Abstract
Objective
To assess blood leucocytes gene profiling during recovery phase of septic shock; to test the relation between encoding gene expression and protein level.
Study design
Gene expression levels were studied at days 0, 1, 7 and 28 (D0, 1, 7 and 28) on a dedicated microarray of 340 genes involved in inflammatory processes.
Settings
16-bed intensive care unit, Lariboisière University hospital.
Patients
Seventeen septic shock patients enrolled when at least one additional organ dysfunction occurred.
Measurements and results
Changes over time were compared with D0 via the ratio Dx/D0. The time-related gene expression study showed significant changes in ten genes. Among them, S100A8 and S100A12 had a reduced expression over time compared with D0, whereas CD74's expression increased. The microarray results were validated by RT-qPCR for four genes. The S100A8 plasma levels decrease along recovery in parallel with the gene expression decrease. The CD74 gene expression evolution significantly correlated with HLA-DR monocyte expression.
Conclusions
These results are the first description of variations in expression of key inflammatory genes in the course of the septic shock recovery period.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Despite a better understanding of pathophysiology, septic shock remains a challenging situation in ICU and retains a high mortality rate [1]. The systemic inflammation has received a lot of attention in the past decades, using animal or cellular models [2–4] or clinical cohorts [5]. Within the past decades, the use of animal and cellular models led to the isolation of important mediators of systemic inflammation, such as TNFα or IL-1β, which were further targeted in protocol-based therapy on clinical cohorts [6–8]. None of the clinical trials evaluating drugs interacting with these factors have been successful. In the complexity of the human septic pathophysiology, the recovery process received little attention in patients who survived septic shock. Taking into account the large number of pathways potentially involved renders the gene-by-gene or protein-by-protein approach obsolete. Multiplex approaches, such as microarray strategies [9], thus appear like powerful alternatives. This technology can indeed be applied to define the kinetic of genes expression during the recovery period. The recently published data on time evolution of healthy volunteers receiving LPS injections provide useful information on the acute modifications of gene expression and their time dependence for recovery [9]. Recently, two studies carried out in human septic patients have been published. In the first study, a biochip dedicated to inflammation was applied to whole blood cell from 8 septic patients, compared with 4 patients without inflammation, questioning the early diagnosis of sepsis, but their results require further validation [4]. In a second study [10], an Affymetrix chip (approximately 14,700 probe sets, whole blood) identified a set of 28 genes efficiently discriminating non-survivors from survivors with a sensitivity of 100% and a specificity of 86%. Several genes overexpressed in survivors participate in innate immunity, i.e. cytokine, chemokine receptor and toll-receptor pathways.
In the present study, we analyzed the gene expression in circulating leucocytes with a microchip dedicated to inflammation pathways. The evolution of gene expression in septic shock patients at days 1, 7 and 28 was compared to day 0 expression, with every patient being his own control.
Materials and methods
Patients
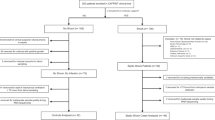
The study concerned patients in septic shock as defined by the ACCP/SCCM consensus [11] and was approved by the Cochin Hospital Ethics Committee (no. CCPPRB 2061). Patients over 18 years old were included after written informed consent from their next of kin. Inclusion criteria were: a documented infection by clinical and/or bacteriological evidence; and septic shock (cardiovascular failure with norepinephrine) with at least one additional sepsis-related organ failure. Exclusion criteria were: long-lasting glucocorticosteroid treatment; recent treatment for cancer; immune or haematological diseases; and a life expectancy < 6 months. Blood samples were initiated within 48 h after reaching the inclusion criteria (D0) and at days 1 (D1), 7 (D7) and 28 (D28). The 28-day outcome (dead or alive) was also recorded (see Table 1 in ESM).
Microarray procedure
Mature granulocytes were eliminated from blood by gradient centrifugation (Histopaque, Sigma, St. Quentin Fallavier, France). Collected cell analysis included morphological characteristics by light scatter and CD66b expression (FACScalibur, BD Bioscience, Le Pont de Claix, France) for evaluating the proportion of immature cells (granulocyte lineage) [12]. Accordingly, the studied cells were at D0 and D1 myeloid cell for 50% and circulating immune mononuclear cells (lymphocytes and monocytes) for the remaining, which became up to 98% at D7 and D28.
Total RNA isolation and microarray preparation are described in the ESM. Hybridisation was performed on the slide Lab-Arraytor human 500-1 cDNA (SIRS-Lab, Jena, Germany). Description of the slide (GPL1936), experimental design and tables of the normalised data for outcome and time evolution were registered in the Gene Expression Omnibus database under the accession numbers GSE5262 and GSE5271. Minimum Information Microarray Experiment (MIAME) criteria were carefully fulfilled (see ESM).
Data measurement
Unpaired t-test was used to generate the list of genes with different levels of expression according to outcome, and ANOVA analysis for comparisons over time. A value of p < 0.05 was considered as statistically significant.
Validation of gene expression by real-time quantitative PCR (RT-qPCR)
Four genes have been selected for validation on the basis of their fold change having a stable trend over time. In addition, their scattering values allowed to well assess a potential correlation. Reverse transcription reactions (High Capacity cDNA Archive kit, Applied Biosystems, Foster City, CA, USA) and TaqMan Gene Expression Assays on ABI PRISM 5700 Sequence Detection System (Applied Biosystems) were performed according manufacturer's specifications. Eukaryotic 18S rRNA was used as an endogenous control and the reference standard sample was Universal Human Reference RNA (Clontech; see ESM).
Plasma S100A8 ELISA
The technique (see ESM) was adapted from a previous publication [13] (from 34 healthy subjects, paired on age and sex; average normal value: 0.26 μm/ml, range 0.052–0.468 μm/ml).
Monocyte HLA-DR expression
Monocyte HLA-DR expression was quantified by flow cytometry (FACScalibur instrument BD Biosciences, Rungis, France) as the number of antibodies per cell (AB/C). Antibodies anti-CD14-FiTC (clone RMO52, Beckman Coulter, Marseille, France), isotype control-PE (Simultest control, BD Biosciences, San Jose, CA, USA.) and anti-HLA-DR-PE (clone L243, BD Biosciences) were used. The average normal value for healthy volunteers was 22,000 (range 11,000–34,000).
Statistical analysis
Clinical characteristics and quantitative results were expressed in median and interquartile range (IQR). Comparison of S100A8 plasma level between survivors and non-survivors at D0 and D1 was performed using the Mann–Whitney test. Changes of plasma S100A8 protein, CD74 gene expression and HLA-DR monocyte expression were tested by Friedman test. Correlations between microarray data with RT-qPCR or protein expression was performed with a Spearman correlation test. A value of p < 0.05 was considered as statistically significant.
Results
The median age of the population was 59 years (IQR 16) with a median Simplified Acute Physiology Score (SAPS II) of 50 (IQR 14), a number of organ failure of 3 (IQR 1) and a Sepsis-related Multiple Organ Failure Score (SOFA) score of 10 (IQR 3) at D0. Three patients (17%) died all within the first week of the study.
Gene expression in leucocytes over 28 days in survivors (n = 14)
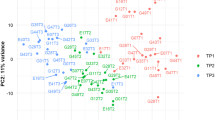
Figure 1 shows the hierarchical clustering representation of gene expression at D1, D7 and D28 referred to D0. Most of the D1/D0 ratios were grouped together as were the later gene expression ratios (D7/D0, D28/D0). Ten genes demonstrated significant expression variations over time in comparison with D0 (p < 0.05; Table 1). CD74 implicated in antigen presentation increased its expression over time until D28. By contrast, the expression of genes encoding proinflammatory proteins, such as S100A8, S100A12, arachidonate pathway (ALOX5AP) and the isoform alpha of IL-3 growth factor receptor, all decreased their expression over time. Table 2 summarizes the variations [fold changes (Dx/D0)] at each time for each gene of interest. D1/D0 comparison in non-survivors vs survivors (clustering representation and list of gene) is shown in Fig. 1 suppl in ESM.
The hierarchical bi-clustering of genes and survivals over 28 days (available data at D1, D7 and D28) referred to D0. The top part illustrates the dendrogram of patient time points. The left part shows the dendrogram of the time evolution of the ten best separated genes set. Each row corresponds to a gene probe and each column to a time point (ratio D1/D0, D7/D0, D28/D0) for one patient. Color intensities of boxes were related to the log10 ratio Dx/D0 in each patient for each gene: increased gene expression in red and decreased gene expression in green. Black boxes correspond to the lack of significant variation
Comparison between microarray and RT-qPCR data in survivors
The real-time PCR data correlated with microarray data for CD74, S100A8, S100A12 and VAMP2 expression (r = 0.62 and p < 0.001, r = 0.58 and p < 0.001, r = 0.81 and p < 0.001, and r = 0.41 and p < 0.05, respectively; see Fig. 2 suppl in ESM).
Relation between monocyte HLA-DR expression and CD74 gene expression in survivors
The CD74 gene expression (microarray data) and the protein HLA-DR expression (Fig. 2) presented the same trend and correlated (r = 0.57, p = 0.0005).
S100A8 plasma measurements in all studied patients (n = 17)
Fig. 3a shows the evolution of S100A8 plasma values along recovery. In addition to the significant difference at day 0 between survival and dead patients (p < 0.01), there was a clear decrease along time evolution (Friedman test, p = 0.001). Fig. 3b shows the correlation between microarray data and the plasma level ratio in survivors (r = 0.89, p < 0.001).
Discussion
The hierarchisation clustering of gene provided a profile of recovery from septic shock in circulating leucocyte. We confirmed the underexpression of CD74 at the early phase of septic shock as previously mentioned by Pachot et al. [14], associated with HLA-DR downregulation at the monocyte surface. The CD74 re-expression along 28 days paralleled the HLA-DR re-expression on monocytes. Modifications of the genes encoding for members of S100α protein family, classically involved in chronic inflammatory disease [15], is reported here for the first time in human septic shock.
Among all studied genes, the CD74 expression increase was the most important over 28 days after septic shock in survivors. CD74 (also named invariant chain, Ii) forms with HLA-DR molecules a complex stocked in the class-II compartment and is essential for the intracellular compartmentalisation of the HLA-DR protein [16]. After degradation in endocytosis vesicules, CD74 will be degraded in class-II associated-invariant peptide (CLIP) and then be exchanged with peptide for presentation to the cell surface [16]. CD74 expression was previously noted to be markedly decreased in the early phase of human septic shock or sepsis [9, 14]. In addition to this confirmation, we showed in the present study (Fig. 2) a recovery of CD74 expression in leucocytes, which paralleled HLA-DR expression recovery at the monocyte surface. Accordingly, this study suggests an additional mechanism to the constantly observed downregulation of HLA-DR expression in monocytes during sepsis or septic shock [17, 18] and the recovery processes.
The increase of S100A8 and S100A12 protein expression has been exclusively described in chronic inflammatory human pathologies such as atherosclerosis and rheumatoid arthritis [19, 20]. These proteins are involved in neutrophil and monocyte activation processes, e.g. adhesion, and have antimicrobial properties [21]. A recent review concluded on growing evidence that S100A8, S100A9 and S100A12 comprise a new group of proinflammatory proteins expressed by phagocytes, which are attractive targets for novel diagnostic and therapeutic approaches in inflammatory diseases [15]. The S100A8 is released by phagocytes by LPS [22, 23] with an amplification by IL-10 and glucocorticoids [24]. The recent publication on a mice model of septic shock, demonstrated the major role of such a protein in sepsis pathophysiology. The knockout mice for S100A8 gene were protected from abdominal sepsis or LPS injection induced death [25]. Although limited by the small number of dead patients, we observed a large difference at day 0 between dead and alive patients, which supports the experimental data. These proteins form complexes (S100A8-S100A9), and their level correlates with inflammatory disorders [15] and prothrombotic response of endothelial cells [26]. These members of calgranulin family of S100 proteins may exert their proinflammatory activities through their interaction with the multiligand receptor for advanced glycation end products (RAGE). In addition to RAGE, recent data demonstrated a S100A8-S100A9 mediated effect on TLR4-MD2 expression in a septic shock mice model [25]. Because of that, this group of proteins might be a new therapeutic target in septic shock. The decreased plasma level trend in survival also fits well with its role in human sepsis. The present study, to our knowledge, is the first that reports variations observed in some transcripts in parallel with variations in protein expression, suggesting a direct relation between transcription and translation. If this result has to be confirmed in a larger population of septic shock patients, it is of greatest interest to use such transcriptomic methods.
The first apparent limitation could relate to the limited number of studied genes (340) provided by our dedicated microchip. Even if a pangenomic chip might give wider information on gene expression, a dedicated chip is also useful. Considering the results published on sepsis or LPS-treated healthy volunteers [9], the number and type of gene of interest are consistent with the number and type of genes whose expression was modified on the dedicated microchip used. Various gene expression profiles were similarly modified on a pangenomic and our dedicated chips, e.g. S100 clusters, VAMP and CD74. In addition, 340 genes' expression evaluation is undoubtedly more informative than a single gene by PCR analysis; however, other important genes' expression modifications in sepsis might be missed. The second limitation concerns the knowledge of cell types collected by gradient centrifugation. It is well known that immature myeloid cells are selected by this separation at the early phase of acute inflammation [12] as we analyzed with flow cytometry. As a consequence, the observed early S100s gene expression is difficult to attribute to a specific category of cells. Thirdly, one has to keep in mind that such white cell gene expression modifications do not cover the modifications occurring in other tissues, such as endothelial and epithelial cells.
Conclusion
This study demonstrates that microarray technique can be applied to circulating leucocytes in septic shock patients, with an adequate validation by RT-qPCR and protein measurements. For the first time, the evolution of gene expression during recovery of septic shock showed significant under- and overexpression of genes known to play a major role in inflammatory response, especially S100 family proteins. The systems involved in the recovery might open new research areas for new therapies.
References
Vincent JL, Sakr Y, Sprung CL, Ranieri VM, Reinhart K, Gerlach H, Moreno R, Carlet J, Le Gall JR, Payen D (2006) Sepsis in European intensive care units: results of the SOAP study. Crit Care Med 34:344–353
Boldrick JC, Alizadeh AA, Diehn M, Dudoit S, Liu CL, Belcher CE, Botstein D, Staudt LM, Brown PO, Relman DA (2002) Stereotyped and specific gene expression programs in human innate immune responses to bacteria. Proc Natl Acad Sci USA 99:972–977
Chinnaiyan AM, Huber-Lang M, Kumar-Sinha C, Barrette TR, Shankar-Sinha S, Sarma VJ, Padgaonkar VA, Ward PA (2001) Molecular signatures of sepsis: multiorgan gene expression profiles of systemic inflammation. Am J Pathol 159:1199–1209
Prucha M, Ruryk A, Boriss H, Moller E, Zazula R, Herold I, Claus RA, Reinhart KA, Deigner P, Russwurm S (2004) Expression profiling: toward an application in sepsis diagnostics. Shock 22:29–33
Bernard GR, Vincent JL, Laterre PF, LaRosa SP, Dhainaut JF, Lopez-Rodriguez A, Steingrub JS, Garber GE, Helterbrand JD, Ely EW, Fisher CJ Jr (2001) Efficacy and safety of recombinant human activated protein C for severe sepsis. N Engl J Med 344:699–709
Abraham E, Wunderink R, Silverman H, Perl TM, Nasraway S, Levy H, Bone R, Wenzel RP, Balk R, Allred RP et al. (1995) Efficacy and safety of monoclonal antibody to human tumor necrosis factor alpha in patients with sepsis syndrome. A randomized, controlled, double-blind, multicenter clinical trial. TNF-alpha MAb Sepsis Study Group. J Am Med Assoc 273:934–941
Fisher CJ Jr, Dhainaut JF, Opal SM, Pribble JP, Balk RA, Slotman GJ, Iberti TJ, Rackow EC, Shapiro MJ, Greenman RL et al. (1994) Recombinant human interleukin 1 receptor antagonist in the treatment of patients with sepsis syndrome. Results from a randomized, double-blind, placebo-controlled trial. Phase III rhIL-1ra Sepsis Syndrome Study Group. J Am Med Assoc 271:1836–1843
Rice TW, Wheeler AP, Morris PE, Paz HL, Russell JA, Edens TR, Bernard GR (2006) Safety and efficacy of affinity-purified, anti-tumor necrosis factor-alpha, ovine fab for injection (CytoFab) in severe sepsis. Crit Care Med 34:2271–2281
Calvano SE, Xiao W, Richards DR, Felciano RM, Baker HV, Cho RJ, Chen RO, Brownstein BH, Cobb JP, Tschoeke SK, Miller-Graziano C, Moldawer LL, Mindrinos MN, Davis RW, Tompkins RG, Lowry SF (2005) A network-based analysis of systemic inflammation in humans. Nature 437:1032–1037
Pachot A, Lepape A, Vey S, Bienvenu J, Mougin B, Monneret G (2006) Systemic transcriptional analysis in survivor and non-survivor septic shock patients: a preliminary study. Immunol Lett 106:63–71
Bone RC, Sibbald WJ, Sprung CL (1992) The ACCP-SCCM consensus conference on sepsis and organ failure. Chest 101:1481–1483
Calvano SE, Greenlee PG, Reid AM, deRiesthal HF, Shires GT, Antonacci AC (1988) Granulocyte contamination of Ficoll-Hypaque preparations of mononuclear cells following thermal injury may lead to substantial overestimation of lymphocyte recovery. J Trauma 28:353–361
Ikemoto M, Tanaka T, Takai Y, Murayama H, Tanaka K, Fujita M (2003) New ELISA system for myeloid-related protein complex (MRP8/14) and its clinical significance as a sensitive marker for inflammatory responses associated with transplant rejection. Clin Chem 49:594–600
Pachot A, Monneret G, Brion A, Venet F, Bohe J, Bienvenu J, Mougin B, Lepape A (2005) Messenger RNA expression of major histocompatibility complex class II genes in whole blood from septic shock patients. Crit Care Med 33:31–38, 236–237
Roth J, Vogl T, Sorg C, Sunderkotter C (2003) Phagocyte-specific S100 proteins: a novel group of proinflammatory molecules. Trends Immunol 24:155–158
Matza D, Kerem A, Shachar I (2003) Invariant chain, a chain of command. Trends Immunol 24:264–268
Monneret G, Finck ME, Venet F, Debard AL, Bohe J, Bienvenu J, Lepape A (2004) The anti-inflammatory response dominates after septic shock: association of low monocyte HLA-DR expression and high interleukin-10 concentration. Immunol Lett 95:193–198
Caille V, Chiche JD, Nciri N, Berton C, Gibot S, Boval B, Payen D, Mira JP, Mebazaa A (2004) Histocompatibility leukocyte antigen-D related expression is specifically altered and predicts mortality in septic shock but not in other causes of shock. Shock 22:521–526
Kerkhoff C, Klempt M, Sorg C (1998) Novel insights into structure and function of MRP8 (S100A8) and MRP14 (S100A9). Biochim Biophys Acta 1448:200–211
PasseyRJ,XuK,HumeDA,GeczyCL (1999) S100A8: emerging functions and regulation. J Leukoc Biol 66:549–556
RavasiT,HsuK,GoyetteJ,SchroderK, Yang Z, Rahimi F, Miranda LP, Alewood PF, Hume DA, Geczy C (2004) Probing the S100 protein family through genomic and functional analysis. Genomics 84:10–22
Koike T, Harada N, Yoshida T, Morikawa M (1992) Regulation of myeloid-specific calcium binding protein synthesis by cytosolic protein kinase C. J Biochem (Tokyo) 112:624–630
Vandal K, Rouleau P, Boivin A, Ryckman C, Talbot M, Tessier PA (2003) Blockade of S100A8 and S100A9 suppresses neutrophil migration in response to lipopolysaccharide. J Immunol 171:2602–2609
Hsu K, Passey RJ, Endoh Y, Rahimi F, Youssef P, Yen T, Geczy CL (2005) Regulation of S100A8 by glucocorticoids. J Immunol 174:2318–2326
Vogl T, Tenbrock K, Ludwig S, Leukert N, Ehrhardt C, van Zoelen MA, Nacken W, Foell D, van der Poll T, Sorg C, Roth J (2007) Mrp8 and Mrp14 are endogenous activators of Toll-like receptor 4, promoting lethal, endotoxin-induced shock. Nat Med 13:1042–1049
Viemann D, Barczyk K, Vogl T, Fischer U, Sunderkotter C, Schulze-Osthoff K, Roth J (2007) MRP8/MRP14 impairs endothelial integrity and induces a caspase-dependent and -independent cell death program. Blood 109:2453–2460
Acknowledgements
This work was supported in part by “Programme Hospitalier Recherche Clinique (PHRC) AORO2006” and by “Quadrienal plan for research from the French Ministry of Research” EA 322.
Author information
Authors and Affiliations
Corresponding author
Additional information
This article is discussed in the editorial available at: http://dx.doi.org/10.1007/s00134-008-1049-0
Electronic supplementary material
Rights and permissions
About this article
Cite this article
Payen, D., Lukaszewicz, AC., Belikova, I. et al. Gene profiling in human blood leucocytes during recovery from septic shock. Intensive Care Med 34, 1371–1376 (2008). https://doi.org/10.1007/s00134-008-1048-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00134-008-1048-1