Abstract
Aims/hypothesis
The transcription factor, forkhead box (FOX)O1, is involved in fatty acid-induced apoptosis in pancreatic beta cells, but the precise mechanism is poorly understood. We aimed to identify which direct downstream targets of FOXO1 are involved in palmitate-induced apoptosis in the pancreatic beta cell line MIN6.
Methods
Chromatin immunoprecipitation (ChIP) coupled to a DNA selection and ligation technique (ChIP–DSL) was used to identify the direct targets of FOXO1. The mRNA level was examined by real-time PCR assay. The ChIP–DSL results were verified using ChIP-PCR and luciferase assay, respectively. The cell apoptosis rate was determined by TUNEL assay and by scoring cells with pycnotic nuclei.
Results
We identified 189 target genes and selected 106 targets for expression analysis in MIN6 cells treated with palmitate. The results showed that six genes were significantly upregulated and four were downregulated. Binding of FOXO1 to the promoters was determined by ChIP-PCR and confirmed by luciferase assay. Among the ten up- and downregulated genes, mRNA expression of A930038C07Rik was significantly decreased and that of Ppa1 was increased in 8-week-old db/db mice. The apoptosis assay showed that overproduction of the protein ‘RIKEN cDNA A930038C07’ (A930038C07Rik) drastically enhanced palmitate-induced apoptosis, while pyrophosphatase (inorganic) 1 (PPA1) partially protected the cells from apoptosis. Knockdown of PPA1, moreover, significantly increased apoptosis.
Conclusions/interpretation
We identified for the first time FOXO1 targets in MIN6 cells treated with palmitate, thus revealing the important roles of A930038C07Rik and PPA1 in palmitate-induced cell apoptosis. These results shed light on the mechanisms of palmitate-induced apoptosis in pancreatic beta cells.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Type 2 diabetes mellitus is characterised by peripheral insulin resistance and defects in insulin secretion [1, 2]. Obesity is a well-known diabetes risk factor, the main feature of which is elevated serum NEFA. There is evidence that excess circulating NEFA plays a key role in abnormal insulin secretion and reduced pancreatic beta cell mass. Chronic elevation of saturated NEFA such as palmitate promotes insulin resistance in target tissues and has detrimental effects on beta cells, resulting in reduced insulin content, abnormally elevated insulin release in the absence of stimuli, diminished capacity to secrete insulin in response to glucose and aggravated beta cell apoptosis [3–7]. Compared with palmitate, unsaturated fatty acids such as oleate are considered to be less cytotoxic and in some cases protective against palmitate toxicity [7–9]. Palmitate-induced apoptosis has been attributed to oxidative stress [10] and endoplasmic reticulum stress [11], as well as other pathways [12]. Nevertheless, the precise mechanisms are not fully understood.
Forkhead box (FOX)O1 is a multifunctional transcription factor that plays an important role in the regulation of cellular processes, including metabolism, cell differentiation, cell cycle arrest, DNA repair and other reactions to cellular stress [13, 14]. In pancreatic beta cells, FOXO1 haploinsufficiency restored beta cell mass and prevented diabetes in Irs2 knockout mice [15]. We also found that the activation of FOXO1 was integrally involved in prostaglandin E2 (PGE2) and dexamethasone (DEX)-induced pancreatic beta cell dysfunction [16–18]. Martinez et al reported that inhibition of FOXO1 reduced palmitate- and endoplasmic reticulum stress-induced apoptosis, and promoted beta cell survival [19], suggesting that FOXO1 may be a key regulator in palmitate-induced apoptosis. Consequently, the identification of specific and direct downstream targets of FOXO1 is crucial for an understanding of the mechanisms of palmitate-induced apoptosis in pancreatic beta cells.
At present, chromatin immunoprecipitation (ChIP) and ChIP-based approaches are widely used to identify DNA elements bound by proteins such as histones or transcription factors. The ChIP-based techniques used to identify the direct targets of a transcription factor include ChIP cloning, ChIP-paired end tag, a ChIP coupled to DNA selection and ligation technology (ChIP–DSL) [20, 21] and several variations of ChIP-on-chip [22]. Oh et al identified 103 direct targets of Caenorhabditis elegans abnormal DAuer formation family member (daf-16) (DAF-16) using ChIP cloning in Caenorhabditis elegans; they also found that 18 of the 33 targets selected by them were associated with life span, fat storage, and diapause [23]. In pancreatic beta cells, only a few direct targets of FOXO1 are known. In this study, we used ChIP–DSL to identify the downstream targets of FOXO1 that are involved in palmitate-induced apoptosis in the pancreatic beta cell line MIN6.
Methods
Reagents
See electronic supplementary material (ESM).
Cell culture
MIN6 cells were grown in DMEM containing 25 mmol/l glucose, 15% FBS (vol./vol.), 100 U/ml penicillin, 100 mg/ml streptomycin, 100 mg/ml l-glutamine and 5 μl/l beta-mercaptoethanol; culture was at 37°C in a humidified atmosphere containing 95% air and 5% CO2 [24]. MIN6 cells were used between passages 21 and 35.
Islet isolation
All animal studies were performed according to guidelines established by the Research Animal Care Committee of Nanjing Medical University, Nanjing, China. A mouse model of type 2 diabetes (db/db mice; BKS.Cg-m +/+ Leprdb/J) was used (Model Animal Research Center of Nanjing University, Nanjing, China). Islet isolation and culturing techniques have been described previously [25].
Treatment of MIN6 cells with palmitic acid
MIN6 cells were incubated for 24, 48 or 72 h in modified DMEM medium with control solution, or with 0.2, 0.3 or 0.4 mmol/l palmitate complexed with 0.5% BSA (wt/vol.). The 0.4 mmol/l fatty acid medium was prepared as previously described [26, 27] with slight modifications. Briefly, palmitate was dissolved in ethanol at a final concentration of 0.2 mol/l. Before treatment of MIN6 cells, an appropriate amount of palmitate was incubated for 1 to 2 h at 37°C, with 10% BSA (wt/vol.); an equal volume of ethanol was incubated with BSA as control. The final concentration of BSA in the medium was 0.5% (wt/vol.). The approximate molar ratio of fatty acids to BSA was 6:1 at 0.4 mmol/l palmitate.
ChIP-PCR assays
Four 10-cm plates of MIN6 cells were incubated with control solution or 0.4 mmol/l palmitate for 12 h and a ChIP assay performed using a kit (Upstate Biotechnology, Lake Placid, NY, USA) according to the manufacturer's protocol. Briefly, the cells were fixed with 1% formaldehyde (wt/vol.) for 10 min at room temperature. Fixation was terminated by adding glycine to 0.125 mol/l and washing twice in PBS. The cells were collected, washed twice with 10 ml cold 1× PBS and lysed with 400 μl SDS lysis buffer (1% SDS (wt/vol.), 10 mmol/l EDTA and 50 mmol/l TRIS–HCl [pH 8.1]) containing 1× protease inhibitor cocktail II (Sigma-Aldrich, St Louis, MO, USA). The lysate was sonicated (VCX130; Sonics & Materials, Newtown, CT, USA) at 40% amplitude (AMPL) and 4°C with eight cycles consisting of a 10 s on pulse and a 20 s off pulse. After centrifugation (10,000×g), the lysates were diluted with dilution buffer (1% Triton X-100 (vol./vol.), 2 mmol/l EDTA, 20 mmol/l TRIS–HCl [pH 8.1] and 150 mmol/l NaCl) and pre-cleared for 1 h at 4°C with 50 μl salmon sperm DNA/protein A-Sepharose. After removal of the Sepharose beads by centrifugation (500×g), the immunoprecipitate was incubated for 2 h at 4°C with anti-FOXO1 antiserum or an equal amount of normal rabbit IgG (Santa Cruz Biotechnology, Santa Cruz, CA, USA), after which the antibody–protein–DNA complexes were precipitated with salmon sperm DNA/protein A-Sepharose. After extensive washing with immunoprecipitation buffer, complexes were eluted from beads. Then, according to the manufacturer's instructions, samples were de-cross-linked at 65°C and treated with proteinase K. The DNA was recovered by phenol/chloroform extraction and ethanol precipitation, and the purified DNA was dissolved in 20 μl double distilled H2O. Then, 2 μl samples were subjected to PCR using the primers listed in ESM Table 1. The reaction conditions were standard with 32 cycles of amplification.
Promoter microarray experiments
The promoter microarray procedure for FOXO1 in MIN6 cells was performed by CapitalBio (Peking, China) using a ChIP–DSL system (M8K; Aviva Systems Biology, San Diego, CA, USA) according to the manufacturer's protocol.
Real-time PCR assay
MIN6 cells were treated with BSA and 0.4 mmol/l palmitate, or infected with adenovirus for 24 h, after which total RNA was extracted with a reagent (Trizol; Invitrogen, Grand Island, NY, USA) according to the manufacturer's protocol. For total RNA from islets, approximately 200 islets were used. mRNA quantification was performed as described previously [17]. The primers used in real-time PCR are listed in ESM Table 2.
Plasmid construction
Promoter fragments of about 1 to 1.5 kb length located before the transcription start sites of the genes A930038C07Rik, Ddx5, Galnt14, Gcdh, Mcm3ap, Pltp and Ppa1 were amplified using the primers listed in ESM Table 3 with appropriate restriction sites at the 5′ end. The PCR products were then respectively digested with MluI/XhoI (A930038C07Rik), XhoI/HindIII (Ddx5), BglII/HindIII (Galnt14) and XhoI/HindIII (Gcdh, Mcm3ap, Pltp and Ppa1), and cloned into the corresponding sites of the vector of pGL3-basic. The overproduction plasmids producing polypeptide N-acetylgalactosaminyltransferase 14 (GALNT14), TNF receptor-associated factor 5 (TRAF5), profilin 1 (PFN1), DEAD (Asp–Glu–Ala–Asp) box polypeptide 5 (DDX5), glutaryl-coenzyme A dehydrogenase (GCDH), RIKEN cDNA A930038C07 (A930038C07Rik) and pyrophosphatase (inorganic) 1 (PPA1) were obtained by cloning the open reading frame (ORF) fragments into pCMV5-MYC with ClaI/BamHI (GALNT14, TRAF5, PFN1, DDX5 and A930038C07Rik) and HindIII/XbaI (GCDH and PPA1), respectively. The overproduction plasmids with a green fluorescent protein (GFP) tag of A930038C07Rik and PPA1were constructed by cloning the same ORF fragments into pEGFP-N2 using XhoI/BamHI and BglII/SalI, respectively.
Luciferase assay
MIN6 cells were plated in 24-well plates 24 h before transfection. At 60% to 70% confluence, each well was transiently transfected with 200 ng pCMV5-MYC or pCMV5-FOXO1, 200 ng luciferase reporter plasmid and 100 ng plasmid expressing the gene that encodes β-galactosidase, as internal control. To do this, a transfection kit reagent (NanoJuice; Merck, Darmstadt, Germany) was used according to the manufacturer's instructions. At 24 h after transfection, cells were collected for the luciferase assay as described previously [25].
Apoptosis analysis
MIN6 cells were grown on glass coverslips in the wells of 24-well plates and incubated for 24, 48 or 72 h in serum-free medium with control, or with 0.2, 0.3 or 0.4 mmol/l palmitate complexed with BSA. Cells were then fixed and permeabilised, and the TUNEL assay performed according to the manufacturer's instructions (In Situ Cell Death Detection Kit; Roche, Basel, Switzerland) [28]. To assess the roles of A930038C07Rik and PPA1 in palmitate-induced apoptosis, the overproduction plasmids or corresponding vectors were transfected into MIN6 cells grown on glass coverslips in the wells of 24-well plates using a transfection kit (NanoJuice; Merck). At 24 h after transfection, the cells were treated with 0.4 mmol/l palmitate for another 72 h, and apoptosis was determined by TUNEL assay and by scoring cells displaying pycnotic nuclei (visualised by staining with the DNA-binding dye, Hoechst 33342 [Roche, Basel, Switzerland] [29]).
Preparation of and infection with adenoviruses
The adenoviruses for Foxo1-specific small interfering RNA (siRNA) (AD-siFoxo1) and for the constitutively active (CA)-Foxo1 were obtained from our laboratory stock. The sequence and preparations have been described in detail previously [16, 18]. MIN6 cells were inoculated on 3.5 cm plates 24 h before infection with adenovirus at a multiplicity of infection of 200. At 2 h after infection, the cells were cultured in fresh medium for another 48 h before collection for total RNA isolation.
Silencing of Ppa1 by RNA interference
The Ppa1-specific siRNA and control siRNA were designed and synthesised by Ribobio (Guangzhou, China). The sequences of the three designed siRNA fragments were as follows: siRNA1: 5′CAGACAAGGAUGUGUUCCA dTdT 3′; siRNA2: 5′GGACGCAGCC AA UUAUAAA dTdT 3′; siRNA3: 5′GGAAUCAGUUGCAUGAACA dTdT 3′. MIN6 cells were transfected with 100 nmol/l siRNA using a transfection kit reagent (NanoJuice; Merck). After 24 h, the cells were treated with 0.4 mmol/l palmitate for another 72 h and apoptosis was determined by scoring cells displaying pycnotic nuclei (visualised by staining with the DNA-binding dye, Hoechst 33342).
Immunoprecipitation and western blot
The 293 cells transfected with pCMV5-FOXO1 in three 6-cm dishes were lysed in 500 μl RIPA buffer (50 mmol/l TRIS–HCl [pH 7.5], 0.1% NP-40 [vol./vol.], 0.5% deoxycholate [wt/vol.], 10 mmol/l EDTA, 150 mmol/l NaCl, 50 mmol/l NaF, 1 mmol/l leupeptin and 0.1 mmol/l aprotinin). After centrifugation (10,000×g), cell lysates were incubated with 5 μl myc or FOXO1 antibody, and 5 μl prewashed agarose A/G beads. Immunoprecipitates were washed four times in lysis buffer and cleared of all supernatant fractions, after which 35 μl 2× SDS sample buffer was added. Samples were subjected to SDS-PAGE and transferred to polyvinyl difluoride (PVDF) (Perkin Elmer, Waltham, MA, USA). Western blot analysis was performed under standard conditions as described previously [18], using the indicated antibodies.
Statistical analysis
Comparisons between two groups were performed using Student's t test. Results are presented as means ± SEM. A value of p < 0.05 was considered statistically significant.
Results
Exposure to NEFA induced beta cell apoptosis in a both time- and dose-dependent manner
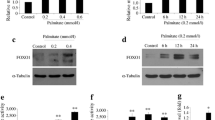
Prolonged exposure to NEFA has deleterious impacts on pancreatic beta cell function, including alterations in insulin secretion and sensitisation to apoptosis [3, 4]. We assessed apoptosis at 24, 48 and 72 h after treatment with 0.2, 0.3 or 0.4 mmol/l palmitate in serum-free medium. The TUNEL assay showed that prolonged incubation (72 h) with palmitate resulted in apoptosis in a significant number of cells with the proportion of apoptotic cells being dose-dependent (Fig. 1, ESM Fig. 1). Moreover, 0.4 mmol/l palmitate induced apoptosis in a significant number of cells at 24 h (p < 0.05), 48 h (p < 0.01) and 72 h (p < 0.01), with the proportion of apoptotic cells increasing with the incubation time. However, 0.2 mmol/l palmitate did not induce significant apoptosis until 72 h. Therefore, 0.4 mmol/l palmitate was used in the subsequent experiments and apoptosis detection was performed after 72 h incubation.
Palmitic acid treatment resulted in a dose- and time-dependent increase in apoptosis in MIN6 cells. Quantification of the percentage of TUNEL-positive MIN6 cells treated for times as shown without (control, white bars), or with 0.2 mmol/l (black bars), 0.3 mmol/l (dark grey bars) and 0.4 mmol/l (light grey bars) palmitate. Data shown are means ± SEM and representative of three separate experiments. *p < 0.05 and **p < 0.01 vs control
Identification of FOXO1 targets using ChIP–DSL
According to a previous report, fatty acid treatment resulted in an increase in FOXO1 activity; moreover, inhibition of FOXO1 reduced palmitate-induced apoptosis in pancreatic beta cells [19]. However, it is still not known which downstream targets of FOXO1 are involved in the signalling pathway of apoptosis. To identify the downstream target genes of FOXO1 in MIN6 cells treated with palmitate, we performed promoter microarray analysis of FOXO1 by the ChIP–DSL method, using a specific FOXO1 antibody (ESM Fig. 2a). V-maf musculoaponeurotic fibrosarcoma oncogene family, protein A (avian) (MAFA), a known direct target of FOXO1 [30], was used to determine the efficiency of the antibody (ESM Fig. 2b). The microarray analysis revealed 189 genes that had an antibody/IgG ratio of >2.0 and may be directly regulated by FOXO1. These genes represent various biological processes, such as metabolism, growth, transport, protein biosynthesis and proteolysis, transcription, apoptosis, signal transduction, RNA splicing, and DNA replication and repair (Fig. 2, ESM Table 4). Several of these genes have been previously suggested to be FOXO1 targets, e.g. the DNA-damage inducible transcript 3 gene, Ddit3 [31], the cyclin-dependent kinase inhibitor 1B (P27) gene, p27 (also known as Cdkn1b) [32], and the Mus musculus inhibitor of kappa light polypeptide enhancer in B cells gene, Ikbkap [33]. Notably, we identified genes with functions that are directly related to the insulin signalling pathway, including: (1) Tbc1d4, which encodes the Rab GTPase-activating protein AS160, a regulator of insulin-stimulated glucose transport in skeletal muscles [34]; and (2) the gene encoding the calcium/calmodulin-dependent protein serine kinase (CASK), which plays a role in connecting the CASK-mediated pathways to the insulin signalling pathways [35]. In addition, we found that Mttp, which encodes the microsomal triacylglycerol transfer protein MTP, is a target of FOXO1. Interestingly, the excessive VLDL production associated with insulin resistance was caused by the inability of insulin to regulate FOXO1, which activates MTP at the transcriptional level [36]. Thus, the ChIP–DSL microarray identified several putative and relevant direct downstream targets of FOXO1.
Effect of NEFA on the expression of FOXO1 target genes both in vitro and in vivo
Microarray analysis showed that 106 of the downstream target genes of FOXO1 were annotated with the basic function and had an antibody/IgG ratio of >3.0. The expression of these genes was examined in MIN6 cells after incubation with 0.4 mmol/l palmitate for 24 h. Of these 106 genes, 86 were successfully amplified and ten of them were found to have altered transcription levels, with six being upregulated and four downregulated. The upregulated genes included A930038C07Rik, Mapt, Pltp, Mcm3ap, Pfn1 and Ppa1; the downregulated genes included Galnt14, Gcdh, Traf5 and Ddx5 (Fig. 3a).
The expression of the downstream targets of FOXO1 was determined in MIN6 cells treated with palmitate and confirmed in db/db mice. (a) MIN6 cells were treated without (control, white bars) or with 0.4 mmol/l palmitate (black bars) for 24 h and the expression of 106 of the 189 FOXO1 target genes was examined by quantitative PCR. Values here are for ten of the above genes. (b) The expression of nine of the ten genes in primary pancreatic islets from db/db mice. Approximately 200 islets were isolated from 4-week-old (white bars), 8-week-old (grey bars) and 12-week-old (black bars) db/db mice and the corresponding normal control mice, respectively. Expression of nine of the ten genes was examined by quantitative PCR. Values (a, b) are means ± SEM and representative of three (a) or more than three (b) separate experiments; β-actin was used as internal control; *p < 0.05 and **p < 0.01 vs control or normal control mice. A930, A930038C07Rik
It has been reported that FOXO1 inhibitors drastically decreased plasma glucose levels in db/db mice [37, 38] and suggested that FOXO1 might be a potential target for treatment of type 2 diabetes mellitus. However, the expression of the above-mentioned targets of FOXO1 in pancreatic islets of db/db mice is poorly understood. Here, the expression of nine of the ten genes was examined in primary islets from 4-, 8- and 12-week-old db/db mice using real-time PCR. The results showed that Pltp expression gradually increased with age. Interestingly, expression of A930038C07Rik and Mapt was significantly decreased, whereas that of Ppa1 was increased in 8-week-old mice (Fig. 3b). On the other hand, the transcription levels of Gcdh and Galnt14 were clearly increased in 4-week-old mice, but reverted to normal in 12-week-old mice (Fig. 3b). However, in contrast to findings in MIN6 cells treated with palmitate, the expression of Mcm3ap, Traf5 and Ddx5 in db/db mice was not significantly altered. These results show that the transcription of FOXO1 target genes, including Pltp, A930038C07Rik, Mapt, Gcdh, Galnt14 and Ppa1, was changed in db/db mice.
Specific binding of FOXO1 to and regulation effect of FOXO1 on its target genes
Next, the specificity of FOXO1 binding to A930038C07Rik, Mapt, Pltp, Mcm3ap, Pfn1, Ppa1, Galnt14, Gcdh, Traf5 and Ddx5 was determined by ChIP-PCR. We found that eight of the ten genes were directly bound to FOXO1 (Fig. 4a) and that the promoter regions of Traf5 and Pfn1 could not be successfully amplified. Notably, binding of FOXO1 to Galnt14, Mapt and Ppa1 was not observed in the control group at our detection limit, and specific binding occurred only after incubation with palmitate (Fig. 4a). To further demonstrate that FOXO1 does indeed regulate its binding targets, luciferase reporter plasmids of Ddx5, Ganlt14, Gcdh, Pltp, Mcm3ap, Ppa1 and A930038C07Rik were successfully constructed and co-transfected with pCMV5-MYC or pCMV5-FOXO1 into MIN6 cells. The plasmid p6xDBE-luc containing six copies of the daf-16 family protein-binding element (DBE) was used as a positive control. The results showed that as the FOXO1 content increased (Fig. 4b), the luciferase activity of A930038C07Rik (1.45 ± 0.05), Ppa1 (1.46 ± 0.06), Pltp (1.88 ± 0.13) and Mcm3ap (2.12 ± 0.25) was significantly increased, and that of Ddx5 (0.73 ± 0.08), Ganlt14 (0.69 ± 0.10) and Gcdh (0.75 ± 0.11) decreased (Fig. 4e).
The binding of FOXO1 to and its regulation of target genes were verified through ChIP-PCR, luciferase assay and real-time PCR in MIN6 cells. (a) ChIP-PCR determined FOXO1 binding to the promoter elements of eight of ten genes whose expression significantly changed after incubation with palmitate. Input chromatin from control and immunoprecipitated DNA was PCR-amplified using primers specific to suspected FOXO1 target regions (ESM Table 1). Sequence enrichment in immunoprecipitated DNA from antibody (Ab, anti-FOXO1 serum) vs IgG chromatin indicated FOXO1 binding within the genomic region. Each experiment was repeated at least three times. Results shown were reproducible for all targets. (b) FOXO1 protein levels and fold changes were determined in the cells co-transfected with pCMV5-FOXO1, or infected with (c) AD-siFoxo1 and (d) (CA)-Foxo1 ADA. (e) MIN6 cells were plated in 24-well plates 24 h before co-transfection with pCMV5-MYC or pCMV5-FOXO1, luciferase reporter plasmids and the plasmid expressing the gene encoding β-galactosidase (internal control). After 24 h, cells were collected for luciferase reporter assays. The luciferase fold change is given as the ratio of the relative luciferase activity obtained from co-transfection with pCMV-FOXO1 vs that obtained with pCMV-MYC. The plasmid containing six copies of DBE p6xDBE-luc was used as positive control. Values are representative of more than three separate experiments. (f) The mRNA expression of the genes shown was determined after infection with AD-siFoxo1 and (g) (CA)-Foxo1 ADA adenovirus for 24 h using quantitative PCR. Values (b–d, f–g) are represented as means ± SEM of three or (e) more than three separate experiments; *p < 0.05 and **p < 0.01 compared with corresponding controls co-transfected with pCMV-MYC (e) or (f, g) infected with control adenovirus. A930, A930038C07Rik
To further confirm that FOXO1 regulates these targets, AD-siFoxo1 and (CA)-Foxo1 ADA were used. When infected with Ad-siFoxo1 (Fig. 4c), mRNA expression of A930038C07Rik (0.55 ± 0.14), Pltp (0.8 ± 0.02), Mcm3ap (0.66 ± 0.16) and Ppa1 (0.79 ± 0.04) was decreased significantly, whereas that of Ddx5 (1.64 ± 0.22), Ganlt14 (1.80 ± 0.25) and Gcdh (1.61 ± 0.21) displayed a pronounced increase (Fig. 4f). Correspondingly, infection with (CA)-Foxo1 ADA (Fig. 4d) significantly increased the mRNA expression of A930038C07Rik (1.65 ± 0.11), Pltp (1.32 ± 0.07), Mcm3ap (1.76 ± 0.14) and Ppa1 (1.81 ± 0.21), while, notably, that of Ddx5 (0.55 ± 0.11), Ganlt14 (0.80 ± 0.08) and Gcdh (0.74 ± 0.08) was decreased (Fig. 4g). All of these events demonstrate that FOXO1 binds to and regulates the target genes A930038C07Rik, Pltp, Mcm3ap, Ppa1, Ddx5, Ganlt14 and Gcdh.
Effect of A930038C07Rik and PPA1 on pancreatic beta cell apoptosis
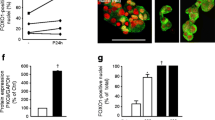
FOXO1 is known to regulate palmitate-induced apoptosis in pancreatic beta cells [19], prompting us to search for the role played by FOXO1 target genes in the process of palmitate-associated apoptosis. We therefore constructed plasmids overproducing GALNT14, TRAF5, PFN1, DDX5, GCDH, A930038C07Rik and PPA1, and examined their effect on palmitate-induced apoptosis in MIN6 cells. The apoptosis assay showed that overproduction of A930038C07Rik drastically enhanced apoptosis and that the proportion of apoptotic cells increased 2.9-fold. Overproduction of PPA1 reduced the proportion of apoptotic cells to 67% and partially protected cells against palmitate-induced apoptosis (Fig. 5a). Overproduction of FOXO1 also caused a 1.5-fold increase in the proportion of apoptotic cells (Fig. 5a). However, overproduction of GALNT14, TRAF5, PFN1, DDX5 and GCDH had no significant effect on palmitate-induced apoptosis (data not shown). To rule out any effect of the transfection rate on the apoptosis assay, the overproduction plasmids of A930038C07Rik and PPA1 were fused with a GFP tag and apoptosis determined by scoring cells that simultaneously displayed pycnotic nuclei and green fluorescence. Surprisingly, after treatment with palmitate, the apoptosis rate reached about 90% in MIN6 cells transfected with A930038C07Rik-GFP (Fig. 5b). However, when we tried to confirm apoptosis after silencing of A930038C07Rik, we were unable to obtain effective silencing of A930038C07Rik, even though several pairs of siRNA fragments for A930038C07Rik were synthesised (data not shown). On the other hand, as overproduction of PPA1 should partially protect MIN6 cells from palmitate-induced apoptosis (Fig. 5a, b), we further determined apoptosis in MIN6 cells with silencing of Ppa1 (Fig. 6a, b). Our results showed that interference of PPA1 production significantly increased palmitate-induced apoptosis in MIN6 cells (Fig. 6c). Surprisingly, the silencing of Ppa1 resulted in obvious apoptosis without palmitate treatment, suggesting an essential role of PPA1 in MIN6 cell survival.
Effect of A930038C07Rik and PPA1 on palmitate-induced apoptosis in MIN6 cells. MIN6 cells were transiently transfected with the pCMV5-MYC vector, pCMV5-FOXO1, and the A930038C07Rik and PPA1 overproduction plasmids, or pEGFP-N2, A930-GFP and PPA1-GFP. After 24 h, cells were incubated with 0.4 mmol/l palmitate for another 72 h. (a) The number of TUNEL-positive cells was determined and divided by the total number of cells stained by Hoechst. About 2,000 cells were scored for each group in one experiment. (b) The cells that displayed pycnotic nuclei and were simultaneously GFP-positive were counted, and about 200 cells were scored in one experiment. Values are means ± SEM and representative of three separate experiments; *p < 0.05 and **p < 0.01 vs the corresponding control
Silencing of Ppa1 significantly increased palmitate-induced apoptosis. (a) The 293 cells were co-transfected with overproduction plasmid PPA1-GFP and Ppa1 siRNAs (siPpa1) or control siRNAs (siNC) respectively. After 24 h, green fluorescence was examined using fluorescence microscopy. Images are representative of three independent experiments. (b) MIN6 cells were transiently transfected without (white bars) or with siRNAs (black bars). After 24 h, mRNA expression of Ppa1 was determined by quantitative PCR. (c) MIN6 cells were transiently transfected with siNC (white bars) or si-Ppa1-2 (black bars) for 24 h and then incubated with 0.4 mmol/l palmitate for another 72 h, followed by staining with Hoechst. Apoptosis was determined by scoring cells displaying pycnotic nuclei. About 2,000 cells were scored for each group in one experiment. Values are means ± SEM and representative of three separate experiments; *p < 0.05 and **p < 0.01 vs (b) the corresponding control or (c) compared with controls transfected with siNC
Discussion
In the present study, we identified for the first time 189 direct targets of the transcription factor, FOXO1, in the pancreatic beta cell line MIN6 using ChIP–DSL technology. The transcriptional levels of 106 targets were examined after incubation with palmitate for 24 h and ten of these targets were significantly altered. Of these ten genes, six were significantly upregulated, while four were downregulated. The ChIP-PCR assay revealed that eight of the ten targets were directly bound to FOXO1. The expression of nine of the ten genes was further examined in primary islets from 4-, 8- and 12-week-old db/db mice; the results showed that: (1) Pltp was gradually upregulated with age; (2) A930038C07Rik and Mapt were significantly downregulated; and (3) Ppa1 was upregulated in 8-week-old mice. TUNEL and pycnotic nuclei assays revealed that the overproduction of A930038C07Rik significantly enhanced palmitate-induced apoptosis in MIN6 cells and that PPA1 partially protected cells from apoptosis.
In pancreatic beta cells, the roles of the transcription factor, FOXO1, have something of a double-edged sword. On the one hand, FOXO1 is integrally involved in PGE2- [16, 17] and DEX-induced [18] islet dysfunction, and palmitate-induced apoptosis [19], and also was able to inhibit beta cell proliferation via suppression of Pdx1 transcription [15]. On the other hand, FOXO1 protects beta cells from oxidative stress by forming a complex with the promyelocytic leukaemia protein (PML) and the NAD-dependent deacetylase sirtuin-1 isoform 2 (SIRT1) to activate production of neurogenic differentiation factor 1 (NEUROD) and MAFA [30]. In this study, we obtained a promoter microarray of FOXO1 using ChIP–DSL technology. The FOXO1 target genes are involved in various biological processes such as metabolism, growth, transport, protein biosynthesis and proteolysis, transcription, apoptosis, signal transduction, RNA splicing, and DNA replication and repair. The identification of FOXO1 targets will help understand the detailed functions of FOXO1 in different signalling pathways.
ChIP coupled with microarray is a powerful tool to determine the in vivo binding profile of transcription factors and thus deduce regulatory circuits in mammalian cells. In contrast to conventional ChIP-on-chip, the ChIP–DSL method involves the use of DSL, permitting robust analysis with considerably fewer materials, and higher specificity and sensitivity [20]. The chip analysis identified several genes that have previously been suggested or found to be FOXO1 targets, e.g. Ddit3 [31], Mttp [36], Ikbkap [33], p27 [32] etc. This suggests that the microarray analysis identified putative and relevant direct downstream targets of FOXO1. Moreover, the ChIP-PCR results revealed that eight of the ten genes were directly bound to FOXO1, confirming the credibility and specificity of the microarray analysis results. The luciferase assay also verified the regulatory effect of FOXO1 on its target genes.
In primary islets from 4-, 8- and 12-week-old diabetic db/db mice, the expression of Pltp gradually increased with age. Plasma phospholipid transfer protein (PLTP) is a lipid transfer glycoprotein that transfers and binds to a number of amphipathic compounds. The amount and activity of PLTP have been reported to be abnormally elevated in type 2 diabetes mellitus and insulin-resistant conditions, a development frequently associated with hypertriacylglycerolaemia and obesity [39]. In this study, we report for the first time that FOXO1 binds to the promoter element of Pltp, but the roles of FOXO1 in PLTP activity remain to be studied. The expression of A930038C07Rik was significantly decreased in 8-week-old mice and increased in 12-week-old mice, while Ppa1 was significantly increased in 8-week-old mice and returned to normal levels in 12-week-old mice. A930038C07Rik encodes a hypothetical protein that has been reported to be a downstream target of T-cell leukaemia homeobox protein 1 (HOX11) function in the developing kidney [40]. The apoptosis assay in MIN6 cells revealed that overproduction of A930038C07Rik drastically enhanced palmitate-induced apoptosis. However, we were unable to obtain effective siRNA fragments for A930038C07Rik and the lack of a commercial antibody for A930038C07Rik also created difficulties during further analysis. PPA1, a cytoplasmic pyrophosphatase, was found to be required for cell growth in yeast [41] and was reported to be essential for life through provision of a thermodynamic pull for biosynthetic reactions [42]. In this study, we found that overproduction of PPA1 partially reduced palmitate-induced cell apoptosis, while silencing of Ppa1 significantly increased MIN6 cell apoptosis in the presence or absence of palmitate, thus indicating the important role of PPA1 in cell survival. PPA1 is an essential enzyme for controlling cellular levels of inorganic pyrophosphate (PPi) [43], and silencing of Ppa1 might result in increased PPi in cells. Indeed, abnormal cellular PPi production has been reported to be associated with disease [44]; however, the question of whether increased PPi is responsible for MIN6 cell apoptosis has yet to be answered. Our study is the first to report on the functions of A930038C07Rik and PPA1 in pancreatic beta cell apoptosis.
In summary, we have, for the first time, successfully identified the direct targets of FOXO1 in pancreatic MIN6 cells treated with palmitate. Further comprehensive sorting and characterisation of these downstream targets will lead to a better understanding of the contribution of FOXO1 signalling pathways to the proliferation and apoptosis of pancreatic beta cells in the development of type 2 diabetes mellitus. Our findings also suggest that ten target genes of FOXO1 are involved in the signalling pathway of palmitate-induced apoptosis. In addition, the roles of A930038C07Rik and PPA1 in beta cell apoptosis have been identified for the first time. Overall, our results provide a useful insight into the mechanism of palmitate-induced apoptosis in pancreatic beta cells.
Abbreviations
- A930038C07Rik:
-
RIKEN cDNA A930038C07
- AD-siFoxO1:
-
Adenovirus for FOXO1-specific small interfering RNA
- CASK:
-
Calcium/calmodulin-dependent serine protein kinase
- ChIP:
-
Chromatin immunoprecipitation
- ChIP–DSL:
-
ChIP coupled to a DNA selection and ligation technology
- DBE:
-
Daf-16 family protein-binding element
- DDX5:
-
DEAD (Asp–Glu–Ala–Asp) box polypeptide 5
- DEX:
-
Dexamethasone
- DSL:
-
DNA selection and ligation
- FOX:
-
Forkhead box
- GALNT14:
-
Polypeptide N-acetylgalactosaminyltransferase 14
- GCDH:
-
Glutaryl-coenzyme A dehydrogenase
- GFP:
-
Green fluorescent protein
- MAFA:
-
V-maf musculoaponeurotic fibrosarcoma oncogene family protein A (avian)
- ORF:
-
Open reading frame
- PFN1:
-
Profilin 1
- PGE2:
-
Prostaglandin E2
- PLTP:
-
Phospholipid transfer protein
- PPA1:
-
Pyrophosphatase (inorganic) 1
- PPi:
-
Inorganic pyrophosphate
- siRNA:
-
Small interfering RNA
- TRAF5:
-
TNF receptor-associated factor 5
References
Fernandez ML (2007) The metabolic syndrome. Nutr Rev 65:S30–S34
Weyer C, Bogardus C, Mott DM, Pratley RE (1999) The natural history of insulin secretory dysfunction and insulin resistance in the pathogenesis of type 2 diabetes mellitus. J Clin Invest 104:787–794
Prentki M, Nolan CJ (2006) Islet beta cell failure in type 2 diabetes. J Clin Invest 116:1802–1812
Newsholme P, Keane D, Welters HJ, Morgan NG (2007) Life and death decisions of the pancreatic beta-cell: the role of fatty acids. Clin Sci (Lond) 112:27–42
Shimabukuro M, Zhou YT, Levi M, Unger RH (1998) Fatty acid-induced beta cell apoptosis: a link between obesity and diabetes. Proc Natl Acad Sci U S A 95:2498–2502
Lupi R, Dotta F, Marselli L et al (2002) Prolonged exposure to free fatty acids has cytostatic and pro-apoptotic effects on human pancreatic islets: evidence that beta-cell death is caspase mediated, partially dependent on ceramide pathway, and Bcl-2 regulated. Diabetes 51:1437–1442
Karaskov E, Scott C, Zhang L, Teodoro T, Ravazzola M, Volchuk A (2006) Chronic palmitate but not oleate exposure induces endoplasmic reticulum stress, which may contribute to INS-1 pancreatic beta-cell apoptosis. Endocrinology 147:3398–3407
Maedler K, Spinas GA, Dyntar D, Moritz W, Kaiser N, Donath MY (2001) Distinct effects of saturated and monounsaturated fatty acids on beta-cell turnover and function. Diabetes 50:69–76
Maedler K, Oberholzer J, Bucher P, Spinas GA, Donath MY (2003) Monounsaturated fatty acids prevent the deleterious effects of palmitate and high glucose on human pancreatic beta-cell turnover and function. Diabetes 52:726–733
Morgan D, Oliveira-Emilio HR, Keane D et al (2007) Glucose, palmitate and pro-inflammatory cytokines modulate production and activity of a phagocyte-like NADPH oxidase in rat pancreatic islets and a clonal beta cell line. Diabetologia 50:359–369
Kharroubi I, Ladriere L, Cardozo AK, Dogusan Z, Cnop M, Eizirik DL (2004) Free fatty acids and cytokines induce pancreatic beta-cell apoptosis by different mechanisms: role of nuclear factor-kappaB and endoplasmic reticulum stress. Endocrinology 145:5087–5096
Cnop M, Welsh N, Jonas JC, Jorns A, Lenzen S, Eizirik DL (2005) Mechanisms of pancreatic beta-cell death in type 1 and type 2 diabetes: many differences, few similarities. Diabetes 54(Suppl 2):S97–S107
Glauser DA, Schlegel W (2007) The emerging role of FOXO transcription factors in pancreatic beta cells. J Endocrinol 193:195–207
Gross DN, van den Heuvel AP, Birnbaum MJ (2008) The role of FoxO in the regulation of metabolism. Oncogene 27:2320–2336
Kitamura T, Nakae J, Kitamura Y et al (2002) The forkhead transcription factor Foxo1 links insulin signaling to Pdx1 regulation of pancreatic beta cell growth. J Clin Invest 110:1839–1847
Meng Z, Lv J, Luo Y et al (2009) Forkhead box O1/pancreatic and duodenal homeobox 1 intracellular translocation is regulated by c-Jun N-terminal kinase and involved in prostaglandin E2-induced pancreatic beta-cell dysfunction. Endocrinology 150:5284–5293
Meng ZX, Sun JX, Ling JJ et al (2006) Prostaglandin E2 regulates Foxo activity via the Akt pathway: implications for pancreatic islet beta cell dysfunction. Diabetologia 49:2959–2968
Zhang X, Yong W, Lv J et al (2009) Inhibition of forkhead box O1 protects pancreatic beta-cells against dexamethasone-induced dysfunction. Endocrinology 150:4065–4073
Martinez SC, Tanabe K, Cras-Meneur C, Abumrad NA, Bernal-Mizrachi E, Permutt MA (2008) Inhibition of Foxo1 protects pancreatic islet beta-cells against fatty acid and endoplasmic reticulum stress-induced apoptosis. Diabetes 57:846–859
Kwon YS, Garcia-Bassets I, Hutt KR et al (2007) Sensitive ChIP–DSL technology reveals an extensive estrogen receptor alpha-binding program on human gene promoters. Proc Natl Acad Sci U S A 104:4852–4857
Garcia-Bassets I, Kwon YS, Telese F et al (2007) Histone methylation-dependent mechanisms impose ligand dependency for gene activation by nuclear receptors. Cell 128:505–518
Deblois G, Giguere V (2008) Nuclear receptor location analyses in mammalian genomes: from gene regulation to regulatory networks. Mol Endocrinol 22:1999–2011
Oh SW, Mukhopadhyay A, Dixit BL, Raha T, Green MR, Tissenbaum HA (2006) Identification of direct DAF-16 targets controlling longevity, metabolism and diapause by chromatin immunoprecipitation. Nat Genet 38:251–257
Miyazaki J, Araki K, Yamato E et al (1990) Establishment of a pancreatic beta cell line that retains glucose-inducible insulin secretion: special reference to expression of glucose transporter isoforms. Endocrinology 127:126–132
Han X, Sun Y, Scott S, Bleich D (2001) Tissue inhibitor of metalloproteinase-1 prevents cytokine-mediated dysfunction and cytotoxicity in pancreatic islets and beta-cells. Diabetes 50:1047–1055
Chavez JA, Knotts TA, Wang LP et al (2003) A role for ceramide, but not diacylglycerol, in the antagonism of insulin signal transduction by saturated fatty acids. J Biol Chem 278:10297–10303
Chavez JA, Summers SA (2003) Characterizing the effects of saturated fatty acids on insulin signaling and ceramide and diacylglycerol accumulation in 3T3-L1 adipocytes and C2C12 myotubes. Arch Biochem Biophys 419:101–109
Gavrieli Y, Sherman Y, Ben-Sasson SA (1992) Identification of programmed cell death in situ via specific labeling of nuclear DNA fragmentation. J Cell Biol 119:493–501
Roehrich ME, Mooser V, Lenain V et al (2003) Insulin-secreting beta-cell dysfunction induced by human lipoproteins. J Biol Chem 278:18368–18375
Kitamura YI, Kitamura T, Kruse JP et al (2005) FoxO1 protects against pancreatic beta cell failure through NeuroD and MafA induction. Cell Metab 2:153–163
Martinez SC, Cras-Meneur C, Bernal-Mizrachi E, Permutt MA (2006) Glucose regulates Foxo1 through insulin receptor signaling in the pancreatic islet beta-cell. Diabetes 55:1581–1591
Dijkers PF, Medema RH, Pals C et al (2000) Forkhead transcription factor FKHR-L1 modulates cytokine-dependent transcriptional regulation of p27(KIP1). Mol Cell Biol 20:9138–9148
Senokuchi T, Liang CP, Seimon TA et al (2008) Forkhead transcription factors (FoxOs) promote apoptosis of insulin-resistant macrophages during cholesterol-induced endoplasmic reticulum stress. Diabetes 57:2967–2976
Cartee GD, Wojtaszewski JF (2007) Role of Akt substrate of 160 kDa in insulin-stimulated and contraction-stimulated glucose transport. Appl Physiol Nutr Metab 32:557–566
Wang Y, Li R, Du D et al (2006) Proteomic analysis reveals novel molecules involved in insulin signaling pathway. J Proteome Res 5:846–855
Kamagate A, Qu S, Perdomo G et al (2008) FoxO1 mediates insulin-dependent regulation of hepatic VLDL production in mice. J Clin Invest 118:2347–2364
Tanaka H, Nagashima T, Shimaya A, Urano Y, Shimokawa T, Shibasaki M (2010) Effects of the novel Foxo1 inhibitor AS1708727 on plasma glucose and triglyceride levels in diabetic db/db mice. Eur J Pharmacol 645:185–191
Nagashima T, Shigematsu N, Maruki R et al (2010) Discovery of novel forkhead box O1 inhibitors for treating type 2 diabetes: improvement of fasting glycemia in diabetic db/db mice. Mol Pharmacol 78:961–970
Tzotzas T, Desrumaux C, Lagrost L (2009) Plasma phospholipid transfer protein (PLTP): review of an emerging cardiometabolic risk factor. Obes Rev 10:403–411
Schwab K, Hartman HA, Liang HC, Aronow BJ, Patterson LT, Potter SS (2006) Comprehensive microarray analysis of Hoxa11/Hoxd11 mutant kidney development. Dev Biol 293:540–554
Kinoshita N, Yamano H, Niwa H, Yoshida T, Yanagida M (1993) Negative regulation of mitosis by the fission yeast protein phosphatase pp a2. Genes Dev 7:1059–1071
Sivula T, Salminen A, Parfenyev AN et al (1999) Evolutionary aspects of inorganic pyrophosphatase. FEBS Lett 454:75–80
Cooperman BS, Baykov AA, Lahti R (1992) Evolutionary conservation of the active site of soluble inorganic pyrophosphatase. Trends Biochem Sci 17:262–266
Terkeltaub RA (2001) Inorganic pyrophosphate generation and disposition in pathophysiology. Am J Physiol Cell Physiol 281:C1–C11
Funding
This work was supported by grants from: (1) the Special Funds for Major State Basic Research Program of China (973 Program, 2011CB504003) and the State Key Program of National Natural Science of China (81130060) to X. Han; (2) the National Natural Science Foundation of China (30900542) and China Postdoctoral Science Foundation (200902527 and 20080441045) to H.Y. Lin; and (3) a project funded by the Priority Academic Program Development of Jiangsu Higher Education Institutions.
Duality of interest
The authors declare that there is no duality of interest associated with this manuscript.
Contribution statement
All authors took part in the conception and design of the study, as well as either drafting or critically revising the manuscript. All authors have approved the final version of the manuscript.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
ESM text
(PDF 10 kb)
ESM Table 1
(PDF 11 kb)
ESM Table 2
(PDF 56 kb)
ESM Table 3
(PDF 32 kb)
ESM Table 4
(PDF 90 kb)
ESM Fig. 1
(PDF 632 kb)
ESM Fig. 2
(PDF 151 kb)
Rights and permissions
About this article
Cite this article
Lin, H.Y., Yin, Y., Zhang, J.X. et al. Identification of direct forkhead box O1 targets involved in palmitate-induced apoptosis in clonal insulin-secreting cells using chromatin immunoprecipitation coupled to DNA selection and ligation. Diabetologia 55, 2703–2712 (2012). https://doi.org/10.1007/s00125-012-2643-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00125-012-2643-9