Abstract
Ten plant species were grown in constructed wetlands (CWs) to remediate water containing 2% (w/v) crude oil. The plant species with better growth and biomass production were Typha latifolia and Cyperus laevigatus, and they were significantly correlated (R2 = 0.91) with hydrocarbon degradation. From T. latifolia and C. laevigatus, 33 hydrocarbon-degrading bacterial strains were isolated from the rhizosphere, and root and shoot interiors. More diversified bacteria were found in the rhizosphere and endosphere of C. laevigatus than those of T. latifolia. The predominant cultural hydrocarbon-degrading bacteria were shown to belong to the genera Pseudomonas, Acinetobacter and Bacillus. In addition to genes involved in hydrocarbon degradation, most of the bacteria displayed multiple plant growth promoting (PGP) activities. This study suggests the importance of selecting suitable bacterial strains with hydrocarbon degradation and PGP activities for improving the efficacy of CWs used in remediating water contaminated with crude oil.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
In industrialized and oil-producing countries, surface and ground water are often contaminated with petroleum hydrocarbons resulting from oil extraction and processing. These hydrocarbons can be toxic to fauna and flora (Li et al. 2016). One way to deal with this problem is to use constructed wetlands (CWs). These specific types of CWs are engineered systems designed to improve wastewater quality by taking advantage of the biogeochemical and biological processes occurring in natural wetlands (Ruan et al. 2006; Singh and Singh 2018). In CWs, both plants and microbial communities are key players in the treatment processes. Plants promote microbial communities by releasing oxygen and low-molecular weight root exudates into the rhizosphere, and by providing a large surface area for microbial attachment (Brix 1997; Wu et al. 2012). The efficiency of CWs in degrading crude oil depends upon the numbers and metabolic activity of microorganisms in the rhizosphere and endosphere of the plants (Liu et al. 2011; Shehzadi et al. 2014). Moreover, hydrocarbon degradation has been reported to correlate with the abundance and expression of alkane-degrading genes (Khan et al. 2013).
In some cases, rhizobacteria colonize plant roots and shoots, where they can degrade contaminants adsorbed to or taken up by the plants (Weyens et al. 2009; Khan et al. 2013). Various plant-associated bacteria contain genes linked to catabolic enzymes that can degrade hydrocarbons (Afzal et al. 2011). Bacterial ability to degrade hydrocarbons is attributed primarily to genes such as alkB and CYP153 (Van Beilen and Funhoff 2007). Furthermore, plant growth stimulating activities and production of phytohormones by microorganisms may stimulate plant growth and biomass production (Glick 2010).
Studies on using plant-bacteria combinations to remediate polluted terrestrial and aquatic environments are increasing (Afzal et al. 2011; Mahmood et al. 2016; Singh and Singh 2018). However, there is still little or no work on the hydrocarbon-degrading bacteria associated with wetland plants in CWs for remediating water contaminated with crude oil. In this study, different plant species were exposed to crude oil contaminated water and rhizo- and endophytic bacteria associated with best growing plants were isolated and characterized using 16 s RNA gene sequencing. Moreover, their hydrocarbon degradation and plant growth-promoting activities were determined.
Materials and Methods
Sixty CW microcosms were prepared and placed in the vicinity of the National Institute for Biotechnology and Genetic Engineering (NIBGE), Faisalabad (31°25′45″N 73°4′44″E), Pakistan, for 13 weeks. Each CW microcosm was prepared from a plastic tank (30 cm long, 26 cm wide, and 40 cm high) containing a tray with media to support growth of the plants (Supplementary Figs. 1 and 2). The supporting media tray was successively filled up from the bottom to the top with the following layers: 2.5 cm of coconut shavings, 10 cm of coarse gravel (ɸ 1–5 cm), 5 cm of fine sand (0.05–2 mm), and 5 cm of loamy soil (ɸ 1–2 mm). The tanks were then filled to a water depth of 30 cm with a mixture of tap water and crude oil (2% oil, w/v) or just tap water for the control tanks. The oil and water were mixed by hand with a glass stirring rod. The crude oil was obtained from an oil-exploring site located at Filmkasar, district Chakwal (33°4′33"N; 72°56′46"E), Pakistan. Seedlings or cuttings of ten plant species (Typha latifolia, Cyperus laevigatus, Urochloa mutica, L. fusca, Cannabis indica, Helianthus annuus, D. spicata, A. viridis, Phragmites australis and Cynodon dactylon) were collected from local drains and ponds, and planted in the trays. Each experiment was run in triplicate. The CW microcosms were loaded with a single exposure to the crude oil (2%, w/v) contaminated water. The plants in the CW microcosms were not fertilized and hence relied on nutrients available in the water and the tray media. Temperature and light were allowed to fluctuate with ambient conditions (1st May to 30th July 2016, Faisalabad, Pakistan) and the average day/night temperatures were 32°C/18°C, and humidity was 53%. When needed, the microcosms were covered with plastic sheet to avoid the addition of rain water in CWs. After 13 weeks, the plants were harvested, a water sample was taken from each microcosm after mixing the oil and water by hand with a glass stirring rod and kept at 4°C until further processing.
From the water samples taken from each microcosm after harvesting, the residual hydrocarbon concentration was determined as described in an earlier study (Afzal et al. 2011). To evaluate the effect of crude oil exposure on plant growth and development, the maximum shoot and root length as well as the total plant biomass of each plant were measured at harvest. The root and shoot lengths were measured using a millimeter scale, and the dry mass of the plants were determined after drying in an oven at 80°C till constant weight had been achieved. One-way analysis of variance (ANOVA) was used to determine the effect of crude oil exposure on the growth of the plants using SPSS software.
At harvest, subsamples of the shoots and roots with attached soil were collected and kept at 4°C until further processing. The hydrocarbon-degrading bacteria from the rhizosphere and endosphere of two of the wetland plant species, T. latifolia and C. laevigatus, were isolated as described in an earlier study (Fatima et al. 2015). The slurries of rhizosphere soil, roots and shoots were plated on a minimal medium containing 1% filtered-sterilized diesel. The plates were incubated at 30°C for 72 h. All the bacterial isolates were distinguished by restriction fragment-length polymorphism (RFLP) analysis as explained previously. On basis of the RFLP analysis, 33 isolates were distinguished and identified by 16S rRNA gene sequencing. A neighborhood-joining phylogenetic tree of the bacterial isolates based on 16S rRNA sequences was generated using the Kimura 2-parameter model in MEGA4. The alkane degradation genes (alkB and CYP153) were detected by PCR (Fatima et al. 2015). Different plant growth promoting (PGP) activities were determined using published protocols. The indole-3-acetic acid (IAA) production activity was determined using Salkowski reagent (Fatima et al. 2015).
Results and Discussion
The total plant biomass as well as the shoot and root length were significantly affected by exposure to crude oil (2%, w/v). The plants grown in CWs spiked with crude oil produced lower biomass and had shorter roots and shoots compared with plants in the control CWs (Fig. 1). High concentrations of hydrocarbons in soil and water can inhibit plant growth as a consequence of the toxic components of crude oil (Kirk et al. 2002; Tara et al. 2014). The very poor growth of A. viridis, C. dactylon, C. indica and H. annuus in the water spiked with crude oil indicates that these plant species are severely affected by crude oil and unlikely to be useful for this remediation purpose. T. latifolia, C. laevigatus, L. fusca, U. mutica and P. australis all grew well, indicating that these species are potential candidate species. However, T. latifolia and C. laevigatus exhibited the highest biomass production and the longest roots and shoots, suggesting that these species are the most suitable candidates among the tested plants. As all of the wetland plants in the CW microcosms were grown for an equal period of time (13 weeks), the plants that showed good growth and fast development in the crude oil-spiked water are recommended for further phytoremediation studies. A similar conclusion was drawn by Ying et al. (Ying et al. 2011), who concluded that plants with large root and shoot biomass possess high petroleum degradation capabilities because of the high biodegradation activities of microorganisms associated with the plants.
The residual amount of hydrocarbon in the water samples collected after 13 weeks showed that all the plant species had potential to remove hydrocarbons from water (Fig. 2). However, the different plant species exhibited different levels of oil removal from water. The hydrocarbon reduction percentage in the tanks with T. latifolia, C. laevigatus, L. fusca, U. mutica, P. australis, D. spicata, C. dactylon, A. viridis, C. indica and H. annuus were 53%, 50%, 48%, 34%, 31%, 23%, 22%, 20%, 20% and 15%, respectively. Hydrocarbon reduction was generally highest in tanks with plants having the highest biomass. Other investigations have presented similarly positive correlations between plant growth and hydrocarbons degradation (Afzal et al. 2011; Sung et al. 2013). Similar results were reported by Ying et al. (Ying et al. 2011), where larger shoot and root biomass stimulated the microbial degradation of hydrocarbons, and higher number of microbes were observed in the plant rhizosphere as compared with non-rhizosphere soil. In the present study, the maximum reduction in hydrocarbon (> 50%) was achieved in tanks planted with T. latifolia and the minimum in tanks planted with H. annuus. The differential ability of plant species to remediate the crude oil contaminated water can probably be attributed to the different numbers and varieties of hydrocarbon-degrading microbes in their rhizosphere and endosphere (Siciliano et al. 2001; Fatima et al. 2015). T. latifolia and C. laevigatus, were selected for the isolation of rhizospheric and endophytic bacteria in our study because of their healthy growth and tolerance to hydrocarbons. 33 strains of culturable bacteria were isolated from the rhizosphere and endosphere of T. latifolia and C. laevigatus, and identified by 16S rRNA gene sequencing (Table 1). The gene sequencing analysis revealed that the majority of the bacteria belonged to the genera Pseudomonas (36%), Acinetobacter (21%) and Bacillus (20%). Acinetobacter, Bacillus, Staphylococus and Pseudomonas were common in the rhizosphere; Pseudomonas and Aeromonas were dominant in the root interior; and Shewanella, Pseudomonas, and Bacillus were dominant in the shoot interior.
All of these genera have been reported to be involved in hydrocarbon degradation (Das and Chandran 2010; Fatima et al. 2015). A higher bacterial diversity was found in the rhizosphere and roots of C. laevigatus compared with those of T. latifolia. Colonization by bacteria was specific with respect to plant species and plant parts (rhizosphere, root and shoot interior) even when they were grown under the same conditions (Siciliano et al. 2001; Fatima et al. 2015).
Alkane hydroxylases, including the integral-membrane non-haem iron monooxygenase (AlkB) and the CYP153 family, are key enzymes in bacterial alkane oxidation. Although both genes have been detected in a number of bacteria and environments (Wang et al. 2010), the knowledge about the diversity of these genes in alkane-degrading bacteria present in crude oil contaminated CWs is almost negligible. In this study, the prevalence of four alkane monooxygenase genes (alkB, alkB1, alkB2 and CYP153) were determined in the isolated bacterial strains using oligonucleotide primers and DNA probes specific for each of the alk genotypes.
From the data (Supplementary Table 1), it is evident that only the Acinetobacter sp. strains (TYRH47, TYRH48, TYRH49, CYRI21, CYRI16, CYRI17 and CYRI19) had all four tested alkane hydroxylase genes. The Pseudomonas sp. strains CYRH25, TYRI40, and CYSI31 had three hydroxylase genes (alkB, alkB2 and CYP153), and the strains CRYI15, CYSI32, CYSI34, CYSI27, TYRH46, and TYRI39 had two hydroxylase genes from different combinations of alkB, alkB1, alkB2 and CYP153. Moreover, (A) salmonicida CYSI30 and (B) sufensis TYRH43 possessed only the alkB gene, and P. alcaliphila CYSI38 did not have any alkane hydroxylase genes. Lastly, PCR analysis revealed that 63% (21/33) of the isolates possessed the P450 gene encoding for cytochrome P450-type alkane hydroxylase (CYP153). Therefore, it can be inferred that specific strains of Acinetobacter and Pseudomonas species may have advantages for survival in crude oil contaminated soil due to the prevalence of multiple alkane hydroxylase genes. Bacteria with multiple alkane hydroxylase genes have been shown to have more potential to tolerate and utilize a wide range of n-alkanes (Van Beilen and Funhoff 2007). Moreover, among 369 alkB-containing and 87 CYP153-containing genomes, 73 and 32 genomes, respectively, were detected to have multiple copies of alkB and CYP153 homologous genes, thereby indicating good potential to degrade alkanes in different environments.
In the present study, we only identified culturable alkane degraders, but this group was likely restricted by the limitations of culturing techniques. As culturable bacteria often represent only a fraction of the total microbial community, it can be assumed that there may be other unculturable bacteria that contributed to the crude oil degradation.
Most of the isolated bacteria exhibited different PGP activities, such as production of IAA, 1-aminocyclopropane-1-carboxylic acid (ACC) deaminase, production of siderophores, zinc solubilization and inorganic phosphate solubilization (Supplementary Table 2). The majority of the bacterial strains produced IAA, with the exceptions of A. junii CYRI16, Acinetobacter sp. CYRI19, Bacillus sp. CYRH22, B. cereus CYRH23, Pseudomonas sp. CYSI34, S. putrefaciens CYSI35, Pseudomonas sp. TYRH40, S. warneri TYRI44, P. gessardii TYRH45 and B. pumilus. However, only a few strains (A. junii CYRI16 and TYRH47, and Bacillus sp. CYRH22) showed phosphate and zinc solubilization activities. Only A. junii TYRH47 had all the tested PGP traits. The most common PGP mechanism in the 33 isolated bacteria seems to be IAA production. Among the strains we isolated, about 70% (23/33) were IAA producers.
The PGP activities of the bacteria associated with T. latifolia and C. laevigatus can be inferred from the high biomass production found in these species when they were exposed to the stressful conditions of crude oil contamination. Several studies have shown that Acinetobacter and Pseudomonas species possess PGP traits and have significant effects on the biomass production of their host plants (Bhattacharyya and Jha 2012; Fatima et al. 2015).
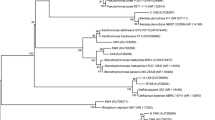
We show the neighborhood-joining phylogenetic tree of the isolates based on 16S rRNA sequences using the Kimura 2-parameter model in MEGA4 (Supplementary Fig. 3). The phylogenetic analysis (16S rRNA based) used Clostridium (NCBI acc # L23477.1) as an out-group. Also, the well-known alkane degrading bacteria Alcanivorax borkumensis was included because of its capability to proliferate on hydrocarbons (Schneiker et al. 2006).
In our phylogenetic tree, the largest group (I) comprised 12 sequences from Pseudomonas strains that were closely related (89%) to A. borkumensis and formed a highly concentrated clade (Supplementary Fig. 3). Group II contained four Acinetobacter strains all isolated from the rhizosphere, group III comprised one Pantoea sp. strain isolated from the root interior, and group IV was composed of one Shewanella strain from the root interior and three Aeromonas strains from the rhizosphere and the root interior. All of these four groups (I–IV) notably shared the nearest ancestral node with A. borkumensis (bootstrap value of 100%). Within these genera, strains of the same genus shared average nucleotide homology of more than 99%. The Bacillus and Staphylococcus strains (group V) were more distantly related to Alcanivorax species and were more divergent (nucleotide distance of 3.4%–3.5%) from the rest of the isolates. This is in contrast to bacterial strains in the two sub-clusters closely related to Alcanivorax, which were found either as endophytic or rhizospheric bacteria. The plant endosphere is a complex micro-ecosystem where different types of microorganisms can grow, and possibly the bacterial colonization of the roots or shoots of the two tested wetland species resulted in the excellent oil biodegradation capacities of these plant species. We conclude from the neighborhood joining phylogenetic analysis that the specific culturable isolates of Pseudomonas, Acinetobacter, Pantoea, Shewanella and Aeromonas species have the common ancestor of A. borkumensis. Similar conclusions have been drawn, where it was found that the most closely related organisms were Acinetobacter sp. and Pseudomonas aeruginosa. Also, no obvious horizontal gene transfer seems to have occurred (Reva et al. 2008).
Different wetland plant species displayed different growth and potential for the cleanup of crude oil contaminated water. A large number of hydrocarbon-mineralizing bacteria were isolated from the rhizosphere and endosphere of T. latifolia and C. laevigatus. These bacteria also possessed multiple PGP activities, a functional trait that deserves further study in CW systems. Most of the bacteria yielded 16S rRNA gene amplicons to the sequences assigned to the genera Alcanivorax (A. borkumensis species). This study provides important information for using local wetland plant species (T. latifolia and C. laevigatus) for plant–microbe (Acinetobacter and Pseudomonas species) joint remediation of crude oil contaminated water using CW technology. Furthermore, we concluded that the investigated wetland plants were also colonized by rhizospheric and endophytic bacteria possessing both alkane hydroxylase genes and plant growth-promoting features, and thus have clear potential to improve phytoremediation of crude oil contaminated water. However, further research is required to assess the role of plant-bacteria partnerships in crude oil degradation using CWs. Aspects like using full-scale wetlands, the intrinsic relationship between other wetland plants species and their associated bacteria, and identification of (unculturable) bacteria can be considered for future studies.
References
Afzal M, Yousaf S, Reichenauer TG, Kuffner M, Sessitsch A (2011) Soil type affects plant colonization, activity and catabolic gene expression of inoculated bacterial strains during phytoremediation of diesel. J Hazard Mater 186:1568–1575
Bhattacharyya PN, Jha DK (2012) Plant growth-promoting rhizobacteria (PGPR): emergence in agriculture. World J Microbiol Biotechnol 28:1327–1350
Brix H (1997) Do macrophytes play a role in constructed treatment wetlands? Water Sci Technol 35:11–17
Das N, Chandran P (2010) Microbial degradation of petroleum hydrocarbon contaminants: an overview. Biotechnol Res Int 2011:941810. https://doi.org/10.4061/2011/941810
Fatima K, Afzal M, Imran A, Khan QM (2015) Bacterial rhizosphere and endosphere populations associated with grasses and trees to be used for phytoremediation of crude oil contaminated soil. Bull Environ Contam Toxicol 94:314–320
Glick BR (2010) Using soil bacteria to facilitate phytoremediation. Biotechnol Adv 28:367–374
Khan S, Afzal M, Iqbal S, Khan QM (2013) Plant-bacteria partnerships for the remediation of hydrocarbon contaminated soils. Chemosphere 90:1317–1332
Kirk JL, Klirnomos JN, Lee H, Trevors JT (2002) Phytotoxicity assay to assess plant species for phytoremediation of petroleum-contaminated soil. Bioremediat J 6:57–63
Li R-L, Liu B-B, Zhu Y-X, Zhang Y (2016) Effects of flooding and aging on phytoremediation of typical polycyclic aromatic hydrocarbons in mangrove sediments by Kandelia obovata seedlings. Ecotoxicol Environ Saf 128:118–125
Liu X, Wang Z, Zhang X, Wang J, Xu G, Cao Z, Zhong C, Su P (2011) Degradation of diesel-originated pollutants in wetlands by Scirpus triqueter and microorganisms. Ecotoxicol Environ Saf 74:1967–1972
Mahmood T, Zhang J, Zhang G (2016) Assessment of constructed wetland in nutrient reduction, in the commercial scale experiment ponds of freshwater prawn Macrobrachium rosenbergii. Bull Environ Contam Toxicol 96:361–368
Reva ON, Hallin PF, Willenbrock H, Sicheritz-Ponten T, Tummler B, Ussery DW (2008) Global features of the Alcanivorax borkumensis SK2 genome. Environ Microbiol 10:614–625
Ruan X, Xue Y, Wu J, Ni L, Sun M, Zhang X (2006) Treatment of polluted river water using pilot-scale constructed wetlands. Bull Environ Contam Toxicol 76:90–97
Schneiker S, dos Santos VAPM, Bartels D, Bekel T, Brecht M, Buhrmester J, Chernikova TN, Denaro R, Ferrer M, Gertler C (2006) Genome sequence of the ubiquitous hydrocarbon-degrading marine bacterium Alcanivorax borkumensis. Nat Biotechnol 24:997–1004
Shehzadi M, Afzal M, Khan MU, Islam E, Mobin A, Anwar S, Khan QM (2014) Enhanced degradation of textile effluent in constructed wetland system using Typha domingensis and textile effluent-degrading endophytic bacteria. Water Res 58:152–159
Siciliano SD, Fortin N, Mihoc A, Wisse G, Labelle S, Beaumier D, Ouellette D, Roy R, Whyte LG, Banks MK (2001) Selection of specific endophytic bacterial genotypes by plants in response to soil contamination. Appl Environ Microbiol 67:2469–2475
Singh T, Singh DK (2018) Assessing the bacterial community structure in the rhizoplane of wetland plants. Bull Environ Contam Toxicol 101:521–526
Sung K, Kim KS, Park S (2013) Enhancing degradation of total petroleum hydrocarbons and uptake of heavy metals in a wetland microcosm planted with Phragmites communis by humic acids addition. Int J Phytorem 15:536–549
Tara N, Afzal M, Ansari TM, Tahseen R, Iqbal S, Khan QM (2014) Combined use of alkane-degrading and plant growth-promoting bacteria enhanced phytoremediation of diesel contaminated soil. Int J Phytorem 16:1268–1277
Van Beilen JB, Funhoff EG (2007) Alkane hydroxylases involved in microbial alkane degradation. Appl Microbiol Biotechnol 74:13–21
Wang L, Wang W, Lai Q, Shao Z (2010) Gene diversity of CYP153A and AlkB alkane hydroxylases in oil-degrading bacteria isolated from the Atlantic Ocean. Environ Microbiol 12:1230–1242
Weyens N, van der Lelie D, Taghavi S, Vangronsveld J (2009) Phytoremediation: plant-endophyte partnerships take the challenge. Curr Opin Biotechnol 20:248–254
Wu T, Hong B, Zhou S, Zhao J, Xia C, Liu H (2012) Residues of HCHs and DDTs in soils and sediments of preconstructing urban wetland. Bull Environ Contam Toxicol 89:563–567
Ying X, Dongmei G, Judong L, Zhenyu W (2011) Plant-microbe interactions to improve crude oil degradation. Energy Procedia 5:844–848
Acknowledgements
This research was supported by the Higher Education Commission, Pakistan (Grant No. 20-3854/R&D/HEC/14).
Author information
Authors and Affiliations
Corresponding author
Electronic Supplementary Material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Hashmat, A.J., Afzal, M., Fatima, K. et al. Characterization of Hydrocarbon-Degrading Bacteria in Constructed Wetland Microcosms Used to Treat Crude Oil Polluted Water. Bull Environ Contam Toxicol 102, 358–364 (2019). https://doi.org/10.1007/s00128-018-2518-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00128-018-2518-y