Abstract
Key message
A candidate nicosulfuron sensitivity gene Nss was identified by combining bulked segregant analysis and RNA-seq. Multiple mutations of this gene were discovered in nicosulfuron-sensitive maize compared with the tolerant.
Abstract
It has been demonstrated that variabilities exist in maize response to nicosulfuron. Two nicosulfuron-sensitive inbred lines (HB39, HB41) and two tolerant inbred lines (HB05, HB09) were identified via greenhouse and field trials. Genetic analysis indicated that the sensitivity to nicosulfuron in maize was controlled by a single, recessive gene. To precisely and rapidly map the nicosulfuron sensitivity gene (Nss), two independent F2 segregating populations, Population A (HB41 × HB09) and Population B (HB39 × HB05), were constructed. By applying bulked segregant RNA-Seq (BSR-Seq), the Nss gene was, respectively, mapped on the short arm of chromosome 5 (chr5: 1.1–15.3 Mb) and (chr5: 0.5–18.2 Mb) using two populations, with 14.2 Mb region in common. Further analysis revealed that there were 43 and 119 differentially expressed genes in the mapping intervals, with 18 genes in common. Gene annotation results showed that a cytochrome P450 gene (CYP81A9) appeared to be the candidate gene of Nss associated with nicosulfuron sensitivity in maize. Sequence analysis demonstrated that two common deletion mutations existed in the sensitive maize, which might lead to the nicosulfuron sensitivity in maize. The results will make valuable contributions to the understanding of molecular mechanism of herbicide sensitivity in maize.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Maize is one of the most important sources of energy consumed in the world because of its productive potential, nutritive value and chemical composition. Maize is planted over 35 million hectares per year in China, which is much more than any other crops. Weeds competition in the fields is one of the most important factors causing yield losses (Ferrero et al. 2017; Massinga et al. 2003). Nicosulfuron is a widely utilized postemergence herbicide in maize fields in China for its broad spectrum of activity against grasses, broadleaf weeds and sedges.
However, with the wide application of nicosulfuron, some maize cultivars were found to be severely injured. It has been reported that maize plants significantly vary in their tolerance to nicosulfuron due to genotypic variability (Meyer et al. 2010). For tolerant maize, nicosulfuron at regular application dosage may cause no injury or small yellow bands on leaves that are transient and does not affect the yield. While for sensitive maize, nicosulfuron would cause severe injury to lipids and change of ascorbate–glutathione cycle, with symptoms ranging from severe stunting of plant growth to death of whole plant (Wang et al. 2018). Herbicide-tolerant differences in plants are attributed to differential metabolism rates (Sprague et al. 1999). Tolerant maize hybrid DK689 was once demonstrated to be able to absorb 60–80% of the applied nicosulfuron within 72 h and metabolize 78–95% of that rapidly (Gallaher et al. 1999).
It is well known that cytochrome P450 monooxygenases (P450s) play an important role in the phase I metabolism of herbicides (Siminszky 2006). There were more than 300 cytochrome P450 genes in maize, which were classified into 10 clans and 44 families (Liu et al. 2018). P450s in maize were demonstrated to be able to metabolize fourteen herbicides of six chemical families through hydroxylations and demethylations (Barrett 1995). The sweet corn sensitivity to different P450-metabolized herbicides had been reported to be regulated by a single recessive cytochrome P450 gene (Nordby et al. 2008; Pataky et al. 2009). Nicosulfuron injury to maize can be reduced by its application combined with isoxadifen, a safener that can enhance the activity of P450 monooxygenases and glutathione S-transferases (Paporisch and Rubin 2017; Sun et al. 2017).
Newly developed BSR-Seq technology based on RNA-Seq data can provide both genetic mapping information and global gene expression information, and it is considered to be one of the most useful methods for mapping monogenic traits (Liu et al. 2012b; Chayut et al. 2015). Through BSR-Seq analysis, an interval on the genetic map which contains the candidate genes could be defined. Moreover, BSR-Seq enables the identification of genes differentially expressed between the mutant and non-mutant pools within the mapping interval, and it also provides clues to study the molecular functions of these genes. Using this approach, many target genes had been identified in different plant species, such as wheat (Li et al. 2018; Wu et al. 2018), cabbage (Huang et al. 2017) and onion (Kim et al. 2015). This study aimed to rapidly identify the candidate genes associated with the nicosulfuron sensitivity of maize using bulked segregant analysis and RNA-seq. In this work, a cytochrome P450 gene was identified to possibly involve in the development of nicosulfuron sensitivity in maize, which would be helpful in illuminating the relationships between nicosulfuron application and sensitivity in maize.
Materials and methods
Plant materials
HB41 and HB39 are nicosulfuron-sensitive maize inbred lines with homozygous recessive Nss, and HB09 and HB05 are nicosulfuron-tolerant types with homozygous dominant Nss (seeds available from Institute of Cereal and Oil Crops, Hebei Academy of Agriculture and Forestry Sciences, China). HB41 (♀) was crossed with HB09 (♂), and HB39 (♀) was crossed with HB05 (♂), to generate F1 populations, respectively. Then, F1 population was self-pollinated to generate two segregating F2 populations (Population A and Population B). The BC1 populations were acquired from a backcross of HB41 and HB39, respectively.
Greenhouse dose–response of parental lines to nicosulfuron
A dose–response experiment on the parental inbred lines (HB05, HB09, HB39 and HB41) was conducted in greenhouse using completely randomized design with three replicates. Five maize seeds were planted 2–3 cm deep in 15-cm plastic pots and thinned to two uniform seedlings after emergence. Day temperature was 30 ± 2 °C and night temperature was 22 ± 2 °C. Relative humidity ranged from 50 to 90%. Maize plants were well watered and fertilized for rapid growth. At V3 stage (seeds were planted for about 14 days), nicosulfuron was sprayed in a volume of 400 L ha−1 with a flat-fan nozzle at 39 cm with spray pressure set at 275 kPa. Application rates were 6.25, 12.5, 25, 50, 100 g a.i. ha−1 for HB39, HB41, and 62.5, 125, 250, 500, 1000 g a.i. ha−1 for HB05, HB09. All treatments were applied with 0.5% (v/v) methylated seed oil. The typical symptoms of maize plant response to nicosulfuron were chlorotic or necrotic leaf and stunting. At 14 days after nicosulfuron application, the percentage of injury leaf area in each plant was visually rated from 0 to 100%, and then the values for each treatment were averaged.
Nicosulfuron sensitivity phenotypic evaluation
The F1, F2 and BC1 populations, as well as their parental lines, were evaluated for responses to nicosulfuron in 2015 and 2016 growing seasons in the field of Dishang Experimental Station of Institute of Cereal and Oil Crops, Hebei Academy of Agriculture and Forestry Sciences. Randomized complete block design and three replications were adopted. In all trials, the maize seeds were planted in 8 m rows spaced 30 cm apart. Commercial formulation of nicosulfuron was applied on the maize plants at four- to five-leaf stages. Nicosulfuron was applied at 60 g a.i. ha−1 with 0.5% (v/v) methylated seed oil. A compressed-air backpack sprayer equipped with a flat-fan nozzle delivering 190 L ha−1 of water at 262 kPa was used for application. Symptoms of injury were evaluated at 7 and 14 days after application, respectively, with the same evaluated method mentioned above. The materials having 0–5%, 5–20% or 20–100% injury leaves were considered as tolerant, moderate or sensitive to nicosulfuron (Williams et al. 2005).
Bulked segregant analysis (BSA) and RNA-seq
Two independent F2 mapping populations (Population A and Population B) were generated for BSR-Seq analysis. Population A consisted of 832 plants and Population B consisted of 891 plants. The aboveground leaf tissues of the two F2 populations were collected at 20 days after planting before nicosulfuron application. The response of each plant in F2 population was investigated at 7, 14 days after treatment of 60 g a.i. ha−1 nicosulfuron, respectively. Plants grown normally without any symptoms were identified as the tolerant individuals, while the plants showing chlorosis, yellow or irregular chlorotic leaf spot and died quickly after the nicosulfuron treatment were identified as the sensitive individuals.
Two extreme pools were constructed using equal amounts of tissues from the same position of plants at same growing stage. Leaf tissue collected from the most 100 nicosulfuron-tolerant individuals and the most 100 sensitive individuals were mixed as tolerant and sensitive pools, respectively. Total RNAs were extracted from the mixed samples using an RNAiso Plus RNA extraction kit (Takara, Dalian, China). The transcriptome sequence was performed on an Illumina HiSeq 4000 platform (China Golden Marker, Beijing) with approximately 5 GB of clean data for each pool, and Q30 percentages were more than 85%.
SNP-index and Δ(SNP-index) analysis
The sequences were mapped against the latest B73 genome (AGPv4.32) using the software Bowtie version 1.1.2 (Langmead et al. 2009). Single nucleotide polymorphism (SNP) callings were processed with SAMtools (Li et al. 2009). Homozygous SNPs between parental lines and high-quality SNPs (minimum 10 total reads at the position of interest, minimum of 4 supporting reads, minimum average quality of 20) were selected for SNP-index analysis. For each genomic position, the proportion of short reads harboring SNPs with the sequence of one of the parents chosen as the reference (the so-called SNP-index) is estimated (Abe et al. 2012). Δ (SNP-index) was calculated by subtracting the SNP-index of the sensitive pool from the tolerant pool. The graph of average values of Δ(SNP-index) was plotted by calculating in a 4 Mb window size and 10 kb window step size.
Differentially expressed genes (DEGs) analysis
RNA sequencing reads were aligned to the B73 reference genome using STAR version 2.4.1d (Dobin et al. 2013), and transcript quantification was performed using kallisto version 0.43.0 (Bray et al. 2016). Differential gene expression analysis was carried out on the kallisto output abundance files using Sleuth (v. 0.28.1) (Pimentel et al. 2017). Identification of DEGs was conducted according to the method previously described (Reiner et al. 2003). The expression fold change was calculated based on the modified expression value between the tolerant and the sensitive pools. Statistical significance of differential expression was determined using multiple testing combined with p value adjusted by false discovery rate (FDR). In this study, genes with adjusted p value less than 0.01 and fold change greater than or equal to 2 were identified as DEGs. Gene ontology (GO) term annotation was conducted for functional classification of DEGs, and further functional enrichment analysis was carried out using AgriGO (plant GO slim, FDR ≤ 0.01) (Du et al. 2010).
Clone, sequence and phylogenetic analysis
Genomic DNA was extracted from the young leaves of maize inbred lines using the CTAB method. Two overlapping primer pairs (F1F: ATAGACACTGGTCAGGCTTAG, F1R: AATCCTATACTT-TGGCGGA; F2F: CTGCGCCTGCACCCG, F2R: CATACACTCGGCGAAAATCAACTA) covering full length of the candidate gene were used for amplification in both nicosulfuron-tolerant and nicosulfuron-sensitive maize inbred lines. PCR conditions were as follows: 95 °C for 5 min; 35 cycles of 95 °C for 30 s, 55 °C for 30 s and 72 °C for 50 s; and a final extension at 72 °C for 10 min. The product was gel purified with AxyPrep Gel DNA Extraction Kit (Axygen, China) and cloned into the pMD19-T vector (TaKaRa, Dalian, China) for sequencing.
Alignment of cytochrome P450 protein sequences was performed by ClustalX program. The phylogenetic tree of plant cytochromes P450 proteins was constructed with the MEGA program (version 4.0) by the neighbor joining (NJ) method with 1000 bootstraps (Tamura et al. 2007).
Results
Genetic analysis of nicosulfuron sensitivity in maize
Dose–response analysis showed that there were significant differences in the nicosulfuron sensitivity symptoms between tolerant (HB05, HB09) and sensitive inbred lines (HB39, HB41) of maize (Fig. 1a), which was further confirmed by our field trials. For HB05 and HB09, no injury symptom showed when nicosulfuron was applied at 125 g a.i. ha−1 (about 2 × recommended use rates). While for HB39 and HB41, the injury ranged from 30 to 50% when nicosulfuron was applied at 6.25 g a.i. ha−1 (about 1/10 × recommended use rates).
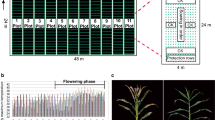
Response of maize to nicosulfuron. a Dose–response of tolerant and sensitive maize to nicosulfuron in greenhouse. b Field responses of the F2 population derived from ‘HB41 × HB09’ after the application of 60 g a.i. ha−1 nicosulfuron. The photographs were taken at 14 days after nicosulfuron treatment. The sensitive plants were totally dead, while the tolerant plants grew normally
The segregation patterns of nicosulfuron sensitivity trait in the F1, F2 and BC1 generations from the crosses of ‘HB39 × HB05’ and ‘HB41 × HB09’ are presented in Table 1. All F1 offsprings were nicosulfuron tolerant, while the F2 generations of ‘HB39 × HB05’ and ‘HB41 × HB09’ segregated 626, 652 tolerant plants and 194, 226 sensitive plants, with χ2 value of 0.79, 0.26 and p value of 0.37, 0.61, respectively (Table 1, Fig. 1b). The two BC1 generations segregated 136, 152 tolerant plants and 118, 144 maize plants, with χ2 value of 1.28, 0.22 and p value of 0.26, 0.64, respectively. The F2 and BC1 generations segregated to tolerant and sensitive in the expected 3:1 and 1:1 ratio. All of these results suggested that nicosulfuron sensitivity in maize was conferred by a single recessive gene. Complementation tests involving the crosses of HB39 and HB41 showed that the F1 progeny was also sensitive to nicosulfuron, which implies that the nicosulfuron sensitivity in HB39 and HB41 is due to the alleles at a common locus.
Preliminary mapping of Nss gene via BSR-Seq
To map the Nss gene in maize, two independent F2 segregating populations, Population A and Population B, were comparatively analyzed using BSR-seq. For each segregating population, leaf tissues from siblings showing nicosulfuron-tolerant and nicosulfuron-sensitive phenotypes were collected and mixed into two separate pools and used for RNA sequencing. RNA-Seq reads were trimmed and aligned to the B73 reference genome. A total of 8.9, 7.6 Gb and 7.6, 7.5 Gb of raw bases were obtained from the tolerant and sensitive bulked samples for A and B, respectively, with the percentage of bases (Q30) over 95%. SNPs were then identified between tolerant and sensitive pools via comparisons of RNA-Seq reads to the B73 reference genome. Via analysis, 83,279 and 73,577 SNPs were identified from Population A and Population B, with 21,485 in common. SNP-index was calculated for each identified SNP. An average SNP-index was computed in a 4-Mb interval using a 10-kb sliding window. By combining the information of SNP-index in the tolerant pool and sensitive pool, Δ(SNP-index) was calculated and plotted against the genome positions (Fig. 2). Associated chromosomal regions were identified with |Δ(SNP-index)| > 0.5 and Fisher’s exact test p value < 0.01. The Nss gene with a high probability of complete linkage disequilibrium was located to a 14.2 Mb region on the short arm of chromosome 5 (chr5: 1.1–15.3 Mb) in the F2 segregating Population A, which contained 706 genes in the filtered gene set. The mapping interval detected in Population B was a little larger, a 17.7 Mb region on the short arm of chromosome 5 (chr5: 0.5–18.2 Mb), which contained 837 genes. The two located regions in Population A and Population B overlapped 14.2 Mb region. Therefore, we concluded that the 1.1–15.3 Mb (14.2 Mb) genomic region in chromosome 5 may harbor the candidate Nss gene.
Δ (SNP-index) graph from BSR-Seq analysis of 10 maize chromosomes. Each chromosome was scanned by using a 4-Mb window containing at least 20 SNPs with a step size of 10 kb. Within each window, the median linkage probability obtained from a Bayesian BSA analysis across all the 20 SNPs was determined and plotted against the middle physical position of the window. a Population A; b Population B. The mapping interval was marked with the shade rectangle
Identification of DEGs and potentially SNPs in the mapping regions
Based on the latest B73 genome (AGPv4.32), a total of 136 and 1918 genes were identified to display significantly differential expression (fold change ≥ 2 and FDR < 0.01) in the tolerant and sensitive pools of Population A and Population B, with 33 genes in common. A total of 2355 and 2682 SNPs were detected in the mapping regions of Population A and Population B, respectively, with 581 SNPs in common. GO slim term analysis showed that DEGs in Population A and Population B were all significantly (p < 0.05) enriched to cellular homeostasis (GO: 0042254) and response to external stimulus (GO: 0009605) within biological process, and catalytic activity within molecular function (GO:0003824) (Fig. 3a). Further analysis showed that there were 43 and 119 DEGs in the 14.2 and 17.7 Mb mapping intervals, respectively, with 18 genes in common (Fig. 3b). A total of 30 potentially deleterious SNPs were detected in the candidate genes in both populations (Table 2). Among these 18 identical DEGs, only one cytochrome P450 gene, designated as Zm00001d013230, was hypothesized to be associated with nicosulfuron metabolism in maize (Table 3). In our previous study, we found that the expression level of Zm00001d013230 in tolerant maize was much higher than that in the sensitive and could be induced by nicosulfuron in both the tolerant and sensitive maize (Liu et al. 2015). Considering that cytochrome P450s are heme proteins and play an important role in detoxification of herbicides, we concluded Zm00001d013230 is the primary candidate for the nicosulfuron sensitivity gene (Nss).
Analysis of differentially expressed genes (DEGs). a GO slim term enrichment analysis of the DEGs in Population A and Popuation B. The size of the dot is relative to the ratio of DEGs number in that GO term. P value values were color-coded. BP, biological process; MF, molecular function; CC, cellular component. b Venn diagram of the DEGs in the two F2 mapping populations (color figure online)
Clone, sequence and phylogenetic analysis of Zm00001d013230
Sequence analysis of Zm00001d013230 in the genome of the maize inbred line B73 revealed that it encodes a cytochrome P450 named as CYP81A9. BLAST results showed that the deduced amino acid sequence of CYP81A9 was highly similar to that of CYP81A12 (84% identity and 78% similarity) and CYP81A21 (84% identity and 77% similarity) from Echinochloa phyllopogon (Iwakami et al. 2014). The CYP81A9 sequence also showed similarity to the other CYP81 family proteins (Fig. 4a), such as CYP81A5, CYP81A6, CYP81A7, CYP81A8 (Pan et al. 2006), CYP81B1 (Cabello-Hurtado et al. 1998), CYP81B2, CYP81C1 (Yamada et al. 2000), CYP81D8 and CYP81D11 (Narusaka et al. 2004). The CYP81A9 gene was then isolated from the genomic DNA of nicosulfuron-tolerant and nicosulfuron-sensitive maize via PCR amplification. The protein sequences involved in 521 amino acids in inbred lines HB05 and HB09 are identical to that in B73 once confirmed to be tolerant to sulfonylurea herbicides (Green and Ulrich 1993). Compared with nicosulfuron-tolerant lines, nicosulfuron-sensitive inbred line GA209 was once reported to contain a 392-base pair insertion that resulted in a frameshift and an open reading frame of only 338 amino acids (Jerald et al. 2008). Compared with the tolerant lines, the conserved cytochrome P450 monooxygenases heme-binding motifs ‘F × ×GPR × C×G’ were identified in HB39 and HB41, and three deletion, one insertion and several point mutations were also confirmed (Fig. 4b).
Analysis of the cytochrome P450 CYP81A9. a The phylogenetic relationship between CYP81A9 and other CYP81 family proteins in plants. Sequences were aligned by ClustalX and analyzed using the MEGA 6.0. Phylogenetic tree was constructed by the parsimony method with 1000 bootstrap replicates. Database accession numbers are as follows: Echinochloa phyllopogon (CYP81A12, BAO73908.1; CYP81A21, BAO73910.1), Zea mays (CYP81A9, ACG28028.1), Oryza sativa (CYP81A5, OSJNBb0048A17.17; CYP81A6, OSJNBb0048A17.18; CYP81A7, OSJNBb0048A17.19; CYP81A8, OSJNBb0048A17.4), Helianthus tuberosus (CYP81B1, CAA04116.1), Nicotiana tabacum (CYP81B2, BAH84782.1; CYP81C1, BAH84784.1) and Arabidopsis thaliana (CYP81D8, OAO99970.1; CYP81D11, OAP05302.1). b Alignment of the deduced amino acid sequences of CYP81A9 from nicosulfuron-tolerant and nicosulfuron-sensitive maize. The red rectangle represents the conserved heme-binding motifs in cytochrome P450 monooxygenases (color figure online)
Discussion
Nicosulfuron is the most frequently utilized postemergence herbicide in maize fields in China because of its high herbicidal activity. Its injury symptoms are often observed in maize fields, such as stunting of the plant, crinkled leaves and bleaching near the whorl. Several types of maize such as sweet corn, waxy corn and popcorn were sensitive to nicosulfuron, which has been reported in the past 20 years (Wang et al. 2018; Williams et al. 2005). In this study, two nicosulfruon-tolerant inbred lines (HB05, HB09) and two sensitive inbred lines (HB39, HB41) were identified among more than 300 inbred lines used for field maize breeding in China. Inheritance analysis results showed that the nicosulfuron sensitivity was conditioned by a single recessive gene, which was consistent with previous researches (Kang 1993; Nordby et al. 2008).
Nicosulfuron sensitivity gene in maize was firstly mapped to the short arm of chromosome 7 via B-A translocational analysis (Moreno 1999). Then, it was located on the short arm of chromosome 5 using microsatellite markers and was reported to condition maize response to multiple P450-metabolized herbicides (Nordby et al. 2008). In the present research, we adopted the newly developed BSR-Seq technology to map the Nss gene in maize using two independent F2 segregating populations. SNP-index analysis showed that the Nss gene was located on almost the same mapping intervals on the short arm of chromosome 5 in both populations, with 14.2 Mb region in common.
Sequencing depth, density of polymorphisms and number of individuals included in the mutant and non-mutant pools are considered to be the most important factors that affect the size and precision of the mapping interval (Liu et al. 2012b). In our study, we collected the tissues from 100 individuals for each pool and adopted two independent F2 segregating populations to increase the probability of locating Nss gene correctly. Precise and thorough evaluation of phenotypes is the prerequisite for mapping candidate gene associated with target trait. Nicosulfuron sensitivity identification of maize plants can be influenced by environmental factors such as temperature, humidity and plant growing stage. We precisely recorded the response of each plant in Population A and B after nicosulfuron treatment. Individuals had no injury symptoms, together with those showed nicosulfuron injury symptoms and finally died were selected for BSR-Seq analysis. The two identical mapping intervals in population A and B indicated that our measurement for nicosulfuron sensitivity in maize was reliable.
Cytochrome P450 monooxygenases are well known for their involvement in the metabolism of xenobiotics including herbicides (Nelson and Werck-Reichhart 2011). In the mapping interval of population A and B, we identified 18 differentially expressed genes in common, including CYP81A9 which was considered to be the candidate gene for conferring nicosulfuron sensitivity in maize. Many CYP81 subfamily genes in other plants have been reported to be involved in herbicide metabolism. Expression of the rice CYP81A6 gene was testified that it confers tolerance to bentazon and sulfonylurea herbicides (Liu et al. 2012a). CYP81B1 in Helianthus tuberosus was confirmed that it can catalyze ringmethyl hydroxylation of chlortoluron (Cabello-Hurtado et al. 1998). CYP81A12 and CYP81A21 in Echinochloa phyllopogon were associated with its resistance to bensulfuron-methyl and penoxsulam (Iwakami et al. 2014).
It has been known that the maize sensitivity to certain P450-metabolized herbicides is largely controlled by a single CYP gene of Nsf1, while only a few reports showed there was a 392-base pair insertion in the Nsf1 gene, which resulted in a non-functional CYP alleles and the sensitivity to multiple P450-metabolized herbicides (Williams et al. 2006; Jerald et al. 2008). Our results showed that the Nss gene appeared to be the mutation of Nsf1 and the 392-base pair insertion was not found in both sensitive inbred lines, so we concluded that there were some other reasons relative to nicosulfuron sensitivity in maize. As sequence alignments showed that there were two common deletion mutations in all the sensitive inbred lines, future work needed to confirm which mutations caused the nicosulfuron sensitivity in maize.
To date, the majority of attention has been focused on target-site herbicide resistance mechanisms in plant, while less is known about the non-target-site herbicide resistance acquisition mechanisms in crops and weeds. Cytochromes P450 monooxygenases played one of the most important role in the non-target-site resistance mechanism, and many CYP genes can be used for herbicide tolerance engineering (Werck-Reichhart et al. 2000). In this study, a candidate cytochrome P450 gene CYP81A9 was identified to be involved in nicosulfuron metabolism in maize, and several mutation sites were also found in nicosulfuron-sensitive maize. It has been demonstrated that maize plant would be killed by spraying nicosulfuron at a regular application dosage when researchers suppressed the expression of CYP81A9 gene via RNA interference technology (Li et al. 2013), which strongly confirmed the close relationship between CYP81A9 and nicosulfuron sensitivity in maize. Further transgenic research would be conducted to elucidate the function of CYP81A9, and it will provide a new insight into its possible interactions with herbicides at the molecular level.
References
Abe A, Kosugi S, Yoshida K, Natsume S, Takagi H et al (2012) Genome sequencing reveals agronomically important loci in rice using MutMap. Nat Biotechnol 30:174–178
Barrett M (1995) Metabolism of herbicides by cytochrome P450 in corn. Drug Metab Drug Interact 12:299–316
Bray NL, Pimentel H, Melsted P, Pachter L (2016) Near-optimal probabilistic RNA-seq quantification. Nat Biotechnol 34:525–527
Cabello-Hurtado F, Batard Y, Salaün J-P, Durst F, Pinot F et al (1998) Cloning, expression in yeast, and functional characterization of CYP81B1, a plant cytochrome P450 that catalyzes in-chain hydroxylation of fatty acids. J Biol Chem 273:7260–7267
Chayut N, Yuan H, Ohali S, Meir A, Yeselson Y et al (2015) A bulk segregant transcriptome analysis reveals metabolic and cellular processes associated with Orange allelic variation and fruit β-carotene accumulation in melon fruit. BMC Plant Biol 15:274
Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C et al (2013) STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29:15–21
Du Z, Zhou X, Ling Y, Zhang Z, Su Z (2010) agriGO: a GO analysis toolkit for the agricultural community. Nucleic Acids Res 38:W64–W70
Ferrero R, Lima M, Davis AS, Gonzalez-Andujar JL (2017) Weed diversity affects soybean and maize yield in a long term experiment in Michigan, USA. Front Plant Sci 8:236
Gallaher K, Mueller TC, Hayes RM, Schwartz O, Barrett M (1999) Absorption, translocation, and metabolism of primisulfuron and nicosulfuron in broadleaf signalgrass (Brachiaria platyphylla) and corn. Weed Sci 47:8–12
Green JM, Ulrich JF (1993) Response of corn (Zea mays L.) inbreds and hybrids to sulfonylurea herbicides. Weed Sci 41:508–516
Huang Z, Peng G, Liu X, Deora A, Falk KC et al (2017) Fine mapping of a clubroot resistance gene in Chinese cabbage using SNP markers identified from bulked segregant RNA sequencing. Front Plant Sci 8:1448
Iwakami S, Endo M, Saika H, Okuno J, Nakamura N et al (2014) Cytochrome P450 CYP81A12 and CYP81A21 are associated with resistance to two acetolactate synthase inhibitors in Echinochloa phyllopogon. Plant Physiol 165:618–629
Jerald KP, Michael DM, Joseph DB, Chris MB, Martin MW (2008) Genetic Basis for varied levels of injury to sweet corn hybrids from three cytochrome P450-metabolized herbicides. J Am Soc Hort Sci 133:438–447
Kang M (1993) Inheritance of susceptibility to nicosulfuron herbicide in maize. J Hered 84:216–217
Kim S, Kim C-W, Park M, Choi D (2015) Identification of candidate genes associated with fertility restoration of cytoplasmic male-sterility in onion (Allium cepa L.) using a combination of bulked segregant analysis and RNA-seq. Theor Appl Genet 128:2289–2299
Langmead B, Trapnell C, Pop M, Salzberg SL (2009) Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10:R25
Li H, Handsaker B, Wysoker A, Fennell T, Ruan J et al (2009) The sequence alignment/map format and SAMtools. Bioinformatics 25:2078–2079
Li J, Yu H, Zhang F, Lin C, Gao J, Fang J, Ding X, Shen Z, Xu X (2013) A built-in strategy to mitigate transgene spreading from genetically modified corn. PLoS ONE 8(12):e81645
Li M, Li B, Guo G, Chen Y, Xie J et al (2018) Mapping a leaf senescence gene els1 by BSR-Seq in common wheat. Crop J 6:236–243
Liu C, Liu S, Wang F, Wang Y, Liu K (2012a) Expression of a rice CYP81A6 gene confers tolerance to bentazon and sulfonylurea herbicides in both Arabidopsis and tobacco. Plant Cell. Tissue Organ Cult (PCTOC) 109:419–428
Liu S, Yeh C-T, Tang HM, Nettleton D, Schnable PS (2012b) Gene mapping via bulked segregant RNA-Seq (BSR-Seq). PLoS ONE 7:e36406
Liu X, Xu X, Li B, Wang X, Wang G et al (2015) RNA-seq transcriptome analysis of maize inbred carrying nicosulfuron-tolerant and nicosulfuron-susceptible alleles. Int J Mol Sci 16:5975–5989
Liu XM, Xu X, Li BH, Yao XX, Zhang HH et al (2018) Genomic and transcriptomic insights into cytochrome P450 monooxygenase genes involved in nicosulfuron tolerance in maize (Zea mays L.). J Integr Agric 17:1790–1799
Massinga RA, Currie RS, Trooien TP (2003) Water use and light interception under Palmer amaranth (Amaranthus palmeri) and corn competition. Weed Sci 51:523–531
Meyer MD, Pataky JK, Williams MM (2010) Genetic factors influencing adverse effects of mesotrione and nicosulfuron on sweet corn yield. Agron J 102:1138–1144
Moreno OJ (1999) Chromosomal Location of Genes for Leafiness (Lfy1) and Susceptibility to Nicosulfuron (Nsf1) in Maize Genome. Dissertation, Louisiana State University
Narusaka M, Seki M, Umezawa T, Ishida J, Nakajima M et al (2004) Crosstalk in the responses to abiotic and biotic stresses in Arabidopsis: analysis of gene expression in cytochrome P450 gene superfamily by cDNA microarray. Plant Mol Biol 55:327–342
Nelson D, Werck-Reichhart D (2011) A P450-centric view of plant evolution. Plant J 66:194–211
Nordby JN, Williams MM, Pataky JK, Riechers DE, Lutz JD (2008) A common genetic basis in sweet corn inbred Cr1 for cross sensitivity to multiple cytochrome P450-metabolized herbicides. Weed Sci 56:376–382
Pan G, Zhang X, Liu K, Zhang J, Wu X et al (2006) Map-based cloning of a novel rice cytochrome P450 gene CYP81A6 that confers resistance to two different classes of herbicides. Plant Mol Biol 61:933–943
Paporisch A, Rubin B (2017) Isoxadifen safening mechanism in sweet corn genotypes with differential response to P450-metabolized herbicides. Pestic Biochem Physiol 138:22–28
Pataky JK, Williams MM, Riechers DE, Meyer MD (2009) A common genetic basis for cross-sensitivity to mesotrione and nicosulfuron in sweet corn hybrid cultivars and inbreds grown throughout North America. J Am Soc Hortic Sci 134:252–260
Pimentel H, Bray NL, Puente S, Melsted P, Pachter L (2017) Differential analysis of RNA-seq incorporating quantification uncertainty. Nat Methods 14:687
Reiner A, Yekutieli D, Benjamini Y (2003) Identifying differentially expressed genes using false discovery rate controlling procedures. Bioinformatics 19:368–375
Siminszky B (2006) Plant cytochrome P450-mediated herbicide metabolism. Phytochem Rev 5:445–458
Sprague CL, Penner D, Kells JJ (1999) Physiological basis for tolerance of four Zea mays hybrids to RPA 201772. Weed Sci 47:631–635
Sun L, Wu R, Su W, Gao Z, Lu C (2017) Physiological basis for isoxadifen-ethyl induction of nicosulfuron detoxification in maize hybrids. PLoS ONE 12:e0173502
Tamura K, Dudley J, Nei M, Kumar S (2007) MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol 24:1596–1599
Wang J, Zhong X, Li F, Shi Z (2018) Effects of nicosulfuron on growth, oxidative damage, and the ascorbate-glutathione pathway in paired nearly isogenic lines of waxy maize (Zea mays L.). Pestic Biochem Physiol 145:108–117
Werck-Reichhart D, Hehn A, Didierjean L (2000) Cytochromes P450 for engineering herbicide tolerance. Trends Plant Sci 5:116–123
Williams MM, Pataky JK, Nordby JN, Riechers DE, Sprague CL et al (2005) Cross-sensitivity in sweet corn to nicosulfuron and mesotrione applied postemergence. HortScience 40:1801–1805
Williams M, Sowinski S, Dam T, Li BL (2006). Map-based cloning of the nsf1 gene of maize. In: 48th Maize genetics conference, p 49
Wu P, Xie J, Hu J, Qiu D, Liu Z et al (2018) Development of molecular markers linked to powdery mildew resistance gene Pm4b by combining SNP discovery from transcriptome sequencing data with bulked segregant analysis (BSR-Seq) in wheat. Front Plant Sci 9:95
Yamada T, Kambara Y, Imaishi H, Ohkawa H (2000) Molecular cloning of novel cytochrome P450 species induced by chemical treatments in cultured tobacco cells. Pestic Biochem Physiol 68:11–25
Acknowledgements
This research was financially supported by the National Natural Science Foundation of China (31501660), the National Key R&D Program of China (2018YFD0200600) and the Technology Research and Development Program of Hebei, China (17226507D).
Author information
Authors and Affiliations
Contributions
Author contribution statement
XL and GW designed the experiments and drafted the manuscript; XL, BB, XX and HZ carried out the experiments and analysis of RNA-seq; BL, ST and JW carried out the phenotyping; GW, YH and JSM conceived the idea of the study and finalized the manuscript. All of the authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare no conflict of interest.
Additional information
Communicated by Michael Gore.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Liu, X., Bi, B., Xu, X. et al. Rapid identification of a candidate nicosulfuron sensitivity gene (Nss) in maize (Zea mays L.) via combining bulked segregant analysis and RNA-seq. Theor Appl Genet 132, 1351–1361 (2019). https://doi.org/10.1007/s00122-019-03282-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00122-019-03282-8