (one-at-a-time), or can be added in a batch sequential manner.
In the literature, there exist various criteria for determining the locations of the infill points (Jones et al. 1998; Williams et al. 2011; Keane 2006; Booker et al. 1999; Audet et al. 2000). Jin et al. (2002) reviewed different one-at-a-time sequential sampling criteria, and illustrated their potential benefits over single stage methods (McKay et al. 1979; Sacks et al. 1989). Among the sequential sampling methods that are not limited to a specific surrogate model types, Kleijnen and Beers (2004) used cross-validation error and Jackknifing variance as infill points criteria (Meckesheimer et al. 2002). This method evaluates the variance between the estimations of the surrogate model and the high fidelity model /experiment and subsequently adds points in the area with the highest variance. Loeppky et al. (2010) explored different batch and one-at-a-time sequential criteria for the Gaussian process model. They pointed out that these criteria perform better under the batch sequential approach, and one-at-a-time augmentation would be likely impractical due to the higher computational cost (especially in the case of physical experiments).
Williams et al. (2011) applied different batch sequential criteria to achieve stability in the fidelity of a Gaussian process (GP) model that is used in the ‘Bayesian Calibration of Computer Models’ (Kennedy and O’Hagan 2000). This GP model is trained using the disagreement between the output values given by experiments and the computer model on infill points; in this case stability implies the phase where adding more infill points has a minimal influence on the accuracy of the GP model (Atamturktur et al. 2011). Atamaturktur et al. (2013) assessed the ability to improve the fidelity of the computer model using different index-based and distance-based batch sequential sampling criteria through a powerful quantitative Predictive Maturity Index (PMI) metric, developed earlier by Hemez et al. (2010). The proposed PMI could guide decision makers to allocate resources (infill points), and track the progress in the improvement of a predictive capability (Atamturktur et al. 2011; Atamturktur et al. 2013).
It can be readily concluded from the research reported in the existing literature that, there is a need for reliable and statistically stable measures of surrogate model fidelity, to facilitate improvement or informed application of surrogates in design and analysis. More specifically, it is necessary for such error quantification approaches to have the following characteristics:
-
(i)
Be independent of the type of model (e.g., Radial basis function, Polynomial response surface, Gaussian processes), so as to allow fair and competitive model selection;
-
(ii)
Provide a predictive estimate of the error of the actual model, instead of the (mean or RMS) error of intermediate surrogates; and
-
(iii)
Provide a measure of error that is minimally sensitive to outlier sample points.
The method presented in this paper seeks to directly incorporate these much-needed characteristics into quantification of error in surrogate models. The objectives of developing a new surrogate model error quantification method are summarized in the next subsection.
1.4 A new predictive and model-independent approach to model error quantification
The method developed in this paper, called Predictive Estimation of Model Fidelity (PEMF), is designed to address two primary objectives. The first objective is to develop a model-independent methodology for quantifying the fidelity of the ‘actual surrogate’ without requiring additional system evaluations. In this context, the ‘actual surrogate’ refers to the surrogate model constructed using all available training points (and not any subset of training points, as used in cross-validation). With this approach, we provide error information for the actual model that is to be used for function analysis or optimization (assuming surrogate model construction is not an end in itself). The second objective is to track the variation of the surrogate model error with an increasing density of training points, thereby creating opportunities for global model selection and model refinement. The investigation of these latter opportunities are however not within the scope of this paper.
The paper is organized as follows: the formulation of the proposed model fidelity quantification approach is described in Section 2; numerical experiments and results are respectively presented in Sections 3 and 4. In Section 5, statistical tests are performed on the PEMF method. Concluding remarks and future work are provided in Section 6.
2 Predictive estimation of model fidelity (PEMF)
2.1 Predictive estimation of model fidelity
The PEMF method determines the model fidelity within the design domain (defined by the user) by analyzing the variation in the model error distribution with increasing number of training points. This measure of fidelity can provide uniquely useful information about the accuracy of the surrogate in regions of interest or the entire design domain, thereby significantly improving the usability of the surrogates in model-based design applications. In the context of surrogate-based design, this information can be directly applied for (i) surrogate model refinement (Atamturktur et al. 2013; Jin et al. 2002; Kleijnen and Beers 2004), (ii) surrogate model selection (Chowdhury et al. 2014a; Gorissen et al. 2009; Martin and Simpson 2005), and (iii) uncertainty analysis (Chowdhury et al. 2014b; Allaire et al. 2012). In these applications, PEMF is either enabling more informed use of the constructed surrogate model or the platform to identify and use the surrogate model that is most suited to the concerned design process. A flowchart describing the PEMF algorithm is illustrated in Fig. 1, and the major steps are described below:
2.1.1 Generating sample data
In this step, a set of experimental designs are generated based on a user-specified distribution or given by a DOE. The expensive system evaluation (simulation or physical experiments) is then performed over the sample data points. The entire set of sample points is represented by {X}. This sample input-output data might also be given by other measured data sources.
2.1.2 Defining the region of interest
The entire set of sample points is divided into inside and outside sets based on the boundaries of the user-defined region of interest, and are represented by {X i n } and {X o u t }, respectively.
2.1.3 Quantifying the variation of the error distribution with sample density
This step is an iterative process, where the number of iterations (N itr) is to be guided by the dimension of the problem, the number of sample points in the inside set, and the observed stability of error variation. In this first development and implementation of PEMF (in this paper) the number of iterations will be fixed (prescribed by the user). At each iteration, sample points are divided into a set of intermediate training data {X T R } and a set of intermediate test data {X T E },i.e.,
where {ß k} represents a k th subset of inside-region sample points. At each iteration t, the size of {ß k} is defined by n t, where n t+1>n t and n 1≥1.
At each iteration, the total number of possible sample combinations is defined by M t where M t≤\({N_{i}} \choose {n^{t}}\); the term N i represents the number of inside-region sample points. In low dimensional problems, all possible subsets of size n t could be used, while in high dimensional problems, a fraction of subsets could be used to avoid intractable computational cost. Intermediate surrogates \({f^{k}_{t}}\), are constructed at the t th i t e r a t i o n for all combinations, k=1,2,...,M t, using the intermediate training points. These surrogates are then evaluated over the intermediate test points.
The median and the maximum errors for each combination (\(E_{med}^{k}\) and \(E_{max}^{k}\)) are then estimated using the RAE values on the intermediate test points (\(e_{1},e_{2},...,e_{m^{t}}\)). Probability distribution functions are used to represent the distribution of the median and the maximum errors over the M t different combinations at each iteration, thereby providing an understanding of the uncertainty directly associated with the surrogate model. A chi-square (χ 2) goodness-of-fit criterion (Haldar and Mahadevan 2000) is used to select the most suitable type of distribution from a list of candidates such as lognormal, Gamma, Weibull, logistic, log logistic, t location scale, inverse Gaussian, and generalized extreme value distribution. The χ 2 criterion is defined based on the error between the observed and the assumed PDF of the distribution. Assuming the observed frequencies of m intervals of the random variable are represented by o 1, o 2, ..., o m , the corresponding theoretical frequencies are represented by t 1, t 2, ..., t m ; and the χ 2 criterion is given by
The mode of the median and the maximum error distributions at each iteration (\(\mathrm {Mo_{med}^{t}}\) and \(\mathrm {Mo_{\max }^{t}}\)) are evaluated to provide a measure of central tendency, and are used to relate the variation of the surrogate error with the sample density.
2.1.4 Predicting the fidelity of the final surrogate
The final surrogate model is constructed using the entire set of training data. Regression models are applied to represent the statistical mode of the median error distribution (Mo m e d ) and that of the maximum error distribution (Mo m a x ) at each iteration as monotonic functions of the number of inside-region training points, n t. These regression functions are called the variation of error with sample density (VESD), and are expressed as
The VESD regression functions are then used to predict the fidelity of the final surrogate model under the condition that a Monotonic Trend (MT) Criterion is satisfied. The MT criterion is defined to statistically test the feasibility of a monotonic decrease of the model fidelity with increasing sample density. If the MT criterion is not satisfied, a stable implementation of the k-fold cross-validation (PEMF-based k-fold) is used instead of the VESD functions to represent the surrogate model fidelity. The formulation and application of the MT criterion is described in the following subsection.
2.2 Constructing and testing functions to represent the variation of error with sample density (VESD)
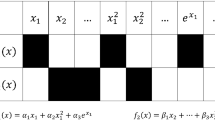
The selection of the type of regression function is critical to quantify the variation of error with training point density. In this paper, two types of regression functions are used to represent the variation of maximum and median errors as functions of the number of inside-region training points. These functions are
- Type 1:
-
Exponential regression function
$$ F(n^{t}) = a_{0}~e^{-a_{1}~(n^{t})} $$(10) - Type 2:
-
Multiplicative regression function
$$ F(n^{t}) = a_{0}~(n^{t})^{-a_{1}} $$(11)
where a 0 and a 1 are regression coefficients known as initial value and rate of decay, respectively. These coefficients are determined using the least square method such that a 0, a 1>0, and n t≥1. The smaller the rate of decay (a 1), the lower the sensitivity of surrogate accuracy to sample density. Numerical experiments exploring linear, polynomial, multiplicative, exponential, and other standard regression functions indicated that exponential and multiplicative functions are the most suitable choice in this context. In this paper, the root mean squared error metric is used to select the best-fit regression model. The choice of these regression functions assume a smooth monotonic decrease of the model error with increasing density or number of training points within the region of interest.
Intuitively, most surrogate modeling scenarios are expected to conform to the monotonically decreasing trend of the model error (with sample density). However, in certain cases, the variation of the estimated error (\(\mathrm {Mo_{med}^{t}}\) and \(\mathrm {Mo_{max}^{t}}\); t=1,2,...,N itr) with sample density (over iterations) may not follow the monotonically decreasing trend. Such cases may arise due to a highly non-uniform distribution of sample points or a highly skewed distribution of nonlinearity of the actual output function over the input space. To consider the possibility of such non-conforming cases, and to avoid unreasonable inaccuracies in error predictions using the fitted VESD function, the feasibility of the monodically decreasing trend of model error is statistically tested using a criterion called the Monotonic Trend (MT) criterion. If the MT criterion is satisfied by the fitted VESD regression function, this VESD function is considered admissible and is used to predict the error in the final surrogate (constructed over all sample points). Otherwise, a stable implementation of k-fold cross-validation, called PEMF-based k-fold cross-validation, is used to represent the error in the final surrogate model. The PEMF-based k-fold cross-validation method is defined as the modal value of the error of the intermediate surrogate constructed using \(\mathrm {n^{N^{itr}}}\) sample points (i.e., the modal error value in the last iteration of PEMF). The median and the maximum error measures of the final surrogate are then respectively given by \(\mathrm {Mo_{med}^{N^{itr}}}\) and \(\mathrm {Mo_{max}^{N^{itr}}}\) (as is further defined in (16)).
In the MT test, we use the Pearson correlation coefficient (Lawrence and Lin 1998) to measure the linear correlation between the log-level and the log-log transformation of Error with respect to the Sample density in the Type 1 and Type 2 VESD functions. In this test, the following standard mathematical properties are employed:
-
(i)
if a (x, y) input/output data set follows an exponential relationship, the transformed (x, log y) data follows a linear relationship, and
-
(ii)
if a (x, y) input/output data set follows a purely p th order power relationship, the transformed (log x, log y) data follows a linear relationship.
In this test, the Pearson coefficient for the Type 1 and Type 2 VESD functions is defined as:
where
In (12), Cov is the covariance, σ is the standard deviation, and \(\rho _{_{\lambda ,\varphi }}\) is a Indicator of Monotonicity (defined by the Pearson Correlation Coefficient). The closer \(\rho _{_{\lambda ,\varphi }}\) is to −1, the stronger is the monotonic (decreasing) relationship between the model error measure and sample density. If the threshold condition value of \(\rho _{_{\lambda ,\varphi }}\) is given by ρ c r , then \(p=\frac {\rho _{cr}}{\sqrt {(1-\rho _{cr}^{2})/d_{F}}}\) follows a t-distribution with the degrees of freedom given by d F =N itr−2. The threshold condition value, ρ c r , for a C c r level of confidence can then be determined by
For a given level of confidence (C c r ), if the Indicator of Monotonicity, \(\rho _{_{\lambda ,\varphi }}\), is not greater than the threshold condition value of ρ c r , the monotonically decreasing characteristics is considered to hold. In that case, the fitted regression function (VESD) is used to predict the fidelity of the final surrogate model (V E S D ({X i n })). Conversely, if the Indicator of Monotonicity (\(\rho _{_{\lambda ,\varphi }}\)) is greater than the threshold condition value of ρ c r , the VESD function is discarded, and instead a stable implementation of k-fold cross-validation (PEMF-based k-fold) is used, as shown below:
2.3 Investigating the stability of PEMF error measures
The individual steps of the PEMF method at each iteration is somewhat analogous to k-fold cross-validation. In k-fold cross-validation, errors are generally evaluated for a specific value of k. In the PEMF method, the errors are evaluated iteratively for different values of k, in order to predict the level of error in the final surrogate. This novel approach gives PEMF the predictive capability that is otherwise lacking in standard k-fold or leave-one-out cross-validation. More importantly, PEMF seeks to formulate and use more stable measures of error compared to those in cross-validation. In k-fold cross-validation, for one specific k, mean errors are used to represent the level of error, as shown in the pseudo code under Algorithm 1 (in the Appendix). In the PEMF method, the median error on each heuristic set of intermediate test points is first estimated, followed by the determination of the statistical mode of the median errors. The modal error value is then used to represent the level of error at any given k. The novel use of the modal value of median errors (and similarly modal value of maximum errors) promotes a monotonic decrease of the error measure with sample density, as opposed to the untraceable noisy variation that is often characteristic of mean or RMS error measures.
To illustrate the potential greater stability of PEMF error measures (with respect to their relation to sample density) compared to conventional measures, a two design variable benchmark problem (Branin-Hoo function (Section 3.2)) is considered. The size of the sample data set, N, is defined to be 500, and the region of interest is set to be the entire design space. The numerical settings of this problem is given by
Based on this definition, at the first iteration, the intermediate surrogates are constructed using 22 sample points, and tested over 478 points, i.e., \(N^{1}_{TR}=22,~and~N^{1}_{TE}=478\), where \(N^{1}_{TR}\) and \(N^{1}_{TE}\) represent the number of training points and test points, respectively. At the last iteration, \(N^{50}_{TR}=120,~and~N^{50}_{TE}=380\).
The variation of the Mode of Median and the Mean of the Mean errors with sample density in different surrogates (Kriging, RBF, and E-RBF) are illustrated in Figs. 2 and 3, respectively. These figures illustrate that the mode of median errors decrease with a practically monotonic trend, with increasing number of training points. Such a monotonic tendency cannot be associated with the Mean of the Mean error, as shown in Figs. 3a–c (due to severe local oscillations). It is also important to consider that model errors may not follow a normal distribution, and that the most probable value of error (modal value) might be of greater practical importance (more insightful) than a mean value. From these perspectives, PEMF error measures are likely to provide a more meaningful understanding of the fidelity of a surrogate model in the context of its use in analysis and design.
3 Application of the PEMF method
3.1 Surrogate models
The effectiveness of the PEMF method in predicting the regional and global errors is explored for application with Kriging, RBF, and E-RBF surrogate model on Branin-Hoo function with two design variables, Perm function with 10 design variables, and Dixon & Price function with 50 design variables. The median and the maximum errors evaluated using PEMF are compared with the actual error evaluated on additional test points. To perform a fair comparison, the estimated median and maximum errors on heuristic subsets of additional test points are used to fit error distributions. The statistical mode of the median and the maximum error distributions (estimated over additional test points) are used as reference values (as shown in Algorithm 2 in the Appendix), to investigate performance of PEMF.
To illustrate the potential greater effectiveness of the PEMF error metric over popular cross-validation error, the mean and maximum errors given by leave-one-out cross-validation are also compared with the actual mean and maximum errors. In this case, the mean and the maximum errors estimated on additional test points (as described in Algorithm 3 in the Appendix) are used as reference values.
To implement the Kriging method, the DACE (design and analysis of computer experiments) package developed by Lophaven et al. (2002) is used. The bounds on the correlation parameters in the nonlinear optimization in Kriging, 𝜃 l and 𝜃 u , are specified to be 0.1 and 20, respectively. In the Kriging surrogate model, the zero-order polynomial function is used as the regression model. To implement the RBF surrogate model, the multiquadric radial basis function (Hardy 1971) is used, where the shape parameter is set to c=0.9. In implementing the E-RBF surrogate model (Mullur and Messac 2005), the shape parameter is set to c=0.9, the λ parameter is set to 4.75, and the order of monomial in the non-radial basis functions is fixed at 2.
3.2 Numerical experiments
PEMF is first applied to the following popular 2D benchmark function:
Branin-Hoo function
The effectiveness of the PEMF method in predicting the global error in high dimensional problems is then explored by applying it to two other analytical benchmark test problems:
-
Perm Function (10 variables)
$$\begin{array}{@{}rcl@{}} &&\!\!\!\!\!\!\!\!\!\!\!\! f(x) = {\sum}_{k=1}^{n}\left\{{\sum}_{j=1}^{k}\left(j^{k}+0.5\right)\left[\left(\frac{x_{j}}{j}\right)^{k}-1\right]\right\}^{2}\\ &&\!\!\!\!\!\!\!\!\!\!\!\! \textit{where}~~x_{i} \in [-n~~n+1],~i=1,...,n \\ &&\!\!\!\!\!\!\!\!\!\!\!\! n=10 \end{array} $$(18) -
Dixon & Price Function (50 variables)
$$\begin{array}{@{}rcl@{}} & f(x) = \left( x_{1} - 1 \right)^{2} + {\sum}_{i=2}^{n} i\left( 2{x_{i}^{2}} - x_{i-1} \right)^{2} \\ & \textit{where}~~x_{i} \in [-10~~10],~i=1,...,n \\ & n=50 \end{array} $$(19)
The numerical settings for the application of PEMF in predicting the median and the maximum errors of surrogate models for the benchmark functions are provided in Table 2, which lists (i) the number of training points, (i i) the number of iterations, (i i i) the number of inside-region training points based on the predefined region’s boundary, and (i v) the size of the training set at each iteration. Two different cases of error quantification are explored for the Branin-Hoo function. In case I, the PEMF method is used for regional error measurement, where the boundaries of the region of interest are defined as: x 1∈[−2 7], x 2∈[3 12]. While in case II, PEMF is used as a global error measurement method to estimate the model fidelity in entire design domain. To implement the MT criterion, the threshold condition value (ρ c r ) in benchmark problems is estimated to be \(\rho _{cr}=\frac {p_{0.95,2}}{\sqrt {(p_{_{0.95,2}}^{2}+2)}}=-0.950\), for a specified level of confidence of C c r =0.95 and N itr=4.
3.3 Performance criteria
The errors evaluated on additional test points, summarized under Algorithm 2 and Algorithm 3 (in the Appendix), are used to compare the performance of PEMF and leave-one-out cross-validation. In this paper, we compare the PEMF method with the popular leave-one-out cross-validation method, particularly with respect to their abilities to predict the quality or fidelity of surrogate models. This comparison is driven by the premise that although these methods are characteristically different the primary ‘end-use’ applications of PEMF and cross-validation methods are similar, namely model testing, model selection, and model refinement. The relative differences between the actual errors and the PEMF errors are evaluated as
The relative difference between the actual errors and the errors given by leave-one-out cross-validation are evaluated as
In this study, the number of additional test points used to compare the error measures is equal to 50 times of the number of inside-region samplepoints (50×N i ).
4 Results and discussion
4.1 Investigating the performance of PEMF (for the Branin-Hoo function)
Figures 4 and 5 show the VESD regression functions constructed to predict the median errors in the surrogates for Cases I and II, respectively. For the sake of illustration, all the regression coefficients in the figures are rounded to two decimal places. The Indicator of Monotonicity, \(\rho _{_{\lambda ,\varphi }}\), for each model is also shown in these figures. The distributions of median errors, and the mode of the median error distributions (Mo m e d ) are shown for each iteration. In these figures, the shaded Δ symbol represents the quantified mode of median errors at each iteration; the hollow Δ symbol represents the predicted mode of median error in the final surrogate.
It is observed from Figs. 4 and 5 that the mode of the error distributions generally decreases with the density of inside-region training points, since the estimated Indicator of Monotonicity in the two cases are less than the pre-estimated threshold (ρ c r ). Based on these observations, the variation of error with training point density can be represented by applying the VESD regression functions described in Section 2.1.1. The predicted median (\(\mathrm {E_{med}^{PEMF}}\)) errors and actual errors in the final surrogates in Cases I and II are provided in Table 3. The PEMF error is within the same order of magnitude as the actual errors , which is a significant achievement in surrogate error quantification. It is also important to note that PEMF provides additional helpful insights into the performance of the surrogates for the concerned problem. For example, in these case studies, it is observed (from Figs. 4 and 6) that the regional accuracy of the Kriging surrogate is significantly more sensitive to the sample density than that of the RBF and E-RBF surrogates.
For the two cases of the Branin-Hoo problem, the VESD functions used to predict the mode of the maximum error, and the Indicator of Monotonicity (\(\rho _{_{\lambda ,\varphi }}\)) are illustrated in Figs. 6 and 7. The mode of the maximum error distributions in the intermediate surrogate models is represented by the solid square symbol; the hollow square symbol represents the predicted mode of maximum error in the final surrogate. It is again observed that the mode of the maximum error distributions decreases with increasing density of inside-region training points and the MT criterion is satisfied in the two cases. The predicted maximum error (\(\mathrm {E_{max}^{PEMF}}\)) and the actual errors evaluated on additional test points (Algorithm 2 in the Appendix) in the final surrogates for Cases I and II are given in Table 4.
4.1.1 Comparing PEMF with cross-validation (for the Branin-Hoo function)
Figures 8 and 9 illustrate how the PEMF error measure and the RAE given by leave-one-out cross-validation compare with the actual error evaluated on additional test points. In these figures, the y-axis represents the relative difference of the error measures compared with the actual errors. These comparisons are given for all three types of surrogate models constructed, i.e., Kriging, RBF, and E-RBF.
The performance criteria defined in Section 3.3 (i.e., R-values in (20) and (21)) are applied to compare the performance of the PEMF method and the cross-validation method with the actual errors in Cases I and II. The comparison results are also provided in Tables 5 and 6. The smaller R-value obtained in each case is shown in boldface. From Figs. 8 and 9 and Tables 5 and 6, it is observed that the measure of surrogate model fidelity provided by the PEMF method is up to two orders of magnitude more accurate than the RAE estimated by leave-one-out cross-validation.
4.2 Application of PEMF to higher dimensional benchmark problems
The VESD m e d and VESD m a x regression functions, and the Indicator of Monotonicity for the two other benchmark problems, the Perm function (with 10 design variables) and the Dixon & Price function (with 50 design variables), are illustrated in Figs. 10, 11, 12 and 13. It is observed that in most of the cases the MT criterion is satisfied, and hence the VESD function can be used to predict the median and maximum errors of the constructed surrogate models. The MT criterion is however not satisfied for the following cases: the estimated median error of the Kriging model constructed for the Perm Function (Fig. 10a), and the estimated median error of the RBF model (Fig. 12b) and the estimated maximum error of the Kriging model (Fig. 13a) constructed for the Dixon & Price Function. Therefore, in these three cases, the PEMF-based k-fold cross-validation (i.e., the modal value of the error in the last iteration of PEMF) are automatically considered to be the error measure of the surrogate model constructed using all sample points.
The comparison of both the PEMF error measure and the RAE given by leave-one-out cross-validation with the actual error evaluated on additional test points are illustrated through bar diagrams in Figs. 14–15. In these figures, the y-axis represents the relative difference of the error measures compared with the actual errors. The comparison results are also provided in Tables 7 and 8. A smaller R-value indicates a more accurate error measure. In these tables, the smaller R-values obtained for each case are shown in boldface. From Figs. 14 and 15, and Tables 7 and 8, it is observed that the PEMF method is up to two orders of magnitude more accurate than the leave-one-out cross-validation for these 10-50 dimensional test problems. These observations again establish the effectiveness of PEMF as a new more accurate approach to quantify surrogate model errors.
5 Statistical test on results of PEMF and leave-one-out cross-validation method
Further statistical tests are performed to illustrate the robustness of the PEMF method. The paired t-test (Montgomery and Runger 2010) is applied to measure the significance of the difference between estimated error measures, and the actual error evaluated on additional test points.
A set of 500 paired sample data is generated using optimal Latin Hypercube, which is defined as X 1, X 2, X 3, ..., X 500. The size of each sample data set is defined to be N i =30, with all sample points denoted as inside-region sample points, i.e., the PEMF method is applied towards global error measurement. In this test, it is assumed that the size of the inside-region training set at each iteration, t, is given by
where the function ⌊x⌋ returns the largest integer less than or equal to x. The differences between the errors estimated using PEMF and the actual error on each pair of observation are defined as
The frequency histogram of the residuals in different types of surrogates (Kriging, RBF, and E-RBF) for the median and maximum errors estimated using PEMF are illustrated in Figs. 16 and 18, respectively. The frequency histogram of the residuals in different types of surrogates (Kriging, RBF, and E-RBF) for the mean and maximum errors estimated using leave-one-out cross validation are illustrated in Figs. 17 and 19, respectively. In these figures, the 10th and the 90th percentiles of the residuals are illustrated using black dashed lines.
It is readily evident from the histograms in Figs. 16 and 18 that the residuals do not necessarily follow a normal distribution. Hence, statistical methods based on a resampling approach such as the Bootstrapping method should be applied to perform the statistical inference (Efron and Tibshirani 1993). In the Bootstrapping method, it is assumed that the statistics of the sampling distribution is the probability of all possible values of the estimated statistics calculated from a sample of size n drawn from a given population (Haukoos and Lewis 2005). To evaluate the 90 % bootstrap confidence intervals (CI), after creating n bootstrapped sets from the original residuals, the mean value for each bootstrapped data set is independently computed. The 90 % CI from the set of computed mean values are then estimated using the normal approximation method. Assuming the normal approximation is normally distributed with sample mean \(\overline {d^{*}}\) and standard deviation \(s^{*}_{D}\) for the n=1000 bootstrapped resamples, a 90 % confidence interval on the difference in means, μ D , is defined as
In this case, t 0.05,n−1 is the upper 0.05 % point of the t distribution with n−1 degrees of freedom. Confidence intervals that are narrower and closer to zero indicate higher level of robustness of the error measurement.
Table 9 presents the 90 % confidence intervals (CI) for residuals of the mode of median and maximum errors estimated using the PEMF method in different surrogates.
Likewise, the statistical test is also performed for leave-one-out cross-validation. In this test, the same 500 paired sample data are used where the differences between the mean and maximum errors estimated by the cross-validation method and the actual error on each pair of observation are given by
The actual errors are errors evaluated on additional test points, as given by Algorithm 3 (in the Appendix).
The frequency histogram of the residuals in different types of surrogates (Kriging, RBF, and E-RBF), for the mean and maximum errors estimated using cross-validation, are illustrated in Figs. 17 and 19, respectively. We again use the normal approximation method to estimate the 90 % bootstrapped confidence intervals on the difference in the means, μ D , for the residuals of errors, which are reported in Table 10.
To compare the PEMF method and the leave-one-out cross-validation method, the distribution of the residuals of each surrogate is described by a box plot. The box plot indicates the deviation of the central tendency from zero (which is the expected value). Figures 20 and 21 respectively illustrate the distribution of residuals of the median and the maximum errors estimated using PEMF and the maximum errors estimated using cross-validation in different surrogates. From the results of the statistical test on PEMF and cross-validation (Figs. 16, 17, 18, 19, 20 and 21 and Tables 9 and 10), it is readily evident that the confidence intervals and the 25th and 75th percentiles for PEMF are significantly closer to zero (up to two orders of magnitude closer) compared to that for cross-validation. These observations show that PEMF is expected to provide far superior robustness in quantifying surrogate model errors compared to standard leave-one-out cross-validation.
6 Conclusion
This paper develops a new approach to quantify the accuracy of surrogate models in a given region of the design domain or the entire design domain. Such an approach is useful for informed decision-making when using surrogate models or metamodels for analysis and optimization. This method, called the Predictive Estimation of Model Fidelity (PEMF), is a model-independent method for predicting the error in the actual surrogate model constructed using all available sample points. In this method, intermediate surrogates are constructed iteratively using multiple heuristic subsets of the available sample points. The remaining sample points are used to evaluate the error in the estimated function at that iteration. In this method, the model error at each iteration is defined by the mode of the median error and the maximum error distributions – this is expected to promote greater stability compared to mean or mean squared error measures. Regression models are then developed to represent the error in the surrogate as a function of the density of training points. These regression models are then extrapolated to predict the fidelity of the final surrogate model under the condition that a Monotonic Trend (MT) criterion is satisfied. The MT criterion is a mechanism to statistically test the feasibility of the monotonic relationship between the modal error values and training sample density. If the MT criterion is not satisfied, the estimated modal error values from the last iteration is used to represent the measure of fidelity of the surrogate model – which is essentially a more stable implementation of k-fold cross-validation. The effectiveness of the new PEMF method is illustrated by applying it to a 2-variable, a 10-variable, and a 50-variable benchmark problems, where the following types of surrogate models were constructed: Kriging, RBF, and E-RBF surrogates. These numerical experiments show that the PEMF method provides up to two orders of magnitude greater accuracy in measuring surrogate model error compared to the relative absolute error estimated by leave-one-out cross-validation. Superior robustness of the PEMF method is also established using additional statistical tests.
At this point, the number of iterations and the initial ratio of the numbers of intermediate training points to test points have to be prescribed by the user. Hence there remains scope for exploring strategies that adaptively determine these two factors as functions of the dimension and strength of the sample data, thereby making PEMF more tractable. The application of PEMF to a wide variety of complex engineering design problems, particularly in the context of model refinement, model selection, and uncertainty analysis will further establish the diverse potential of this novel approach to quantify the fidelity of surrogate models.
References
Allaire D, He Q, Deyst J, Willcox K (2012) An information-theoretic metric of system complexity with application to engineering system design. J Mech Des 134(10):100,906
Atamturktur S, Hemez F, Williams B, Tome C, Unal C (2011) A forecasting metric for predictive modeling. Comput Struct 89(23):2377–2387
Atamturktur S, Williams B, Egeberg M, Unal C (2013) Batch sequential design of optimal experiments for improved predictive maturity in physics-based modeling. Struct Multidiscip Optim 48(3):549–569
Audet C, Dennis JE, Moore DW, Booker A, Frank PD (2000) A surrogate-model-based method for constrained optimization. In: 8th symposium on multidisciplinary analysis and optimization. Long Beach
Booker AJ, Dennis JE, Frank P, Serafini DB, Torczon V, Trosset MW (1999) A rigorous framework for optimization of expensive functions by surrogates. Struct Optim 17(1):1–13
Bozdogan H (2000) Akaike’s information criterion and recent developments in information complexity. J Math Psychol 44:62–91
Chowdhury S, Mehmani A, Messac A (2014a) Concurrent surrogate model selection (cosmos) based on predictive estimation of model fidelity. In: ASME 2014 international design engineering technical conferences (IDETC). Buffalo
Chowdhury S, Mehmani A, Tong W, Messac A (2014b) A visually-informed decision-making platform for model-based design of wind farms. In: 15th AIAA/ISSMO multidisciplinary analysis and optimization conference. Atlanta
Efron B, Tibshirani R (1993) An introduction to the bootstrap, vol 57. CRC press
Forrester A, Keane A (2009) Recent advances in surrogate-based optimization. Progress Aerospace Sci 45(1-3):50–79
Goel T, Stander N (2009) Comparing three error criteria for selecting radial basis function network topology. Comput Methods in Appl Mech Eng 198(27):2137–2150
Goel T, Haftka RT, Shyy W, Queipo NV (2007) Ensemble of surrogates. Struct Multidiscip Optim 33(3):199–216
Goel T, Haftka RT, Shyy W (2009) Comparing error estimation measures for polynomial and kriging approximation of noise-free functions. Struct Multidiscip Optim 38(5):429–442
Gorissen D, Dhaene T, Turck FD (2009) Evolutionary model type selection for global surrogate modeling. J Mach Learn Res 10:2039–2078
Gunn SR (1998) Support vector machines for classification and regression. Tech. rep., ISIS - 14. NASA Langley Research Center, Hampton, VA
Haldar A, Mahadevan S (2000) Probability, reliability, and statistical methods in engineering design. Wiley
Hardy RL (1971) Multiquadric equations of topography and other irregular surfaces. J Geophys Res 76:1905–1915
Haukoos JS, Lewis RJ (2005) Advanced statistics: bootstrapping confidence intervals for statistics with ŞdifficultŤ distributions. Acad Emerg Med 12(4):360–365
Hemez F, Atamturktur S, Unal C (2010) Defining predictive maturity for validated numerical simulations. Comput Struct 88(7):497–505
Jin R, Chen W, Simpson TW (2000) Comparative studies of metamodeling techniques under multiple modeling criteria. AIAA 1(4801)
Jin R, Chen W, Sudjianto A (2002) On sequential sampling for global metamodeling in engineering design. In: ASME 2002-design engineering technical conferences and computers and information in engineering conference. Montreal
Jones D, Schonlau M, Welch W (1998) Efficient global optimization of expensive black-box functions. J Global Optim 13(4):455–492
Keane AJ (2006) Statistical improvement criteria for use in multiobjective design optimization. AIAA Journal 44(4):879–891
Kennedy MC, O’Hagan A (2000) Predicting the output from a complex computer code when fast approximations are available. Biometrika 87(1):1–13
Kleijnen J (1975) Statistical techniques in simulation. Publishing House Statistics, New York
Kleijnen J, Beers WV (2004) Application-driven sequential designs for simulation experiments: Kriging metamodelling. J Oper Res Soc 55:876–883
Lawrence I, Lin K (1998) A concordance correlation coefficient to evaluate reproducibility. Biometrics, pp 255–268
Lehmensiek R, Meyer P, Muller M (2002) Adaptive sampling applied to multivariate, multiple output rational interpolation models with application to microwave circuits. Int J RF and Microwave Comput Aided Eng 12(4):332–340
Loeppky JL, Moore LM, Williams B (2010) Batch sequential designs for computer experiments. J Stat Plan Infer 140(6):1452–1464
Lophaven SN, Nielsen HB, Sondergaard J (2002) Dace - a matlab kriging toolbox, version 2.0. Tech. Rep, IMM-REP-2002-12. Informatics and mathematical modelling report, Technical University of Denmark
Martin JD, Simpson TW (2005) Use of kriging models to approximate deterministic computer models. AIAA J 43(4):853–863
McKay M, Conover W, Beckman R (1979) A comparison of three methods for selecting values of input variables in the analysis of output from a computer code. Technometrics 21(2): 239–245
Meckesheimer M, Booker AJ, Barton RR, Simpson TW (2002) Computationally inexpensive metamodel assessment strategies. AIAA J 40(10):2053–2060
Mehmani A, Zhang J, Chowdhury S, Messac A (2012) Surrogate-based design optimization with adaptive sequential sampling. In: 53rd AIAA/ASME/ASCE/AHS/ASC Structures, Structural Dynamics And Materials Conference. Hawaii
Montgomery DC, Runger GC (2010) Applied statistics and probability for engineers. Wiley, Hoboken
Mullur A, Messac A (2005) Extended radial basis functions: more flexible and effective metamodeling. AIAA J 43(6):1306–1315
Nguyen HM, Couckuyt I, Knockaert L, Dhaene T, Gorissen D, Saeys Y (2011) An alternative approach to avoid overfitting for surrogate models. In: Proceedings of the 2011 winter simulation conference, pp 2765–2776
Queipo N, Haftka RT, Shyy W, Goel T, Vaidyanathan R, Tucker P (2005) Surrogate-based analysis and optimization. Progress Aerospace Sci 41(1):1–28
Sacks J, Welch W, Mitchell T, Wynn H (1989) Design and analysis of computer experiments. Stat Sci 4(4)
Simpson T, Korte J, Mauery T, Mistree F (2001) Kriging models for global approximation in simulation-based multidisciplinary design optimization. AIAA J 39(12):2233–2241
Sugiyama M (2006) Active learning in approximately linear regression based on conditional expectation of generalization error. J Mach Learn Res 7:141–166
Viana FAC, Haftka RT, Steffen V (2009) Multiple surrogates: How cross-validation errors can help us to obtain the best predictor. Struct Multidiscip Optim 39(4):439–457
Viana FAC, Pecheny V, Haftka RT (2010) Using cross validation to design conservative surrogates. AIAA J 48(10):2286–2298
Williams B, Loeppky JL, Moore LM, Macklem MS (2011) Batch sequential design to achieve predictive maturity with calibrated computer models. Reliab Eng Syst Saf 96(9):1208–1219
Yegnanarayana B (2004) Artificial neural networks. PHI Learning Pvt. Ltd
Zhang J, Chowdhury S, Messac A (2012) An adaptive hybrid surrogate model. Struct Multidiscip Optim 46(2):223–238
Zhang J, Chowdhury S, Mehmani A, Messac A (2014) Characterizing uncertainty attributable to surrogate models. J Mech Des 3(136):031,004
Acknowledgments
Support from the National Science Foundation Awards CMMI-1100948 and CMMI-1437746 is gratefully acknowledged. Any opinions, findings, and conclusions or recommendations expressed in this paper are those of the authors, and do not necessarily reflect the views of the NSF.
The surrogate model codes provided by Dr. Jie Zhang are gratefully acknowledged.
Author information
Authors and Affiliations
Additional information
Parts of this manuscript have been presented at the 54th AIAA/ASME/ASCE/AHS/ASC Structures, Structural Dynamics and Materials Conference, in April, 2013, at Boston, Massachusetts - Paper Number: AIAA 2013-1751.
Appendix A
Appendix A
1.1 A.1 Standard k-fold cross-validation

1.2 A.2 Quantifying the mode of median and maximum errors, estimated on additional test points (for performance testing of PEMF)

1.3 A.3 Quantifying the mean and maximum error estimated on additional test points (for performance validation of cross-validation)

Rights and permissions
About this article
Cite this article
Mehmani, A., Chowdhury, S. & Messac, A. Predictive quantification of surrogate model fidelity based on modal variations with sample density. Struct Multidisc Optim 52, 353–373 (2015). https://doi.org/10.1007/s00158-015-1234-z
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00158-015-1234-z