Abstract
Key message
Both SH and BHA weedy rice genotypes evolved independently and have distinct genomic composition. Different genetic mechanisms may be responsible for their competitiveness and adaptation to diverse environmental conditions.
Abstract
Two major types of weedy rice are recognized in the USA based on morphology: straw-hull (SH) and black-hull awned (BHA) weedy rice. We performed whole-genome resequencing of a SH weedy rice ‘PSRR-1’, a BHA weedy rice ‘BHA1115’, and a japonica cultivar ‘Cypress’ to delineate genome-wide differences and their relevance to genetics and evolution of weedy attributes. The high-quality reads were uniformly distributed with 82–88% genome coverage. The number of genotype-specific SNPs and InDels was highest in Cypress, followed by BHA1115 and PSRR-1. However, more genes were affected in BHA1115 compared with other two genotypes which is evident from the number of high-impact SNPs and InDels. Haplotype analysis of selected genes involved in domestication, adaptation, and agronomic performance not only differentiated SH from BHA weedy rice and supported evolution of weedy rice through de-domestication, but also validated the function of several genes such as qAn-1, qAn-2, Bh4, Rc, SD1, OsLG1, and OsC1. Several candidate genes were identified for previously reported seed dormancy and seed shattering QTLs. The SH and BHA weedy rice have distinct genomic composition, and the BHA weedy rice likely diverged earlier than SH weedy rice. The accumulation of plant development, reproduction, and defense-related genes in weedy rice possibly helped them to compete, survive, and spread under a wide range of environmental conditions by employing novel and diverse mechanisms. The genomic resources will be useful for both weed management and rice improvement by exploring the molecular basis of key agronomic, adaptive, and domestication attributes.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Weedy rice (Oryza sativa f. spontanea Rosh.), also known as red rice, is a major problem in rice-growing areas in the southern USA and other parts of the world, where farmers practice direct seeding. It outcompetes rice plants due to its aggressive growth habit leading to reduction in yield and contamination of harvests with its undesirable dark pigmented grains also reduces quality and marketability. In the southern rice-growing areas of the USA alone, reduction in rice yield can be as high as 80% with an annual economic loss over $50 million due to red rice infestation (Estorninos et al. 2005; Gealy et al. 2012). Its high seed shattering, intense seed dormancy, and superior ability to compete for water, nutrient, and sunlight compared to cultivated rice is mainly responsible for the persistence and proliferation (Estorninos et al. 2005; Burgos et al. 2006). Its phenotypic resemblance to cultivated rice and ability to persist in soil has made eradication of weedy rice challenging. After the adoption of herbicide-resistant Clearfield varieties, both farmers and scientists are concerned about the flow of resistant alleles to weedy rice populations (Shivrain et al. 2009).
In the USA, two major morphologically differentiated weedy rice ecotypes are prevalent: the straw-hull (SH) and the black-hull awned (BHA) types. SH weedy rice resembles most cultivated rice varieties with light brown hull without awns, whereas BHA type is phenotypically closer to Oryza rufipogon, a wild progenitor of Asian rice with black hulls and long awns (Londo and Schaal 2007). Awn, an extension of the tip of the lemma of spikelet, is an important domestication trait present in BHA weedy and wild rice, but is absent in cultivated and SH weedy rice. The SH weedy rice differs from BHA weedy rice in a number of traits such as plant height, heading date, tiller number, and awn length (Shivrain et al. 2010). For example, SH plants are shorter with fewer tillers and typically flower earlier compared to BHA plants in the southern USA. Wide range of variation for these traits is observed within each group.
The origin of weedy rice continues to be a scientific puzzle for last several decades. There are several hypotheses proposed regarding the origin of weedy rice. Two of the earlier hypotheses suggested evolution of weedy rice resulting from ongoing selection and adaptation of wild rice to unfavorable environment (De Wet and Harlan 1975), and hybridization between cultivated rice and O. rufipogon (Londo and Schaal 2007). A third hypothesis suggested origin of weedy rice from indica or japonica rice cultivars (Londo and Schaal 2007; Vaughan et al. 2008) or from indica x japonica hybrids (Qiu et al. 2014). This hypothesis has been supported by several molecular studies. Analysis of Korean weedy rice populations revealed genetic closeness to cultivated rice suggesting their evolution from indica/indica or japonica/japonica hybridization (He et al. 2017). Based on comparison at the mitochondrial genome level, weedy rice was more likely to have originated from cultivated rice instead of wild rice (Tong et al. 2017). The de-domestication hypothesis is gaining acceptance based on whole-genome sequence analyses of weedy rice from China and USA (Li et al. 2017; Qiu et al. 2017). A recent population genomics study, however, suggested evolution of weediness through a semi-domestication process which happened with the breeding effort to improve the cultivated rice (Sun et al. 2019). The SH and BHA weedy rice were believed to have evolved independently from indica and aus varieties, respectively (Reagon et al. 2010). Since neither indica nor aus was cultivated in North America, evolution of weedy rice through de-domestication process seems most plausible (Thurber et al. 2014). Distinct phenotypic differences between SH and BHA weedy rice with different ancestors indicate their unique genetic composition. Since the evidences gathered in studies conducted so far led to competing hypotheses regarding the evolution of weedy rice, there is need for assessment of genome-wide variation in additional weedy ecotypes.
Weedy rice is an excellent model not only for evolutionary genetics studies (Qiu et al. 2017) but also for genetic dissection of weedy traits (Subudhi et al. 2012, 2014). Analysis of weedy rice genome may allow discovery of beneficial alleles for yield and adaptive attributes which can be exploited to breed rice varieties with improved yield and stress adaptability in a rapidly changing global climate (He et al. 2017; Stewart 2017). As few genetic loci were responsible for the transformation of cultivated rice into weedy rice (Li et al. 2017), genomic regions that have undergone selection should be targeted for an in-depth functional genetic analysis to discover beneficial variants from weedy rice for use in rice breeding programs. Analysis of genetic variation on a genome-scale in cultivated, weedy, and wild accessions of rice using high-throughput next-generation sequencing (NGS) technologies has provided insights into the molecular processes underlying weedy rice evolution (Li et al. 2017; Qiu et al. 2017). The high-impact single-nucleotide polymorphisms (SNPs) and insertions/deletions (InDels) present within coding regions of a genome are assumed to be disruptive with regard to gene function. In addition, DNA sequence variations present in upstream regions regulate gene function by inducing or repressing gene expression. Our earlier study involving a SH weedy rice, an indica, and a japonica rice cultivar indicated that majority of high-impact DNA polymorphisms resulted in stop codon gain and genes carrying weed-specific DNA polymorphisms were involved in protein modification/phosphorylation, protein kinase activity, and protein/nucleotide binding (Chai et al. 2018). Therefore, discovery of high-impact SNPs and InDels related to gene function may provide insights into the molecular basis of the phenotypic variations in important agronomic, domestication, and stress adaptation attributes within weedy rice groups as well as between weedy and cultivated rice.
In this study, we performed whole-genome resequencing of a cultivated rice, a SH weedy rice, and a BHA weedy rice with many contrasting domestication traits. First, we assessed the genomic variability among them by discovering SNPs and InDels. Secondly, we also analyzed sequence variation in genes controlling some key weedy, agronomic, and abiotic stress tolerance attributes among them and identified candidate genes for seed dormancy and seed shattering.
Materials and methods
Plant materials
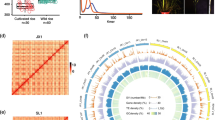
Three genotypes were used in this study: a SH weedy rice ‘PSRR-1’, a BHA weedy rice ‘BHA1115’, and a japonica rice cultivar ‘Cypress’ (Fig. 1) which were abbreviated as PSRR, BHA, and CPRS, respectively. CPRS is a high yielding, early maturing long grain cultivar developed at the Louisiana State University (LSU) Agricultural Center, LA, USA (Linscombe et al. 1993). It is a non-dormant, non-shattering, and awnless cultivar. PSRR-1 was purified by single plant selection for two generations from a mixture of weedy rice seeds collected from the LSU Rice Research Station at Crowley, LA, USA (Subudhi et al. 2012). It is straw-hulled, medium grain type, and awnless with high degree of seed dormancy and shattering. BHA is a black-hulled, highly dormant weedy rice from Arkansas with high degree of seed shattering and long awn, obtained from Dr. David Gealy of the United States Department of Agriculture’s Dale Bumpers Rice Research Center, Stuttgart, AR, USA. Seed dormancy of all three genotypes was performed through germination test described earlier (Subudhi et al. 2012) except for drying 6 days at room temperature to reduce moisture content.
Whole-genome sequencing
Genomic DNA was extracted from leaves of 14-day-old seedlings using Qiagen DNeasy kit (Qiagen Inc., Valencia, CA, USA). DNA quality and quantity were measured by Bioanalyzer 2100 (Agilent Technologies, Singapore) and Qubit 2.0 Fluorometer (Invitrogen Life Technologies, Eugene, OR, USA), respectively. The libraries were prepared using Illumina TruSeq DNA sample preparation kit (Illumina, USA). The paired-end sequencing of ‘PSRR’ and ‘CPRS’ was performed in Illumina HiSeq 2000 platform at the Virginia Bioinformatics Institute, Blacksburg, VA, and ‘BHA’ was sequenced in Illumina HiSeq 2500 platform at the Novogene Corporation Inc., Sacramento, CA, USA. The generated raw data were filtered using an in-built standard Illumina pipeline. The raw read sequences of PSRR-1, BHA1115, and Cypress were deposited in the NCBI’s Sequence Read Archive (SRA) under the accession numbers PRJNA413818, PRJNA599347, and PRJNA598851, respectively.
Reads mapping to the reference and filtering
The FASTQ files were analyzed by the NGS QC toolkit (v2.3.3) (Patel and Jain 2012) to eliminate low-quality reads and reads containing adapter or primer sequences. The high-quality reads were mapped to the reference nuclear genome of Nipponbare (IRGSP 1.0) (http://rapdb.dna.affrc.go.jp/download/archive/irgsp1/IRGSP-1.0_genome.fasta.gz) using BWA-MEM algorithm of Burrows–Wheeler Alignment (BWA) software (v0.7.15) with default parameter setting (Li 2013). The mapping output was processed and filtered using SAMtools (v1.3.1) (Li et al. 2009) to keep the reads mapping to a single location on the reference genome and to remove alignments with mapping quality (MAPQ) smaller than 30. Prior to variant calling, local realignment of reads around InDels was performed using the Genome Analysis Toolkit (GATK) (v3.7) (https://software.broadinstitute.org/gatk/).
Identification and analysis of SNPs and InDels
The realignment files were subjected to analysis by GATK’s Unified Genotyper to discover variants of the individual genotype. The SNPs and InDels were separated using GATK’s Select Variants tool, followed by filtering using stringent criteria including depth of coverage >10. The bi-allelic and homozygous SNPs and InDels were selected for further analysis. The SNPs and InDels clusters were filtered if three of them were present in any 10 bp window using GATK’s VariantFiltration program. After performing filtering procedure, SNPs and InDels between two genotypes (e.g., BHA/CPRS, PSRR/CPRS and BHA/PSRR) were identified by selecting unique variants using GATK’s CombineVariants and SelectVariants program. Similarly, genotype-specific SNPs and InDels were identified from the intersection of combined variants. For example, if CPRS has ‘T’ allele and PSRR and BHA have ‘A’ allele in a certain position of genome, ‘T’ allele is considered as CPRS-specific allele in that position. To analyze the distribution of SNPs and InDels per 1 Mb interval across rice genome, SNPs substitution type, and InDels length, VCFtools (v0.1.13) was used (Danecek et al. 2011). The genomic distribution of genotype-specific SNPs and InDels identified on each rice chromosome was visualized using Circos (v0.69-4) (Krzywinski et al. 2009).
The SnpEff (v.4.2) (Cingolani et al. 2012) program was used to annotate the identified genotype-specific SNPs and InDels using the rice7 gene models (http://downloads.sourceforge.net/project/snpeff/databases/v4_2/snpEff_v4_2_rice7.zip), and to predict variant effects (high, moderate, low, modifier) in the genic and other genomic regions. For determining the regions with significant deviation from the expected SNP rate, box-whisker plots were constructed. The outliers for genes with non-synonymous SNPs higher than the expected number were calculated using five-number summary of the box and whisker plot.
Gene ontology (GO) analysis of the genes containing high-impact SNPs and InDels was performed by singular enrichment analysis (SEA) tool using Web-based agriGO v2.0 (Tian et al. 2017) with the following parameter settings: (i) Fisher’s exact test with Benjamini-Yekutieli (FDR under dependency) multiple test adjustment method and (ii) significance level α = 0.05. MSU7.0 gene ID (LOC_Os06g29340) was used as the reference gene.
Copy number variants (CNVs) detection
The copy number variants were called using three algorithms: LUMPY (Layer et al. 2014), cn.MOPS (Klambauer et al. 2012), and CNVnator (Abyzov et al. 2011). CNVs were first called using CNVnator with bin size of 130 bp, 150 bp, and 110 bp for CPRS, BHA, and PSRR, respectively, with default values on other parameters. CNVnator output was passed as input into LUMPY using the -bedpe option. Resulting CNVs were used to create a set of copy number variable region (CNVRs) by merging CNVs across samples using svtools (Larson et al. 2019). CNVs with < 1 kbp in length and QUAL score < 100 were removed. To reduce the false positive calls, CNVs were called using cn.MOPS with window length of 500 bp, DNAcopy segmentation algorithm, and default values in other parameters. The CNVRs that overlapped in analyses with both LUMPY and cn.MOPS were retained for succeeding analyses.
Identification of variants in agronomically important genes
To identify the DNA variants (SNPs/InDels) of the cloned genes for the seed dormancy and seed shattering traits in the US weedy rice, two major seed dormancy-related QTLs (qSD7-1CR and qSD7-2CR) and a major shattering QTL qSH4CR were selected from an earlier study involving CPRS x PSRR RIL population (Subudhi et al. 2012, 2014). Since there were no QTL mapping studies for seed dormancy and seed shattering in crosses involving BHA, QTL intervals from the CPRS x PSRR population were used to identify the candidate genes. Several other genes controlling domestication and agronomically important traits were also selected to identify the variants. The start and end position of these QTLs were determined based on the genomic coordinates of flanking markers (McCouch et al. 2002) using BLASTn search against Nipponbare reference genome (http://rapdb.dna.affrc.go.jp/tools/blast) (Table S1). The SNPs and InDels were identified in the candidate genes located in the seed dormancy and seed shattering QTL intervals. Similarly, the SNPs and InDels in the CDS regions of other genes were identified among the three genotypes. The putative functions of candidate genes of the above-mentioned traits were collected from both Rice Genome Annotation Project (http://rice.plantbiology.msu.edu/analyses_search_locus.shtml) and Rice Annotation Project Database (RAP-DB) (http://rapdb.dna.affrc.go.jp/).
Results
Genome assembly and discovery of genotype-specific SNPs and InDels
About 95% of the reads were high-quality reads, and 70-80% of those, which mapped to unique location of the reference rice nuclear genome, were considered for subsequent downstream analysis (Table 1). The reads were distributed uniformly with 82-88% genome coverage and sequencing depth of 18-24-fold. A total of 3,181,137 SNPs and 818,732 InDels were identified in the three genotypes relative to Nipponbare (Table S2). The read depth of the identified SNPs and InDels ranged from 11 to >50. Compared with Nipponbare, the number of both SNPs and InDels was fewer in cultivated rice ‘CPRS’ than both weedy rice genotypes (Table S2). The number of SNPs and InDels was comparable in all three comparisons with highest number of SNPs in BHA/CPRS and highest number of InDels in PSRR/CPRS (Table 2, Table S3). Finally, we identified DNA polymorphisms specific to PSRR (554,338 SNPs and 157,544 InDels), BHA (689,257 SNPs and 146,769 InDels) and CPRS (733,233 SNPs and 188,021 InDels) (Table S4). The density of genotype-specific DNA polymorphisms per Mb was 1,491 (SNPs) and 422 (InDels) for PSRR, 1,850 (SNPs) and InDels (391) for BHA, and 1,977 (SNPs) and 506 (InDels) for CPRS (Table S4). All genotype-specific SNPs and InDels were listed (Tables S5A, S5B, S6A, S6B, S7A and S7B).
Genomic distribution of SNPs and InDels
The number of PSRR and BHA-specific SNPs was highest on chromosome 1, whereas it was chromosome 3 for CPRS (Fig. 2a, Table S4). The number of CPRS-specific SNPs was higher in chromosomes 2, 3, 7, 8, and 9 compared to both weedy rice genotypes. In case of InDels, a larger number of PSRR and BHA-specific InDels were present on chromosome 1 and CPRS-specific InDels on chromosome 2 and 3 (Fig. 2b, Table S4). The SNPs and InDels were unevenly distributed over the genome (Fig. 2c, d). There were 89 PSRR-specific, 160 BHA-specific, and 188 CPRS-specific high-density (≥ 2000 SNP/Mb) SNP regions (Fig. 2c, Tables S5A, 6A, S7A). Following stringent criteria of ≥ 4000 SNPs per 1 Mb, 5, 6, and 7 SNP-rich regions were identified for PSRR, BHA, and CPRS, respectively. The PSRR, BHA, and CPRS-specific SNP-rich regions were detected on 2 chromosomes (Chr 1: 14–16 Mb and 17–18 Mb regions; and Chr 6: 7–9 Mb, 12–13 Mb, and 16–17 Mb regions), four chromosomes (Chr 1: 1–2 Mb and 8–9 Mb regions; Chr 2: 3–4 Mb region; Chr 4: 17–18 Mb and 22–23 Mb regions; and Chr 5: 7–8 Mb region), and six chromosomes (Chr 1: 11–12 Mb region; Chr 3: 14–15 Mb region; Chr 6: 20–21 Mb region; Chr 7: 24–25 Mb region; Chr 9: 14–15 region and 19-21 Mb regions; and Chr 12: 27-27.5 Mb region), respectively (Fig. 2c). Using the criteria of ≤ 100 SNPs per 1 Mb, SNP-poor regions known as ‘SNP deserts’ were identified. Four PSRR-specific SNP deserts were on chromosome 1 (43-43.2 Mb region), chromosome 3 (36–36.4 Mb region), and chromosome 4 (15–16 Mb and 26–27 Mb regions). Six BHA-specific SNP deserts were identified on chromosome 1 (16–17 Mb and 20–22 Mb regions), chromosome 6 (2–3 Mb, 12–13 Mb, and 15–16 Mb regions), and chromosome 10 (23–23.2 Mb region). Nine CPRS-specific SNP deserts were on chromosome 1 (8–9 Mb, 15–17 Mb, and 20–22 Mb regions), chromosome 4 (15–16 Mb and 26–27 Mb regions), chromosome 6 (2–3 Mb, 12–13 Mb, and 14–17 Mb regions), and chromosome 10 (23–23.2 Mb region) (Fig. 2c). There were 22 PSRR-specific, 13 BHA-specific, and 52 CPRS-specific high-density (≥ 800 InDels/Mb) InDels regions, whereas a total of 16 PSRR-specific, 20 BHA-specific, and 15 CPRS-specific low-density InDel regions or InDel deserts (<100 InDels/Mb) were identified (Fig. 2d; Tables S5B, S6B, S7B).
Number and distribution of genotype-specific SNPs (a) and InDels (b) identified on the rice chromosomes. Total number of SNPs and InDels identified on each chromosome are displayed in the bar graphs. Genotype-specific SNPs and InDels were indicated as PSRR-specific, BHA-specific, and CPRS-specific. Distribution of SNPs (c) and InDels (d) identified on each chromosome (1 Mb window size) are demonstrated on the Circos diagram. The outermost circle represents rice chromosomes (chr 1–12) in different colors, and three inner circles represent distribution of SNPs/InDels specific to BHA (I), PSRR (II), and CPRS (III). Different range of SNPs and InDels are shown by different colors of different types of glyph
Analysis of SNPs and InDels
The SNPs identified in each of the three pair-wise comparisons were classified as transitions (A/G and C/T) and transversions (A/C, A/T, C/G, and G/T) based on nucleotide substitutions type (Fig. 3a). The frequency of transition (Ts) or transversion (Tv) SNPs and the ratio of Ts:Tv were of similar magnitude in all three comparisons (Table S8). The highest length of insertions (36-bp) and deletions (57-bp) were found in BHA/PSRR and BHA/CPRS, whereas the length of insertions and deletions were up to 29-bp and 45-bp in PSRR/CPRS, respectively (Fig. 3b). A major portion of InDels was of single nucleotide, followed by di- to hexa-nucleotides, and the remaining InDels were of ≥7 bp long in all three pairs (Table S9).
Frequency of different types of substitution in the identified SNPs and length distribution of InDels. a Frequency of substitution types of SNPs in PSRR/CPRS, BHA/CPRS and BHA/PSRR. b Length distribution of InDels. Number of InDels (y-axis) of different lengths (x-axis, in bp) is presented in the bar graph. PSRR/CPRS, BHA/CPRS and BHA/PSRR indicate InDels identified between PSRR and CPRS, BHA and CPRS, and BHA and PSRR cultivars
Annotation of SNPs and InDels
There were 27–29% SNPs and 23–24% InDels in genic regions (Fig. 4). The remaining SNPs and InDels were detected in other genomic regions, which included 2 kb upstream (promoter), 2 kb downstream, and intergenic regions. Within the entire genome, only 11–12% SNPs were identified in the CDS regions of all three genotypes (Fig. 4a, b). But only 4–5% of InDels were identified in the CDS regions (Fig. 4c, d). Among the SNPs in genic regions, 39–43% were in the CDS regions and 22–25% were non-synonymous in all three genotypes. The number of non-synonymous SNPs was higher than that of synonymous SNPs. Among the InDels in the genic regions, 16–20% were in the CDS regions. In addition, the 3′-UTR regions contained 1.8–2.2 times more SNPs and 1.3-1.6 times more InDels than the 5′-UTR regions.
Annotations of genotype-specific SNPs and InDels. Distribution of PSRR-specific, BHA-specific, and CPRS-specific SNPs in different genomic (a) and genic regions (b). Distribution of PSRR-specific, BHA-specific, and CPRS-specific InDels in different genomic (c) and genic regions (d) was shown. The number of non-synonymous and synonymous SNPs identified with the coding region was shown
The high-impact SNPs and InDels result from disruption of splice sites, loss of start codon, loss of stop codon, gain of stop codon, and frame shift mutations. There were 2256 and 3698 CPRS-specific high-impact SNPs and InDels, respectively, affecting 4523 genes that could distinguish CPRS from PSRR and BHA weedy rice (Fig. 4; Tables S5, S6, S7). There were 4182 genes impacted by 1992 and 3507 PSRR-specific high-impact SNPs and InDels, respectively, whereas 4720 genes were affected by 2640 and 3686 BHA-specific high-impact SNPs and InDels, respectively. The PSRR-specific and BHA-specific high-impact SNPs and InDels made them unique from each other.
Non-synonymous SNPs are moderate (non-disruptive) impact SNPs that are responsible for changing protein effectiveness. In the CDS regions, there were 35,962 PSRR-specific, 46,949 BHA-specific, and 47,218 CPRS-specific non-synonymous SNPs involving 14,283, 16,996, and 17,437 genes, respectively (Fig. 3b; Tables S5A, S6A and S7A). The range for the number of BHA-, CPRS-, and PSRR-specific non-synonymous SNPs per gene was 1–59, 1–58, and 1–119, respectively. The number of non-synonymous SNPs per kb of genic region had wide distribution with a range of 0.11–38.76 (PSRR-specific), 0.06-42.42 (BHA-specific), and 0.06-42.86 (CPRS-specific). Using the box and whisker plot, a total of 1159; 1306; and 1442 genes were classified as outliers as they had higher density of PSRR-specific (> 6.86), BHA-specific (> 7.75) and CPRS-specific (> 7.09) non-synonymous SNPs/Kb of genic region than expected (Fig. S1).
Gene ontology (GO) analysis of genotype-specific polymorphisms
GO analysis was performed using PSRR-, BHA-, and CPRS-specific high-impact SNPs and InDels to get further information about their involvement in biological processes, molecular functions, and cellular components. A total of 47, 45, and 57 GO terms were significant for biological processes and molecular functions in case of PSRR-, BHA-, and CPRS-specific genes, respectively (Fig. 5, Table S10). The genes from all three genotypes were involved in 21 GO terms under biological process and 62–65% of genes were represented in the cellular process category. Similarly, the genes from all three genotypes represent 23 GO terms under molecular function and 80–83% of genes were involved in binding.
Enrichment of the gene ontology term in the genes harboring high-impact SNPs/InDels specific to PSRR, BHA, and CPRS. The biological process (A) and molecular function (B) gene ontology terms significantly enriched (P ≤ 0.05) are displayed in x-axis and the percent of genes of each gene ontology term are shown in y-axis. GO terms for the biological process are: (a) DNA metabolic process, (b) RNA-dependent DNA replication, (c) DNA replication, (d) Nucleobase, nucleoside, nucleotide and nucleic acid metabolic process, (e) nitrogen compound metabolic process, (f) DNA integration, (g) cellular macromolecule metabolic process, (h) Macromolecule metabolic process, (i) programmed cell death, (j) apoptosis, (k) death, (l) cell death, (m) cellular process, (n) cellular metabolic process, (o) defense response, (p) primary metabolic process, (q) cellular macromolecule biosynthetic process, (r) macromolecule biosynthetic process, (s) response to stress, (t) metabolic process, (u) response to stimulus, (v) multi-organism process, (w) pollen–pistil interaction, (x) cell recognition, (y) recognition of pollen, (z) reproductive process, (aa) pollination, (ab) reproduction, (ac) cellular biosynthetic process, and (ad) biosynthetic process. GO terms for the molecular function are as follows: (a) RNA-directed DNA polymerase activity, (b) DNA polymerase activity, (c) nucleotidyl transferase activity, (d) nucleic acid binding, (e) RNA binding, (f) transferase activity, transferring phosphorus-containing groups, (g) binding, (h) transferase activity, (i) ribonuclease H activity, (j) endonuclease activity, (k) endoribonuclease activity, producing 5’-phosphomonoesters, (l) endoribonuclease activity, (m) endonuclease activity, active with either ribo- or deoxyribonucleic acids and producing 5’-phosphomonoesters, (n) nuclease activity, (o) ribonuclease activity, (p) hydrolase activity, acting on ester bonds, (q) DNA binding, (r) catalytic activity, (s) ATP binding,(t) adenyl ribonucleotide binding, (u) sugar binding, (v) purine ribonucleotide binding, (w) ribonucleotide binding, (x) adenyl nucleotide binding, (y) purine nucleoside binding, (z) nucleoside binding, and (aa) purine nucleotide binding
There were seven significant biological process GO terms (multi-organism process, pollen-pistil interaction, cell recognition, recognition of pollen, reproductive process, pollination, and reproduction) and two molecular function GO terms (sugar binding and purine nucleotide binding), which are enriched only in cultivated rice (Fig. 5, Fig. S2). There were only 22 genes, which were common among the above seven GO terms. These were protein kinases, S-locus specific genes, and carbohydrate binding proteins which may be involved in plant reproduction process. Many of these genes overlapped with the genes representing GO term ‘sugar binding’ of molecular function. There were 283 genes under the ‘purine nucleotide binding’ category which are involved in resistance to biotic and abiotic stresses.
There was only one GO term ‘response to stimulus’ which was significantly enriched in BHA and CPRS compared to PSRR. Most of these genes under this category were related to disease resistance. Furthermore, there was clear difference with respect to one biological GO term, metabolic process and two molecular function GO terms, purine ribonucleotide binding and ribonucleotide binding which are enriched in PSRR and CPRS but not in BHA. Majority of these genes under the ‘metabolic processes’ category were retrotransposons in addition to receptor kinases and gibberellin receptors.
Since weedy rice are known for higher level of stress tolerance compared to cultivated rice, we focused on the biological process GO term ‘response to stress’ which was common in all three genotypes. This GO term contained a total of 82, 98, and 95 genes, of which 44, 58, and 66 genes were private in PSRR, BHA, and CPRS, respectively (Fig. 5, Fig. S2). Most of them were disease resistant protein, heat shock protein, universal stress protein domain containing protein, peroxidase precursor, and jacalin-like lectin, etc.
CNV discovery
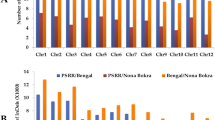
A total of 5,011 CNVRs overlapped between cn.MOPs and CNVnator-LUMPY methods. Filtering out the CNVRs without copy number changes resulted in 1,886 CNVRs. Sizes of the CNVRs ranged from 1.5 to 585 kb, with an average of 19 kb. The CNVRs occupied a total of 36.5 Mb or 8.5% of the rice genome. Among the CNVRs, 1,705 showed copy number loss (deletion) and 181 showed copy number gain (duplication). A total of 3,666 genes from MSUv7 annotation of the reference genome overlapped the detected CNVRs (Table S11). The distribution of CNVs revealed that copy number loss was more pronounced in both BHA and PSRR compared with CPRS in which both CNV types were represented in high numbers (Fig. 6). Among all the genes listed in Table 3, only two genes, qAn-1 (LOC_Os04g28280) and Ptr (LOC_Os12g18729), showed copy number loss in PSRR, whereas there was no copy number variation in BHA and CPRS.
Variants in genes controlling domestication and stress adaptation
The variants in genes controlling several domestication and stress adaptation traits were identified in all three genotypes (Table 3). The synonymous SNPs were not considered for this analysis. In case of An-1 (Luo et al. 2013), non-synonymous SNPs and a 3-bp deletion differentiated awned BHA from awnless CPRS and PSRR. A 1-bp deletion causing a frame shift mutation and a 12-bp deletion were present only in PSRR and CPRS, respectively. For An-2/LABA1 (Gu et al. 2015; Hua et al. 2015), one 1-bp deletion resulted in a premature stop codon in PSRR and CPRS but not in awned BHA. Both PSRR and CPRS had a 22-bp deletion and two non-synonymous SNPs in the black hull color gene Bh4 (Zhu et al. 2011). For the pericarp color gene Rc (Sweeney et al. 2006), a 14-bp deletion and two non-synonymous SNPs differentiated cultivated rice from the weedy rice. No mutation was found in all three genotypes in the shattering genes, Sh4 (Li et al. 2006), Sh-h (Ji et al. 2010), and SHAT1 (Zhou et al. 2012), whereas a stop gain mutation was found in both CPRS and BHA in case of qSH1 (Konishi et al. 2006). For the closed panicle gene OsLG1 (Ishii et al. 2013), there were three non-synonymous SNPs and a 3-bp deletion present in PSRR compared with other two genotypes.
For the semi-dwarfing gene SD1 (Spielmeyer et al. 2002), a 383-bp deletion distinguished CPRS from PSRR and BHA. A 10-bp deletion causing frameshift in the OsC1 gene (Saitoh et al. 2004) was present in both weedy rice genotypes. In case of the Phr1 gene responsible for phenol reaction (Yu et al. 2008), a 1-bp insertion was responsible for a frameshift mutation in CPRS. Seven non-synonymous SNPs were identified in the Ptr blast gene (Zhao et al. 2018), differentiating PSRR from both CPRS and BHA. For the cold tolerance gene COLD1 (Ma et al. 2015), a non-synonymous mutation distinguished all three genotypes. In case of the cold tolerant gene CTB4a (Zhang et al. 2017) and drought resistance genes, MODD (Tang et al. 2016) and PYL9 (Zhao et al. 2016), 2 non-synonymous SNPs were identified between cultivated rice and weedy genotypes. We identified a weedy rice-specific non-synonymous SNP and an 18-bp deletion in PSRR for the low-temperature germinability QTL qLTG-1 (Fujino et al. 2008). There were three weedy rice-specific non-synonymous SNPs in the Salt Intolerance 1 (SIT1) gene (Li et al. 2014). The sequence analysis of SLR1 gene (Fukao and Bailey-Serres 2008) showed the same SNP in CPRS and PSRR. In case of submergence tolerance gene SUB1A (Xu et al. 2006), a C to A mutation induced a stop codon in CPRS and BHA, resulting in a truncated SUB1A protein. There was no sequence variation among all three genotypes for the plant architecture gene Prog1 (Tan et al. 2008), and thermo-tolerance gene ERECTA (Shen et al. 2015).
Candidate genes for seed dormancy and seed shattering
Among the three genotypes, both weedy rice genotypes exhibited deep dormancy and high shattering (Fig. 1). The major shattering QTL qSH4CR identified earlier (Subudhi et al. 2014) was used to identify the candidate gene(s) in the US weedy rice genotypes. Based on the genomic coordinates of the flanking markers, a total of 12 genes containing weedy rice-specific SNPs and InDels were identified within the QTL qSH4CR region. By removing hypothetical proteins, expressed proteins, and transposon proteins from the QTL region, 3 candidate genes were identified within the QTL qSH4CR region (Table S12). Similarly, a total of 69 and 121 genes containing weedy rice-specific SNPs and InDels were present within the seed dormancy QTL qSD7-1CR and qSD7-2CR intervals, respectively. Based on the criteria discussed above, 28 and 41 candidate genes were identified for the qSD7-1CR and qSD7-2CR, respectively (Table S13).
Discussion
The whole-genome sequence analysis revealed an interesting fact: Both PSRR and BHA were not only unique in their genomic composition but were also genetically distinct from the cultivated rice. Although each weedy rice showed similar level of SNPs and InDels when compared with CPRS or Nipponbare, higher number of SNPs and lower number of InDels observed in BHA compared to PSRR suggests the importance of SNPs in their differentiation. Greater level of polymorphisms observed between japonica type and weedy rice was expected because BHA and SH weedy rice were genetically closer to aus and indica types, respectively (Chai et al. 2018; Reagon et al. 2010). However, lower level of cultivated rice-specific SNPs was observed in BHA compared to PSRR suggesting evolution of BHA earlier than SH weedy rice (Li et al. 2017). It is also supported by our observation on the genotype-specific variants and number of genes carrying them. The genotype-specific variants affected 4720 genes in BHA compared to 4,120 genes in PSRR. Li et al. (2017) identified 178 and 307 candidate genes for weed-specific adaptation in SH and BHA, respectively, based on analysis of the whole-genome sequences from 18 SH and 20 BHA weedy rice along with 145 published Oryza genome sequences. Identification of fewer genes associated with weediness adaptation may be due to increasing power of contrast by using large number of SH, BHA, and other Oryza accessions. The occurrence of higher number of Ts compared to Tv observed in this study is in agreement with earlier studies (Subbaiyan et al. 2012; Jain et al. 2014). But higher number of Ts and Tv observed in BHA/CPRS compared to PSRR/CPRS supports earlier evolution of BHA compared with PSRR. Another line of evidence suggesting early divergence of BHA in the crop evolution process is our observation of wild type traits and the haplotypes of the corresponding awn and Bh4 genes in BHA compared with PSRR.
Nonrandom distribution of SNPs and InDels over the rice genome led to identification of polymorphism-rich and polymorphism-poor regions, confirming earlier reports in rice, wild rice, and weedy rice (Feltus et al. 2004; Wang et al. 2009; Subbaiyan et al. 2012; Krishnan et al. 2014; Li et al. 2017; Chai et al. 2018). Krishnan et al. (2014) suggested that limited sequence variation in a 5 Mb region on chromosome 5 observed between Australian wild rice and cultivated rice may be due to natural selection before domestication. In a comparative study involving indica and japonica subspecies, Feltus et al. (2004) indicated that introgression between them may have led to SNP-poor chromosomal regions. The SNP deserts in this study were much smaller compared to those studies (Feltus et al. 2004; Krishnan et al. 2014), but the number of CPRS-specific SNP deserts was more than those from BHA and PSRR. The reduced genetic variations in these SNP/InDel poor regions may be the result of multiple introgression events between weedy and cultivated rice during the weedy rice evolution which is consistent with the de-domestication process (Li et al. 2017).
Although weedy rice has better biotic and abiotic stress tolerance (Borjas et al. 2016; Burgos et al. 2006; Jia and Gealy 2018), there was no differential enrichment in the GO term ‘response to stress’ between weedy and cultivated rice which is in agreement with an earlier study (Li et al. 2017). There were 44-66 private genes related to biotic and environmental stress tolerance in each genotype, which may be expressed differentially in stress environments to differentiate weedy and cultivated rice. The significantly enriched genes in CPRS compared with weedy rice included largely the protein kinase and S-locus genes, which are related to plant development, defense, reproduction, and self-incompatibility (Dievart and Clark 2004; Xing et al. 2013). The genes involved in sugar binding played a positive role on plant growth and development through sugar metabolism and energy production during photosynthesis (Sheen et al. 1999). In addition, the genes associated with purine nucleotide binding activity are required for repair of some sort of DNA damage during vegetative growth in plants (Alani et al. 1989). Therefore, our observation supports the earlier findings that suggested contribution of balancing selection for self-incompatibility and resistances genes toward evolution of weedy rice through the de-domestication process (Qiu et al. 2017). In this study, only one GO term ‘genes for response to stimulus’ was enriched in BHA and CPRS relative to PSRR. Many of these were disease resistance and LRR motif enriched genes with role in plant development and defense. In addition, genes involved in lipid metabolic process, carbohydrate metabolic process, protein serine/threonine kinase activity were found to be specific to PSRR and CPRS, thus distinguishing them from BHA. Our study suggests that weedy rice populations might have evolved through accumulation of plant development, reproduction, and defense-related genes which helped them to compete, survive, and spread under a wide range of environmental conditions by employing novel and diverse mechanisms.
Since rice was domesticated from its wild progenitor through artificial selection for many desirable traits, we selected few cloned genes involved in domestication, adaptation, and agronomic performance to examine the DNA polymorphisms or haplotypes (Table 3) among the three genotypes to assess genetic differences as well as evolutionary path leading to crop domestication. The role of some genes could be validated by correlating the variants with the phenotypic differences among them. BHA had a unique haplotype for the underlying genes qAn-1, qAn-2/LABA1, and Bh4 (Zhu et al. 2011; Gu et al. 2015; Hua et al. 2015), which differentiated it from PSRR and CPRS with respect to awn and hull characteristics. Similarly, the variants identified in Rc, OsC1, and SD1, separated CPRS from BHA and PSRR. The OsLG1 haplotype of PSRR with open panicle was different from BHA and CPRS with closed panicles. In case of the Phr1 gene, 1-bp deletion causing frameshift was identified in both weedy rice genotypes instead of the 18-bp deletion that differentiates japonica cultivars from indica cultivar (Yu et al. 2008).
This study delineated the genomic differences between both weedy rice genotypes by analyzing haplotypes of several genes involved in biotic and abiotic stress tolerance. Copy number variations have been proposed to be associated with biotic and abiotic stress tolerance genes in plants (Oh et al. 2012; Dolatabadian et al. 2017). Although the type of copy number variations distinguished cultivated rice CPRS from both weedy rice BHA and PSRR, it was difficult to associate the stress tolerance with copy number variations, since these weedy rice used in this study have not been characterized for various biotic and abiotic stress tolerance. Therefore, evaluation of these genotypes for stress tolerance may enhance the usefulness of these identified variants of these selected genes for rice improvement.
With respect to the domestication-related genes such as Prog1, SHAT1, Sh-h, and Sh4, presence of cultivar like haplotype in all three genotypes supported the de-domestication hypothesis (Li et al. 2017). Since none of the shattering genes could distinguish high shattering PSRR and BHA from non-shattering CPRS through our analysis, it reinforces idea of a novel genetic mechanism for seed shattering despite the overlapping of Sh4 with the QTLs identified in mapping populations involving PSRR (Subudhi et al. 2014). There was disagreement with the earlier finding (Li et al. 2017) with regard to qSH1 because all three genotypes carried 3 distinct variants for the qSH1. The whole-genome scanning of PSRR, an indica genotype Nona Bokra, and a japonica cultivar Bengal in this QTL interval identified eight candidate genes which are not known to be involved in shattering (Chai et al. 2018). In this study, we compared the high-impact DNA polymorphisms present in the QTL interval in non-shattering genotype ‘CPRS’ and both high shattering weedy rice and identified three candidate genes (GTP cyclohydrolase I, TLD family protein, and Tafazzin) for further investigation (Table S12). GTP cyclohydrolase I was reported to be involved in folate synthesis pathway impacting rice seed metabolism (Blancquaert et al. 2013). The other two candidates were involved in protein modification and transferase activity based on ontology classification (http://rice.plantbiology.msu.edu/) but were not investigated in plants.
To explore the genetic basis of seed dormancy in weedy rice, candidate genes in two major QTLs (qSD7-1CR and qSD7-2CR) were examined (Table S13). Since the qSD7-1CR interval did not include the Rc gene, which controlled both seed dormancy and pericarp color pleiotropically (Gu et al. 2011), involvement of other gene(s) could not be ruled out. However, the candidate gene, LOC_Os07g09590, which encodes a basic helix-loop-helix (bHLH) protein like Rc gene, may be an ideal candidate for seed dormancy. Two out of 11 candidate genes, LOC_Os07g10490 (zeta-carotene desaturase) and Rc, were earlier suggested (Chai et al. 2018). In the qSD7-2CR interval, there were 41 potential candidate genes including Sdr4 (Sugimoto et al. 2010) carrying high-impact DNA polymorphisms. Although Sdr4 co-localized with a major QTL for seed dormancy, its association with seed dormancy could not be validated due to sequence inconsistency between weedy genotype and cultivated rice (Subudhi et al. 2012). The candidate genes identified in this study should be investigated for genetic dissection of seed dormancy in future.
Development of an effective weedy rice management strategy is complicated because weeds are adapted to wide range of environments due to genetic variability and phenotypic plasticity (Rathore et al. 2016). The genome-wide differences between PSRR and BHA revealed in this study suggest that SH and BHA weedy rice may have different adaptive mechanisms and therefore will require different management and control strategy (Qiu et al. 2017). Since diversity in functional traits is not sufficient to devise strategies for weedy rice management, molecular understanding of weedy traits and their evolution will be helpful.
Since weedy rice harbors a number of desirable agronomic and adaptive traits (Borjas et al. 2016; Burgos et al. 2006; Jia and Gealy 2018; Ziska and McClung 2008), it could be a valuable genetic resource for rice improvement. Our study showed that about one-third of the SNPs and one fourth of InDels occurred in the genic regions and most moderate and high-impact variants were in the conserved CDS regions. Therefore, these variants will be useful to differentiate cultivated rice from weedy wild rice as well as to conduct molecular genetic analysis of important agronomic and adaptive traits.
Availability of data and materials
All relevant data are provided as tables and figures with the paper and in the supplementary materials.
Abbreviations
- SH:
-
Straw hulled
- BHA:
-
Black-hulled awned
- CPRS:
-
Cypress
- QTL:
-
Quantitative trait locus
- SNP:
-
Single-nucleotide polymorphism
- InDel:
-
Insertions and deletions
- RIL:
-
Recombinant inbred lines
- CDS:
-
Coding sequences
- UTR:
-
Untranslated regions
- GO:
-
Gene ontology
- NGS:
-
Next-generation sequencing
References
Abyzov A, Alexander E E, Urban AE, Snyder M, Gerstein M (2011) CNVnator: an approach to discover, genotype and characterize typical and atypical CNVs from family and population genome sequencing. Genome Res. https://doi.org/10.1101/gr.114876.110
Alani E, Subbiah S, Kleckner N (1989) The yeast RAD50 gene encodes a predicted 153-kD protein containing a purine nucleotide-binding domain and two large heptad-repeat regions. Genetics 122:47–57
Blancquaert D, Van Daele J, Storozhenko S, Stove C, Lambert W, Van Der Straeten D (2013) Rice folate enhancement through metabolic engineering has an impact on rice seed metabolism, but does not affect the expression of the endogenous folate biosynthesis genes. Plant Mol Biol 83:329–349
Borjas AH, De Leon TB, Subudhi PK (2016) Genetic analysis of germinating ability and seedling vigor under cold stress in US weedy rice. Euphytica 208:251–264
Burgos NR, Norman RJ, Gealy DR, Black H (2006) Competitive N uptake between rice and weedy rice. Field Crops Res 99:96–105
Chai C, Shankar R, Jain M, Subudhi PK (2018) Genome-wide discovery of DNA polymorphisms by whole genome sequencing differentiates weedy and cultivated rice. Sci Rep 8:14218
Cingolani P, Platts A, Wang LL et al (2012) A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly (Austin) 6:80–92
Danecek P, Auton A, Abecasis G, 1000 Genomes Project Analysis Group et al (2011) The variant call format and VCFtools. Bioinformatics 27:2156–2158
De Wet JMJ, Harlan JR (1975) Weeds and domesticates: evolution in the man-made habitat. Econ Bot 29:99–108
Dievart A, Clark SE (2004) LRR-containing receptors regulating plant development and defense. Development 131:251–261
Dolatabadian A, Patel DA, Edwards D, Batley J (2017) Copy number variation and disease resistance in plants. Theor Appl Genet 13:2479–2490
Estorninos LE, Gealy DR, Gbur EE, Talbert RE, McClelland MR (2005) Rice and red rice interference. II. Rice response to population densities of three red rice (Oryza sativa) ecotypes. Weed Sci 53:683–689
Feltus FA, Wan J, Schulze SR, Estill JC, Jiang N, Paterson AH (2004) An SNP resource for rice genetics and breeding based on subspecies indica and japonica genome alignments. Genome Res 14:1812–1819
Fujino K, Sekiguchi H, Matsuda Y, Sugimoto K, Ono K, Yano M (2008) Molecular identification of a major quantitative trait locus, qLTG3-1, controlling low-temperature germinability in rice. Proc Natl Acad Sci USA 105:12623–12628
Fukao T, Bailey-Serres J (2008) Submergence tolerance conferred by Sub1A is mediated by SLR1 and SLRL1 restriction of gibberellin responses in rice. Proc Natl Acad Sci USA 105:16814–16819
Gealy DH, Agrama H, Jia MH (2012) Genetic analysis of a typical U.S. red rice phenotypes: indications of prior gene flow in rice fields? Weed Sci 60:451–461
Gu XY, Foley ME, Horvath DP et al (2011) Association between seed dormancy and pericarp color is controlled by a pleiotropic gene that regulates abscisic acid and flavonoid synthesis in weedy red rice. Genetics 189:1515–1524
Gu B, Zhou T, Luo J et al (2015) An-2 encodes a cytokinin synthesis enzyme that regulates awn length and grain production in rice. Mol Plant 8:1635–1650
He Q, Kim KW, Park YJ (2017) Population genomics identifies the origin and signatures of selection of Korean weedy rice. Plant Biotechnol J 15:357–366
Hua L, Wang DR, Tan L et al (2015) LABA1, a domestication gene associated with long, barbed awns in wild rice. Plant Cell 27:1875–1888
Ishii T, Numaguchi K, Miura K et al (2013) OsLG1 regulates a closed panicle trait in domesticated rice. Nat Genet 45:462
Jain M, Moharana KC, Shankar R, Kumari R, Garg R (2014) Genomewide discovery of DNA polymorphisms in rice cultivars with contrasting drought and salinity stress response and their functional relevance. Plant Biotechnol J 12:253–264
Ji H, Kim SR, Kim YH et al (2010) Inactivation of the CTD phosphatase-like gene OsCPL1 enhances the development of the abscission layer and seed shattering in rice. Plant J 61:96–106
Jia Y, Gealy D (2018) Weedy red rice has novel sources of resistance to biotic stress. Crop J 6:443–450
Klambauer G, Schwarzbauer K, Mayr A, Clevert DA, Mitterecker A, Bodenhofer U, Hochreiter S (2012) cn.MOPS: mixture of Poissons for discovering copy number variations in next-generation sequencing data with a low false discovery rate. Nucl Acid Res 40(9):e69
Konishi S, Izawa T, Lin SY, Ebana K, Fukuta Y, Sasaki T, Yano M (2006) An SNP caused loss of seed shattering during rice domestication. Science 312:1392–1396
Krishnan G, Waters DLE, Henry RJ (2014) Australian wild rice reveals pre-domestication origin of polymorphism deserts in rice genome. PLoS ONE 9:e98843
Krzywinski M, Schein J, Birol I, Connors J, Gascoyne R, Horsman D, Jones SJ, Marra MA (2009) Circos: an information aesthetic for comparative genomics. Genome Res 19:1639–1645
Larson DE, Abel HJ, Chiang C et al (2019) svtools: population-scale analysis of structural variation. Bioinformatics 35(22):4782–4787
Layer RM, Chiang C, Quinlan AR, Hall IM (2014) LUMPY: a probabilistic framework for structural variant discovery. Genome Biol 15:R84
Li H (2013) Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv:13033997 [q-bioGN]
Li C, Zhou A, Sang T (2006) Rice domestication by reducing shattering. Science 311:1936–1939
Li H, Handsaker B, Wysoker A et al (2009) The sequence alignment/map format and SAMtools. Bioinformatics 25:2078–2079
Li C-H, Wang G, Zhao J-L et al (2014) The receptor-like kinase SIT1 mediates salt sensitivity by activating MAPK3/6 and regulating ethylene homeostasis in rice. Plant Cell 26:2538–2553
Li LF, Li YL, Jia Y, Caicedo AL, Olsen KM (2017) Signatures of adaptation in the weedy rice genome. Nat Genet 49:811
Linscombe SD, Jodari F, McKenzie KS, Bollich PK, White LM, Groth DE, Dunand RT (1993) Registration of ‘Cypress’ rice. Crop Sci 33:355
Londo JP, Schaal BA (2007) Origins and population genetics of weedy red rice in the USA. Mol Ecol 16:4523–4535
Luo J, Liu H, Zhou T et al (2013) An-1 encodes a basic helix-loop-helix protein that regulates awn development, grain size, and grain number in rice. Plant Cell 25:3360–3376
Ma Y, Dai X, Xu Y et al (2015) COLD1 confers chilling tolerance in rice. Cell 160:1209–1221
McCouch SR, Teytelman L, Xu Y et al (2002) Development and mapping of 2240 new SSR markers for rice (Oryza sativa L.). DNA Res 9:199–207
Oh DH, Dassanayake M, Bohnert HJ, Cheeseman JM (2012) Life at the extreme: lessons from the genome. Genome Biol 13:241
Patel RK, Jain M (2012) NGS QC Toolkit: a toolkit for quality control of next generation sequencing data. PLoS ONE 7:e30619
Qiu J, Zhu J, Fu F et al (2014) Genome re-sequencing suggested a weedy rice origin from domesticated indica-japonica hybridization: a case study from southern China. Planta 240:1353–1363
Qiu J, Zhou Y, Mao L et al (2017) Genomic variation associated with local adaptation of weedy rice during de-domestication. Nat Commun 8:15323
Rathore M, Singh R, Kumar B, Chauhan BS (2016) Characterization of functional trait diversity among Indian cultivated and weedy rice populations. Sci Rep 6:24176
Reagon M, Thurber CS, Gross BL, Olsen KM, Jia Y, Caicedo AL (2010) Genomic patterns of nucleotide diversity in divergent populations of US weedy rice. BMC Evol Biol 10:180
Saitoh K, Onishi K, Mikami I, Thidar K, Sano Y (2004) Allelic diversification at the C (OsC1) locus of wild and cultivated rice: nucleotide changes associated with phenotypes. Genetics 168:997–1007
Sasaki A, Ashikari M, Ueguchi-Tanaka M et al (2002) A mutant gibberellin-synthesis gene in rice. Nature 416:701–702
Sheen J, Zhou L, Jang JC (1999) Sugars as signaling molecules. Curr Opin Plant Biol 2:410–418
Shen H, Zhong X, Zhao F et al (2015) Overexpression of receptor-like kinase ERECTA improves thermotolerance in rice and tomato. Nat Biotechnol 33:996
Shivrain VK, Burgos NR, Gealy DR, Smith KL, Scott RC, Mauromoustakos A, Black H (2009) Red rice (Oryza sativa) emergence characteristics and influence on rice yield at different planting dates. Weed Sci 57:94–102
Shivrain VK, Burgos NR, Scott RC, Gbur EE, Estorninos LE, McClelland MR (2010) Diversity of weedy red rice (Oryza sativa L.) in Arkansas, USA in relation to weed management. Crop Prot 29:721–730
Spielmeyer W, Ellis MH, Chandler PM (2002) Semidwarf (sd-1), “green revolution” rice, contains a defective gibberellin 20-oxidase gene. Proc Natl Acad Sci USA 99:9043–9048
Stewart CN Jr (2017) Becoming weeds. Nat Genet 49:654
Subbaiyan GK, Waters DL, Katiyar SK, Sadananda AR, Vaddadi S, Henry RJ (2012) Genome-wide DNA polymorphisms in elite indica rice inbreds discovered by whole-genome sequencing. Plant Biotechnol J 10:623–634
Subudhi PK, Parco A, Singh PK et al (2012) Genetic architecture of seed dormancy in U.S. weedy rice in different genetic backgrounds. Crop Sci 52:2564–2575
Subudhi PK, Singh PK, DeLeon T et al (2014) Mapping of seed shattering loci provides insights into origin of weedy rice and rice domestication. J Hered 105:276–287
Sugimoto K, Takeuchi Y, Ebana K et al (2010) Molecular cloning of Sdr4, a regulator involved in seed dormancy and domestication of rice. Proc Natl Acad Sci USA 107:5792–5797
Sun J, Ma D, Tang L et al (2019) Population genomic analysis and de novo assembly reveal the origin of weedy rice as an evolutionary game. Mol Plant 12:632–647
Sweeney MT, Thomson MJ, Pfeil BE, McCouch S (2006) Caught red-handed: Rc encodes a basic helix-loop-helix protein conditioning red pericarp in rice. Plant Cell 18:283–294
Tan L, Li X, Liu F et al (2008) Control of a key transition from prostrate to erect growth in rice domestication. Nat Genet 40:1360–1364
Tang N, Ma S, Zong W et al (2016) MODD mediates deactivation and degradation of OsbZIP46 to negatively regulate ABA signaling and drought resistance in rice. Plant Cell 28:2161–2177
Thurber CS, Reagon M, Olsen KM, Jia Y, Caicedo AL (2014) The evolution of flowering strategies in US weedy rice. Am J Bot 101:1737–1747
Tian T, Liu Y, Yan H, You Q, Yi X, Du Z, Xu W, Su Z (2017) agriGO v2.0: a GO analysis toolkit for the agricultural community, 2017 update. Nucl Acids Res 45:W122–W129
Tong W, He Q, Park YJ (2017) Genetic variation architecture of mitochondrial genome reveals the differentiation in Korean landrace and weedy rice. Sci Rep 7:43327
Vaughan DA, Lu BR, Tomooka N (2008) Was Asian rice (Oryza sativa) domesticated more than once? Rice 1:16–24
Wang L, Hao L, Li X, Hu S, Ge S, Yu J (2009) SNP deserts of Asian cultivated rice: genomic regions under domestication. J Evol Biol 22:751–761
Xing S, Li M, Liu P (2013) Evolution of S-domain receptor-like kinases in land plants and origination of S-locus receptor kinases in Brassicaceae. BMC Evol Biol 13:69
Xu K, Xu X, Fukao T et al (2006) Sub1A is an ethylene-response-factor-like gene that confers submergence tolerance to rice. Nature 442:705–708
Yu Y, Tang T, Qian Q et al (2008) Independent losses of function in a polyphenol oxidase in rice: differentiation in grain discoloration between subspecies and the role of positive selection under domestication. Plant Cell 20:2946–2959
Zhang Z, Li J, Pan Y et al (2017) Natural variation in CTB4a enhances rice adaptation to cold habitats. Nat Commun 8:14788
Zhao Y, Chan Z, Gao J et al (2016) ABA receptor PYL9 promotes drought resistance and leaf senescence. Proc Natl Acad Sci USA 113:1949–1954
Zhao H, Wang X, Jia Y et al (2018) The rice blast resistance gene Ptr encodes an atypical protein required for broad-spectrum disease resistance. Nat Commun 9:2039
Zhou Y, Lu D, Li C et al (2012) Genetic control of seed shattering in rice by the APETALA2 transcription factor shattering abortion1. Plant Cell 24:1034–1048
Zhu BF, Si L, Wang Z et al (2011) Genetic control of a transition from black to straw-white seed hull in rice domestication. Plant Physiol 155:1301–1311
Ziska LH, McClung A (2008) Differential response of cultivated and weedy (red) rice to recent and projected increases in atmospheric carbon dioxide. Agron J 100:1259–1263
Acknowledgements
This work is supported by the National Institute of Food and Agriculture, U.S. Department of Agriculture (Grant No. 2006-35320-16555). This manuscript is approved for publication by the Director of Louisiana Agricultural Experiment Station, USA as Manuscript Number 2020-306-34605.
Author information
Authors and Affiliations
Contributions
PKS conceived and designed the experiment. MSI conducted the experiment and wrote the manuscript. MSI and SC analyzed data. PKS critically revised the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
On behalf of all authors, the corresponding author states that there is no conflict of interest.
Additional information
Communicated by Takuji Sasaki.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Islam, M.S., Coronejo, S. & Subudhi, P.K. Whole-genome sequencing reveals uniqueness of black-hulled and straw-hulled weedy rice genomes. Theor Appl Genet 133, 2461–2475 (2020). https://doi.org/10.1007/s00122-020-03611-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00122-020-03611-2