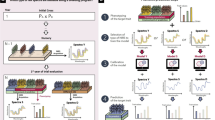
Abstract
Key message
Using NIR and NMR predictions of quality traits overcomes a major barrier for the application of genomic selection to accelerate improvement in grain end-use quality traits of wheat.
Abstract
Grain end-use quality traits are among the most important in wheat breeding. These traits are difficult to breed for, as their assays require flour quantities only obtainable late in the breeding cycle, and are expensive. These traits are therefore an ideal target for genomic selection. However, large reference populations are required for accurate genomic predictions, which are challenging to assemble for these traits for the same reasons they are challenging to breed for. Here, we use predictions of end-use quality derived from near infrared (NIR) or nuclear magnetic resonance (NMR), that require very small amounts of flour, as well as end-use quality measured by industry standard assay in a subset of accessions, in a multi-trait approach for genomic prediction. The NIR and NMR predictions were derived for 19 end-use quality traits in 398 accessions, and were then assayed in 2420 diverse wheat accessions. The accessions were grown out in multiple locations and multiple years, and were genotyped for 51208 SNP. Incorporating NIR and NMR phenotypes in the multi-trait approach increased the accuracy of genomic prediction for most quality traits. The accuracy ranged from 0 to 0.47 before the addition of the NIR/NMR data, while after these data were added, it ranged from 0 to 0.69. Genomic predictions were reasonably robust across locations and years for most traits. Using NIR and NMR predictions of quality traits overcomes a major barrier for the application of genomic selection for grain end-use quality traits in wheat breeding.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Grain end-use quality traits, such as milling yield, dough rheology, baking, and noodle traits are among the most important in wheat breeding. However, these traits are difficult to breed for as their assays require flour quantities that can only be obtained late in the breeding cycle, and are expensive. These traits are therefore an ideal target for genomic selection, where traits are predicted for candidate wheat lines early in the breeding cycle using genome-wide marker effects for such traits (e.g. Guzman et al. 2016). As for other traits, the key parameters determining the accuracy of genomic predictions for grain end-use quality traits will be the size of the reference (or training) population where the marker effects are estimated, the extent of linkage disequilibrium between markers and the mutations (QTL) affecting the trait, and the heritability of the trait (Daetwyler et al. 2008; Goddard 2009). Traits with low heritability require much larger reference populations to achieve the same accuracy of prediction as traits with moderate or high heritability.
The heritability of grain end-use quality traits in wheat that have been reported encompass a wide range. For example, O’Brien and Ronalds (1987) reported broad-sense heritabilities in Australian wheat varieties from 0.15 for farinograph measured dough development time, to 0.88 for grain hardness, with an average heritability of 0.54 across 15 traits measured. Pearson et al. (1981) reported that, in early segregating generations (F2, and F3) of a genetically diverse wheat population, pearling resistance, Pelshenke time and 1000 kernel weight had high heritabilities (0.72–0.80), flour yield had an intermediate heritability (0.57) while grain protein content had a low heritability (0.19). Baker et al. (1971) assessed the heritability of 25 grain end-use quality traits in Canadian hard red spring wheat cultivars. Heritabilities ranged from 0.26 for diastatic activity to 0.89 for pigment content, with average heritabilities of milling, flour, farinograph, extensograph and baking traits of 0.66, 0.69, 0.71 and 0.72, respectively. All these estimates are broad-sense heritabilities, while the key parameter for determining the accuracy of genomic selection for cumulative additive genetic gain, is the narrow-sense (additive component) heritability. By using a diallele cross of parents and assessing grain end-use quality characteristics in their F2 progeny, Barnard et al. (2002) were able to determine narrow-sense heritabilities for a range of traits, and compare these to broad-sense heritability estimates. Narrow-sense heritability estimates were on average half that of the broad-sense heritabilities, and ranged from 0 for falling number to 0.71 for 1000 kernel mass. The average narrow-sense heritability was 0.3. While lower than broad-sense heritabilities, 0.3 is more than sufficient to derive genomic breeding values with useful accuracy, as has been demonstrated in species ranging from loblolly pine, rice, cassava, meat and wool sheep and dairy cattle (Resende et al. 2012; Spindel et al. 2015; Ly et al. 2013; Daetwyler et al. 2012; Wiggans et al. 2011).
While the heritability of grain end-use quality traits is unlikely to be the limiting factor in genomic prediction of end-use quality, small reference population size may be. This size of reference set required depends on the genetic diversity, or effective population size of the selection candidates. If the goal is to predict within bi-parental populations, quite small reference populations may be sufficient. Heffner et al. (2011) compared the phenotypic and genomic prediction accuracy of genetic value for nine different grain quality traits within two bi-parental soft winter wheat (Triticum aestivum L.) populations, with only 96 lines in each population. They observed accuracies in cross-validation ranging from 0.36 for softness to 0.68 for pre-harvest sprouting, with an average of 0.52 across traits, using a BLUP method of genomic prediction (e.g. Meuwissen et al. 2001). This is a useful level of accuracy; however, genomic predictions derived from one bi-parental population are very unlikely to work in another, that is the prediction equation will be unique to each set of crosses. In order to derive genomic predictions that are accurate across more diverse material, much larger reference sets will be required. For example, Battenfield et al. (2016) assessed the accuracy of genomic prediction for grain end-use quality traits in advanced lines of the CIMMYT spring bread wheat breeding program. Using a much larger reference population (reference set = 3659 lines grown from 2009 to 2014, validation set = 1345 lines grown in 2014), they achieved similar accuracies, which ranged from 0.26 for grain hardness to 0.65 for mixograph mix time.
Assembling large reference sets for grain end-use quality traits is particularly challenging, due to their assays requiring large amounts of flour and being expensive. Historically, such data have been collected only on a limited number of lines. Consequently, predictors of grain end-use quality, such as near infrared (NIR) (e.g. Delwiche et al. 1998) and nuclear magnetic resonance (NMR) (e.g. Chambers et al. 1989) may provide a solution. These techniques require much smaller quantities of flour and can have substantially lower cost. One potential approach to building a reference set large enough for accurate genomic predictions of end-use quality would be to develop predictions for these traits based on NIR and/or NMR, assess a large number of lines with these assays, and combine with available end-use quality data (from industry standard assays) in a multi-trait analysis.
Our aim was to assess the accuracy of genomic prediction for end-use quality (19 traits) that could be achieved with this approach, using as a reference a large number of diverse wheat accessions, grown across multiple sites and multiple years, that had either industry end-use assays (398 accessions), or NIR and NMR predictions of these traits (2076 accessions).
Materials and methods
Germplasm
The reference population for genomic predictions included 2076 bread wheat accessions from
-
bread wheat accessions representing worldwide germplasm from Australian, Canadian and USA varieties with known end-use characteristics,
-
Australian released varieties from the Australian winter cereal collection with known grain quality and end-use characters,
-
Dow AgroSciences breeding germplasm, and
-
a number of synthetic derivatives (derived from backcrossing a subset of the primary synthetics, described in van Ginkel and Ogbonnaya 2006, to adapted Australian wheat varieties) lines for T. aestivum × Aegilops triuncialis crosses.
In 2012–2013, 920 accessions from the above were grown in rows under rain-fed field conditions at Horsham, Victoria, Australia. In 2013–2014, 1500 accessions were grown under irrigated and rain-fed conditions at Horsham Victoria, Australia.
The validation set for genomic predictions included subsets of the above (defined below), as well as separate validations released varieties grown in national National variety trials (NVT, http://www.nvtonline.com.au/). The NVT trails included multiple evaluation sites across Australia.
The location and number of lines included in the reference and validation at each location are presented in Fig. 1.
Location of grow-out for accessions evaluated for end-use quality traits. Sites other than Horsham were Australian National Variety trial sites. The size of the circle at each location and in each colour represents the number of accessions evaluated in that year. Actual numbers of accessions at each site in each year are given in Supplementary Table 1
Genotypes
All accessions in the reference set were genotyped with the Illumina 90 K wheat array, as described in Wang et al. (2014). After filtering to remove SNP with high missing data, low minor allele frequency control, filtering on quality score, and assessment of accuracy of imputation of missing genotypes, 51 208 genetically mapped SNP remained for genomic predictions. Missing genotypes (10% of the data) were imputed with Beagle v3 (Browning and Browning 2009). Default parameters were used, and the map used for the Beagle imputation was the genetic map described by Wang et al. (2014). Missing genotypes were imputed with an average r 2 of 0.85.
Phenotypes
End-use quality traits
398 accessions were evaluated for 19 end-use quality assays, including grain traits, milling traits, dough rheology traits, and baking traits, Table 1. The traits were evaluated using the methods described in Panozzo et al. (2014) and AACC 76-21 (AACC International Approved Methods, General Pasting Method for Wheat or Rye Flour or Starch Using the Rapid Visco Analyser (2000)), and CCD 07-06 (2010) Yellow Alkaline Noodles, Supplement Official Testing Methods of the Cereal Chemistry Division. The accessions evaluated included the varieties obtained from the Australian National Variety Trials, and a subset of the accessions grown at Horsham. Collectively, these accessions covered the spectrum of wheat quality classes.
NIR traits
The grain samples were cleaned over a 2.0-mm sieve. The reflectance spectra, Log (1/R), were collected on the clean grain using a NIR Systems XDS (Foss Pacific Pty Ltd, Denmark) equipped with transport module. Spectra were recorded across a range of 400–2498 nm with 0.5-nm wavelength increment. Diffuse reflectance readings of a ceramic tile were referenced before and after the sample scan. The wheat samples were equilibrated to 21 °C for 24 h prior to analysis.
The collected Vis–NIR spectra were corrected for scatter with standard normal variance (SNV) and de-trending. Mathematical treatments included the following: for the grain and milling traits, first-order derivative with gap of four and smooth size of four data points on NIR spectra; for dough rheology and baking traits, second-order derivative with gap of five and smooth size of five data points on NIR spectra; for flour colour and noodle colour stability traits, first-order derivative with gap of four and smooth size of four data points on Vis–NIR spectra. Calibrations were developed by using modified partial least squares (mPLS) algorithm and cross-validation technique. WinISI software V 4.6.11.14874 (Infrasoft International LLC, USA) was used for all data processing. The r 2 of the NIR prediction equation, assessed with cross-validation, and standard errors in the reference population, is given in Table S1.
NMR traits
Grain samples (50 individual undamaged seed) were ground (and a subsample (25 mg) extracted in deuterated solvent [CD3OD–D2O (1 mL, 80:20, v/v)]). The sample was vortexed, then sonicated for 10 min and centrifuged at 13,000 rpm for 5 min at 20 °C. The supernatant (600 μL) was transferred to an NMR tube. Proton spectra were obtained on a Bruker 700 MHz instrument equipped with a cryoprobe. A Bruker pulse sequence was used 18 ppm spectral range with 80 scans collected after 8 dummy scans. A line broadening of 0.3 Hz was applied to all spectra prior to Fourier transformation. Spectra were manually phased, and baseline corrected in Topspin 3.1. Samples were referenced to residual methanol (3.31 ppm). Data were analysed in MATLAB R2014b (The Mathworks, Inc.) using PLStoolbox (Ver 9.2, Eigenvector Research). The data were pre-processed removing solvent signal and normalising to total signal area prior to genetic algorithm variable selection, using the align peaks and align spectra packages. Calibrations were developed from the lab quality set (398 accessions) using partial least squares regression (SIMPLS package) on autoscaled data with venetian blinds cross-validation (using the crossval package in the PLStoolbox). The venetian blinds cross-validation procedure divides the data up into n folds, then runs n cross-validations with a fold left out at each point. Predictions were tested against 20% of the calibration data which was withheld from initial modelling (e.g. fivefold cross-validation). Mean square error was the criteria used to assess predictions. Permutation testing (50 iterations) was also employed to ensure statistical robustness (using the Permutetest package). Resultant calibrations were then applied to the spectra from the remaining accessions to predict end-use quality data. The spectra were complex, containing a mix of both small and large water-soluble carbohydrates, organic acids, amino acids and phenolics. Starch was not extracted using this methodology. The r 2 of the NMR prediction equation, assessed with cross-validation, and standard errors in the reference population, is given in Table S2.
Genomic heritability and genotype by environment interactions
The model fitted to the end-use quality data was
where y is a vector of quality phenotypes, 1 n is a (number of phenotypes × 1) vector of ones, days heading , days grain filling , days physiological maturity , were fixed effects (covariates), respectively, representing the number of days the accession took to flower in the location and year the phenotype was measured in, the number of days the accession took to complete grain filling in the location and year the phenotype was measured in, and the number of days the accession took to reach physiological maturity in the location and year the phenotype was measured in, location is a fixed effect for the location of growing, year is the fixed effect for year (2011, 2012 or 2013) the accession was grown in, hard or soft is a fixed effect for hard versus soft wheats, and e is a random error term. Accession, accession × location, accession by year, and accession × location × year were fitted as random effects, assumed normally distributed with a mean of zero, and variance the genetic variance (\(\sigma_{\it{G}}^{2}\)) and genotype × location variance (\(\sigma_{G \times L}^{2}\)), genotype by year variance (\(\sigma_{G \times Y}^{2}\)), and genotye by location by year variance (\(\sigma_{G \times L \times Y}^{2}\)). The co(variances) between accessions were assumed to be zero in the first analysis, to calculate broad-sense heritabilities, and G in the analysis to estimate genomic heritabilities. The (accession × accession) matrix G was constructed from the 51208 SNP genotypes as described by VanRaden (2008). Broad-sense heritability was calculated as \(H^{2} = \frac{{\sigma_{G}^{2} }}{{\sigma_{G}^{2} + \frac{{\sigma_{G \times L}^{2} }}{L} + \frac{{\sigma_{G \times Y}^{2} }}{Y} + \frac{{\sigma_{G \times Y \times L}^{2} }}{LY} + \frac{{\sigma_{E}^{2} }}{LYR}}},\) where \(\sigma_{G}^{2}\) and \(\sigma_{G \times L}^{2} ,\sigma_{G \times Y}^{2} ,\sigma_{G \times Y \times L}^{2} ,\sigma_{E}^{2}\) were estimated assuming no covariance between accessions (Holland et al. 2003), and L, Y and R are the number of lines, number of years and R is the number of replications. The genomic heritability was estimated in the same way, but \(\sigma_{G}^{2}\) was calculated assuming the relationships among the accessions were G (with an additive model of SNP action). Variance components were estimated with ASREML (Gilmour et al. 2009).
Multi-trait analysis to incorporate NIR and NMR data
The NIR and NMR data were included by considering the predicted values for each quality trait from either NIR or NMR as a correlated trait with the quality trait as measured by industry end-use assay. NIR and NMR data were considered separately. The model fitted was
where y 1 and y 2 are phenotypes (trait 1 is end-use quality assay measurement, trait 2 is either the NIR or NMR predictor of the trait), I 1 and I 2 are identity matrices, µ 1 and µ 2 are the vector of intercepts of end-use quality assay and NIR or NMR predictor, respectively, X 1 and X 2 are design matrices for fixed effects, b 1 and b 2 are vectors of fixed effects relating to each trait (e.g. days heading, days physiological maturity, days grain filling, location and year), Z 1 and Z 2 are the design matrices that relate accessions with phenotypes, g 1 and g 2 are genomic breeding values for accessions for end-use quality assay and NIR or NMR predictors, respectively, and e 1 and e 2 are vectors of random residuals for end-use quality and NIR or NMR predictors. It was assumed that \(\left[ {\begin{array}{*{20}c} {\varvec{g}_{{\mathbf{1}}} } \\ {\varvec{g}_{{\mathbf{2}}} } \\ \end{array} } \right]\) \(\sim {\text{MVN}}(0,\varvec{G T})\), where T = \(\left[ {\begin{array}{*{20}c} {\varvec{\sigma}_{{\varvec{g}{\mathbf{2}}}}^{{\mathbf{2}}} } & {\varvec{\sigma}_{{\varvec{g}{\mathbf{12}}}}^{{}} } \\ {\varvec{\sigma}_{{\varvec{g}{\mathbf{12}}}}^{{}} } & {\varvec{\sigma}_{{\varvec{g}{\mathbf{2}}}}^{{\mathbf{2}}} } \\ \end{array} } \right]\varvec{ }\), the variance–covariance matrix of end-use assay and NIR or NMR predictor, G is the genomic relationship matrix between accessions as described above, and \(\begin{array}{*{20}c} {\varvec{e}_{{\mathbf{1}}} } \\ {\varvec{e}_{{\mathbf{2}}} } \\ \end{array} \varvec{ }\sim \varvec{ }{\text{MVN}}\left( {{\mathbf{0}},\varvec{I \otimes R}} \right),\varvec{ }{\text{where}}\;\varvec{ R} = \varvec{ }\left[ {\begin{array}{*{20}c} {\varvec{\sigma}_{{\varvec{e}{\mathbf{1}}}}^{{\mathbf{2}}} } & {\varvec{\sigma}_{{\varvec{e}{\mathbf{12}}}}^{{}} } \\ {\varvec{\sigma}_{{\varvec{e}{\mathbf{12}}}}^{{}} } & {\varvec{\sigma}_{{\varvec{e}{\mathbf{2}}}}^{{\mathbf{2}}} } \\ \end{array} } \right]\varvec{ }\) and MVN is multi-variate normal. Variance components were estimated with ASREML (Gilmour et al. 2009).
Genomic predictions
For the single-trait analysis (end-use assay data only, measured using industry standard assays), the accuracy of genomic prediction was assessed in two ways. First, the accessions grown at Horsham in 2013 (116 with end-use quality data) were excluded from the reference set. Genomic estimated breeding values (GEBV) were calculated for these accessions using model (1) above (that is their trait data were not included in the analysis, but genomic estimated breeding values for these accessions were estimated as they were included in the genomic relationship matrix). The correlation of the GEBV and phenotype (corrected for fixed effects, y*) was taken as the accuracy of genomic prediction. This correlation was performed for hard wheats (n = 74 in the validation) and soft wheats (n = 42) separately, to avoid a high correlation due to just predicting hard or soft wheat type. This approach calculates the accuracy of genomic prediction for a single location in a single year.
A second approach aimed to assess how robust the accuracy of genomic prediction was across years and locations. For each of the Australian national variety trials (which test elite varieties each year, and defined by year and location) in Fig. 1, phenotypes for each trait for all accessions in that trial were predicted (for more information on national variety trials, including which lines were grown in each location and in each year, see http://www.nvtonline.com.au). The numbers of accessions at each site in each year are given in Supplementary Table 1. When each trial was predicted, data for accessions in that trial were completely omitted from the reference used to calculate the genomic breeding values, such that the validation sets were always independent of the reference set.
For multi-trait predictions, the validation set was always the accessions grown at Horsham in 2013 with end-use quality data (116 accessions). Genomic estimated breeding values (GEBV) for end-use quality were calculated for these accessions using model (2) above. The NIR data or NMR data contributed through the genetic correlation with the end-use quality data, and NIR and NMR data for other accessions—the NIR and NMR for the 116 validation lines were completely excluded from the analysis. This correlation was performed for hard wheats (n = 74 in the validation) and soft wheats (n = 42) separately, to avoid a high correlation due to just predicting hard or soft wheat type. A potential source of over prediction is that the 116 accessions were included in the derivation of the NIR and NMR prediction equations. To assess the impact of this, we performed additional analyses (accuracy of GEBV) with NIR data from 2012 only, that did not include the 116 accessions in the derivation of the prediction equation.
Even though we attempted to correct for the effect of phenology in our model by fitting heading date, grain filling period and age at physiological maturity, it could be argued that with such a wide range of phenology, part of the accuracy of our genomic predictions is actually derived from predicting phenology and its subsequent effect on end-use quality, rather than the variation in end-use quality that would be expected in lines with similar phenology. To test this, we calculated the accuracy of genomic prediction, r (GEBV, y*), in subsets of lines that were within 10 days of flowering of one other.
Results
Broad-sense heritabilities, genomic heritabilities and G × L
The proportion of variance in the end-use quality traits captured by accession was high (0.60–0.80) for most classes of traits (Table 1). The proportion of variance explained by accession × location (G × L) effects was generally low (0.0–0.13), and none were significantly different to zero.
The SNP captured 10–93% of the total phenotypic variance, and 66–95% of the accession variance (Table 1). It is important to note that this is close to an estimate of the total genetic variance that is additive genetic variance, not an estimate of the proportion of additive genetic variation captured by the SNP. We cannot easily estimate the latter as is done in human populations (e.g. Yang et al. 2010) because our population includes a significant number of related individuals, so the heritability will be more like the heritability that would be estimated from a pedigree (de los Campos et al. 2015).
Genomic predictions for quality traits
When the reference population included only the accessions evaluated for end-use quality using industry standard assays, the accuracies of genomic prediction were quite variable (compare Fig. 2a from Horsham 2013 validation only with Fig. 2b, averaged across NVT and Horsham sites). Accuracies in both validations were highest for baking traits and noodle traits, and lowest for traits such as starch damage and ash content. Accuracy of prediction was similar for hard and soft wheats (for example, averaged across traits, accuracy of prediction with the Horsham 2013 trial as the validation was 0.19 for hard wheats and 0.23 for soft wheats).
Accuracy of genomic prediction (a). Validation set is only hard and soft wheats in 2013 Horsham trial, and (b). Accuracy of the average across validation set where each trial is a validation set (either Horsham 2012 or 2013, or National Variety trials in 2011 and 2012). Accuracies were calculated within wheat type (hard or soft) then averaged. Error bars denote standard errors, calculated as the standard deviation of the accuracy across locations divided by the square root of the number of locations
Multi-trait genomic predictions including NIR data
Two sets of NIR-predicted trait values were used, one from 2012 only, and one from predictions derived with combined 2012 and 2013 data. There is a potential issue of confounding with the predictions from the combined 2012 and 2013 data, as the validation accessions (116 from Horsham 2013 field trial) were included in the derivation of the NIR prediction equation (but not the genomic prediction equation). However, the 2012 data alone obviously have many fewer accessions in the reference population than the 2012 and 2013 data (950 versus 1500 accessions). Even with only the 2012 NIR and end quality assay data used as a reference (which has no confounding due to calibration of the NIR prediction equation in the validation set), the accuracy of genomic predictions for nearly all end-use quality traits was increased, compared with a reference based on just the end-use quality measured using industry standard assays (146 accessions in this reference), Fig. 3. It is important to note that there was still considerable variability across traits in how much the accuracies were improved with the addition of the NIR prediction data. When the 2012 and 2013 data were used as a reference (excluding the validation set data), the increase was even greater, although a proportion of this increase may be due to the confounding mentioned above. The increase in accuracy of genomic predictions as a result of including the NIR data in the reference set reflected the genetic correlations between the NIR trait predictions and the trait itself (Table 2). For example, for grain protein, the genetic correlation between the NIR-predicted values and assayed grain protein was 0.81, and therefore, there was a large increase in the accuracy of genomic prediction when the NIR data are included. In contrast, for Maximum strength, the genetic correlation between the NIR-predicted values and assayed trait was 0.2, and including NIR did not improve the accuracy of genomic prediction.
Accuracies of genomic prediction for quality traits for accessions in the 2013 Horsham field trial validation, using end-use trait data only, NIR predictions of trait phenotype derived from 2011 to 2012 data only, and NIR predictions of trait phenotype derived from 2011, 2012 and 2013 data (multi-trait analysis). Accessions in the validation that were evaluated in other locations and in other years were removed from the reference set, that is they were unique to the validation. a For hard wheats, and b for soft wheats. Standard errors on the accuracies are approximately 0.11 for hard wheats and 0.15 for soft wheats. In some cases, accurate NIR predictions of an end-use quality trait could only be made using combined 2011, 2012 and 2013 data, so prediction for 2011 and 2012 data is missing
Multi-trait genomic predictions including NMR data
The increase in accuracy of genomic prediction as a result of including the NMR predictions was greatest for grain protein and loaf volume, 20 and 21% increase, respectively (Fig. 4). Including NMR data increased the accuracy of prediction by on average 6.5% for hard wheats (Fig. 4a) and 6.7% for soft wheats (Fig. 4b).
Accuracies of genomic prediction for quality traits for accessions in the 2013 Horsham field trial validation, using end-use trait data only, and NMR predictions of trait phenotype derived from 2011, 2012 and 2013 data (multi-trait analysis). Accessions in the validation that were evaluated in other locations and in other years were removed from the reference set, that is they were unique to the validation. a For hard wheats and b for soft wheats
The increase in accuracy of genomic prediction from including either NIR or NMR data was similar (Table 3), although the subset of traits predicted was slightly different (Table 2). The average accuracies for grain traits, dough rheology traits and baking traits are at useful levels for wheat breeding programs, as discussed below.
We did run a tri-variate analysis for some traits, including NIR prediction, NMR prediction and end-use quality assay as separate traits, however, this did not improve the accuracy of genomic prediction (data not shown).
Analysis within phenology groups
For the validation set grown at Horsham in 2013 with end-use quality data (116 accessions), phenology was very variable (261–313 days). It could be argued that with such a wide range of phenology, part of the accuracy of our genomic predictions is actually derived from predicting phenology and its subsequent effect on end-use quality, rather than the variation in end-use quality that would be expected in lines with similar phenology. We calculated the accuracy of genomic prediction, r (GEBV, y*), in subsets of lines that were within 10 days of flowering of one other, Table 4. The average accuracy from these subsets was quite close to the accuracy calculated across the lines, suggesting that the model is correctly removing the effect of phenology on the trait, and that the GEBV for end-use quality are reasonably independent of phenology.
Discussion
Grain end-use quality traits are among the most important determining the farm-gate value of a wheat variety, and therefore are a major target of wheat breeding programs. However, they are also among the hardest traits to improve because their assays are typically expensive, require large amounts of flour, and cannot be measured until late in the breeding cycle. Here, we have demonstrated that useful accuracies of genomic prediction can be obtained for dough rheology, baking and grain traits, using reference populations that have been evaluated for these traits with NIR and NMR. Using NIR and NMR predictions of quality traits overcomes a major barrier for the application of genomic selection for grain end-use quality traits in wheat breeding—namely that the size of reference populations that can be assembled for these traits has been limited by the cost of these end-use assays. The accuracy of genomic predictions reported here is sufficiently high (greater than 0.5 for many traits) to allow breeders to select for many quality traits earlier in the breeding cycle to accelerate genetic gain. It is worth noting that we have measured accuracy here as the correlation of GEBV and phenotype in the reference population. As the GEBV only attempt to predict the genetic component (breeding value) of the phenotype, the upper bound of these accuracies for each trait is the square root of their genomic heritabilities (Table 1). So the accuracies of predicting breeding value \(\frac{{r({\text{GEBV}},y^{*} )}}{{\sqrt {h^{2} } }}\), are in many case substantially higher than the accuracy of predicting phenotype.
The accuracies of genomic prediction we observed are broadly similar to those that have been previously reported for quality traits in wheat when both the reference population and validation population included diverse sets of accessions (although the diversity of the validation set is unlikely to have been as diverse as ours). For example, Battenfield et al. (2016) reported an average across quality traits of 0.5, which is similar, though higher than our 0.4 when our reference set included NIR- or NMR-predicted phenotypes. Charmet et al. (2014) reported accuracies of 0.7 for grain test weight within populations, though this dropped to zero when across population prediction was attempted (reduced accuracy of genomic prediction across locations compared within a location was also reported for Fusarium head blight resistance in wheat, Rutkoski et al. 2013). Our results for across population prediction for grain test weight were 0.36 (Fig. 2b). The fact that we observed better across population prediction may reflect the fact that we had a greater number of locations and years in the reference population (Fig. 1) in the reference set (e.g. 9 compared with 3 in Charmet et al. 2014). The relative robustness of our predictions across locations and across years for most traits (see standard errors in Fig. 2b) may also reflect this composition of our reference population (multiple locations, multiple years). Lado et al. (2013) also observed that the best predictions between environments were obtained when data from different years were used in the reference sets, for prediction of yield, thousand-kernel weight, number of kernels per spike, and heading date for wheat varieties. The accuracies we observed for genomic prediction of end-use quality traits are lower however than reported by Heffner et al. (2011), in which a bi-parental mapping population was considered. The higher accuracy of prediction in that study likely reflected the fact that within a bi-parental population, there is a very high degree of relationship, and limited variation, between reference and validation sets.
The reference set we used here encompassed a very broad range of germplasm, from current elite varieties to accessions representing world-wide germplasm and synthetic wheats. It could be argued that the results of genomic prediction in such material would not be representative of what could be achieved in current breeding programs with elite germplasm. However, we have validated our genomic predictions in material from the National Variety Trails (NVT), in which only elite current varieties are represented, and demonstrate that there is still reasonable accuracies in these validations. So, we consider the results of these validations (NVT) to be indicative of what could be achieved in breeding program with elite germplasm.
One question is where does the accuracy of our genomic predictions come from? Accuracy of genomic prediction can be derived from markers in high linkage disequilibrium (LD) with QTL, where the linkage disequilibrium persists across the reference and validation populations, or it can be derived from linkage, where large chromosome blocks from parents in the validation, for example, are passed onto progeny, or some combination of linkage disequilibrium and linkage (e.g. Habier et al. 2013). We investigated the extent of linkage disequilibrium in our population, and found that there was significant LD that extended over several centimorgans, Fig. 5. This is consistent with other studies investigating the extent of LD in wheat, and with a relatively small effective population size for this crop (Chao et al. 2010). This extensive LD will contribute substantially to the accuracy of genomic predictions observed here, although the small proportion of close relatives in the reference and validation population would undoubtedly contribute as well.
Linkage disequilibrium (r 2) in the reference population. r 2 was calculated between all pairs of markers on a chromosome, then averaged in bins of 0.001 cM. Map positions were from Wang et al. (2014)
For a small number of quality traits, the accuracy of our genomic predictions was limited (e.g. Dough Breakdown, Dough Stability, Starch Damage, % Screenings). One possible explanation for these observations could be low heritability, and/or that the proportion of genetic variation explained by the SNP was limited. However, this was not the case—the genomic heritabilities for these traits were 0.61, 0.56, 0.67 and 0.74, respectively. Another possibility is that there are several QTL of large effect for these traits, and these QTL are segregating in some validation populations and not others (since our validation and reference sets are very diverse). For these traits, even larger reference populations may be required, such that the reference populations capture all segregating QTL in a range of validation populations, and these reference populations are sufficiently large that the QTL effects are estimated accurately (particularly if they have low minor allele frequencies). It is interesting to note that the addition of NIR and NMR did improve accuracies of genomic prediction for these traits (Figs. 3, 4), so an expanded reference set of NIR or NMR predictions would be useful to improve accuracies for these traits.
Here, we have focussed on the use of NIR/NMR predictions of quality phenotypes to enlarge the reference population for genomic predictions. This is a useful approach if the aim is to select plants at a very early stage in their development for future breeding—that is a large number of seeds are produced from crossing, DNA is extracted from the seed or seedlings, and those with the highest genomic breeding values are selected to breed the next generation. An alternative application is to combine the NIR/NMR predictions with the genomic prediction to come up with a joint prediction of performance of a particular variety. Many authors have pointed out this possibility, in both crops and other species, and in some cases demonstrated an improvement in accuracy of prediction, particularly using metabolomics data (Gartner et al. 2009; Vazquez 2016; Ward et al. 2015; Guo et al. 2016; Riedelsheimer et al. 2012; Xu et al. 2016). In our case, the NIR/NMR predictions were based on small quantities of flour, so making such combined predictions was not as useful as if the NIR/NMR predictions of quality could be made from single seeds. Potential to do this will be explored in future work.
In the approach used here, the NIR and NMR spectra were first processed to obtain a prediction for each quality trait, then these predictions were used as phenotypes in a multi-trait genomic prediction (where these phenotypes were considered to be different traits to the actual quality trait, but potentially with a correlation between the actual trait and NIR/NMR-predicted trait). An alternative would be to use features of the NIR/NMR data directly as predictors. This approach is appealing, as a SNP may be associated with a large effect on a spectra feature (but a smaller effect on the actual quality trait), making it easier to detect, and include in the genomic prediction model. However, a very large number of traits might have to be analysed simultaneously, although dimension reduction techniques such as PLS (as used here) should be useful. These approaches will be explored in future work.
We have demonstrated that assembling reference populations with NIR and NMR predictions of end-use quality traits will be a cost effective way to derive genomic predictions for selection candidates with useful accuracy for wheat breeding programs. How should these genomic predictions, and the NIR/NMR predictions, be applied in wheat breeding programs? With genomic prediction accuracies of 0.4 for many traits, one strategy would be to use the genomic predictions to remove 50–60% of progeny that result from crossing from further evaluation at an early stage (from testing seedling or even seed prior to sowing). Then very accurate predictions for quality traits could be made for the remaining lines at harvest by combining the genomic predictions and NIR or NMR predictions of quality, for example, in the F2s. Only those F2s with excellent predicted quality would be continued in further generations of selfing, at each stage of which quality could again be predicted with NIR or NMR.
The predicted quality traits, and the genotypes of the lines, could be added back into the reference population for deriving genomic predictions at each stage, further increasing the accuracy of predictions for the quality traits. At some stage in the future, accuracies may be high enough to confidentially select a small set of lines for evaluation as potential release varieties. However, a very large reference population of lines with NIR- or NMR-predicted quality phenotypes would be required to achieve this.
Finally, it is worth pointing out that a major advantage of the genomic selection approach is that in the near future wheat breeders will have access to predictions of performance for a new cross for quality, yield (e.g. Lado et al. 2013; Poland et al. 2012) and disease resistance (e.g. Rutkoski et al. 2013; Daetwyler et al. 2014), simultaneously, from the same DNA test, enabling wheat cycle times to be substantially accelerated.
Author contribution statement
BJH wrote the paper and analysed the genotype and phenotype data, JP, CKW, ALC and SR performed the phenotype analysis including NIR and NMR and contributed to paper writing, JT, DW, MJH and HD performed the genotype data analysis and contributed to paper writing, SK performed the field trial and contributed to paper writing, GCS designed the experiment and contributed to paper writing.
References
AACC-76-21 (2000) General pasting method for wheat or rye flour or starch using the Rapid Visco Analyser, AACC International. Approved methods of analysis, 10th edn. AACC International, St Paul
Baker RJ, Tipples KH, Campbell AB (1971) Heritabilities of and correlations among quality traits in wheat. Can J Plant Sci 51:441–448
Barnard A, Labuschagne M, van Niekerk H (2002) Heritability estimates of bread wheat quality traits in the Western Cape province of South Africa. Euphytica 127:115
Battenfield SD, Guzmán C, Gaynor RC, Singh RP, Peña RJ, Dreisigacker S, Fritz AK, Poland JA (2016) Genomic selection for processing and end-use quality traits in the cimmyt spring bread wheat breeding program. Plant Genome 9. doi:10.3835/plantgenome2016.01.0005
Browning BL, Browning SR (2009) A unified approach to genotype imputation and haplotype-phase inference for large data sets of trios and unrelated individuals. Am J Hum Genet 84:210–223
CCD 07-06 (2010) Yellow alkaline noodles. Supplement Official Testing Methods of the Cereal Chemistry Division, 4th edn. Royal Australian Chemical Institute, North Melbourne
Chambers J, McKevitt NJ, Scudamore KA, Bowman CE (1989) Non-destructive determination of the moisture content of individual wheat grains by nuclear magnetic resonance. J Sci Food Agric 49:211–224
Chao S, Dubcovsky J, Dvorak J, Luo MC, Baenziger SP, Matnyazov R, Clark DR, Talbert LE, Anderson JA, Dreisigacker S, Glover K, Chen J, Campbell K, Bruckner PL, Rudd JC, Haley S, Carver BF, Perry S, Sorrells ME, Akhunov ED (2010) Population-and genome-specific patterns of linkage disequilibrium and SNP variation in spring and winter wheat (Triticum aestivum L.). BMC Genom 11:727
Charmet G, Storlie E, Oury FX, Laurent V, Beghin D, Chevarin L, Lapierre A, Perretant MR, Rolland B, Heumez E, Duchalais L, Goudemand E, Bordes J, Robert O (2014) Genome-wide prediction of three important traits in bread wheat. Mol Breed 34:1843–1852
Daetwyler HD, Villanueva B, Woolliams JA. (2008) Accuracy of predicting the genetic risk of disease using a genome-wide approach. PLoS One 3:e3395
Daetwyler HD, Swan AA, van der Werf JH, Hayes BJ (2012) Accuracy of pedigree and genomic predictions of carcass and novel meat quality traits in multi-breed sheep data assessed by cross-validation. Genet Sel Evol 44:33
Daetwyler HD, Bansal UK, Bariana HS, Hayden MJ, Hayes BJ (2014) Genomic prediction for rust resistance in diverse wheat landraces. Theor Appl Genet 127:1795–1803
de los Campos G, Sorensen D, Gianola D (2015) Genomic heritability: what is it? PLoS Genet 11:e1005048
Delwiche SR, Graybosch RA, Peterson CJ (1998) Predicting protein composition, biochemical properties, and dough-handling properties of hard red winter wheat flour by near-infrared reflectance. Cereal Chem J 75:412–416
Dowell FE, Maghirang EB, Xie F, Lookhart GL, Pierce PO, Seabourn BW, Bean SR, Heffner EL, Jannink J, Sorrells ME (2011) Genomic selection accuracy using multifamily prediction models in a wheat breeding program. Plant Genome J 4:65–75
Gartner T et al (2009) Improved heterosis prediction by combining information on DNA-and metabolic markers. PLoS One 4:e5220
Gilmour AR, Gogel B, Cullis B, Thompson R, Butler D (2009) ASReml user guide release 3.0. VSN International Ltd, Hemel Hempstead
Goddard M (2009) Genomic selection: prediction of accuracy and maximisation of long term response. Genetica 136:245–257
Guo ZG, Magwire MM, Basten CJ, Xu ZY, Wang DL (2016) Evaluation of the utility of gene expression and metabolic information for genomic prediction in maize. Theor Appl Genet 129:2413–2427
Guzman C, Peña RJ, Singh R, Autrique E, Dreisigacker S, Crossa J, Rutkoski J, Poland J, Battenfield S (2016) Wheat quality improvement at CIMMYT and the use of genomic selection on it. Appl Transl Genom 11:3–8
Habier D, Fernando RL, Garrick DJ (2013) Genomic BLUP decoded: a look into the black box of genomic prediction. Genetics 194:597–607
Holland JB, Nyquist WE, Cervantes-Martinez CT (2003) Estimating and interpreting heritability for plant breeding: an update. Plant Breed Rev 22:9–112
Lado N, Matus I, Rodríguez A, Inostroza L, Poland J, Belzile F, del Pozo A, Quincke M, Castro M, von Zitzewitz J (2013) Increased genomic prediction accuracy in wheat breeding through spatial adjustment of field trial data. G3 3:2105–2143
Ly D, Hamblin M, Rabbi I, Melaku G, Bakare M, Gauch HG et al (2013) Relatedness and genotype × environment interaction affect prediction accuracies in genomic selection: a study in Cassava. Crop Sci 53:1312–1325
Meuwissen TH, Hayes BJ, Goddard ME (2001) Prediction of total genetic value using genome-wide dense marker maps. Genetics 157:1819–1829
O’Brien L, Ronalds JA (1987) Heritabilities of small-scale and standard measures of wheat quality for early generation selection. Aust J Agric Res 38:801–808
Panozzo JF, Walker CK, Partington DL, Neumann NC, Tausz M, Seneweera S, Fitzgerald GJ (2014) Elevated carbon dioxide changes grain protein concentration and composition and compromises baking quality. A FACE study. J Cereal Sci 60:461–470
Pearson DC, Rosielle AA, Boyd WJR (1981) Heritabilities of five wheat quality traits for early generation selection. Aust J Exp Agric 21:512–515
Poland J, Endelman J, Dawson J, Rutkoski J, Wu S et al (2012) Genomic selection in wheat breeding using genotyping-by-sequencing. Plant Genome J 5:1–11
Resende MF Jr, Muñoz P, Resende MD, Garrick DJ, Fernando RL, Davis JM, Jokela EJ, Martin TA, Peter GF, Kirst M (2012) Accuracy of genomic selection methods in a standard data set of loblolly pine (Pinus taeda L.). Genetics 190:1503–1510
Riedelsheimer C et al (2012) Genomic and metabolic prediction of complex heterotic traits in hybrid maize. Nat Genet 44:217–220
Rutkoski JE, Poland J, Jannink JL, Sorrells ME (2013) Imputation of unordered markers and the impact on genomic selection accuracy. G3 3:427–439
Spindel J, Begum H, Akdemir D, Virk P, Collard B, Redoña E, Atlin G, Jannink JL, McCouch SR (2015) Genomic selection and association mapping in rice (Oryza sativa): effect of trait genetic architecture, training population composition, marker number and statistical model on accuracy of rice genomic selection in elite, tropical rice breeding lines. PLoS Genet 11:e1004982
Van Ginkel M, Ogbonnaya F (2006) Invited paper: using synthetic wheats to breed cultivars better adapted to changing production conditions “Groundbreaking Stuff”. Edited by Neil Turner and Tina Acuna. In: Proceedings of the 13th Australian Agronomy Conference. 10–14 September 2006 Perth, Western Australia
VanRaden PM (2008) Efficient methods to compute genomic predictions. J Dairy Sci 91:4414–4423
Vazquez AI (2016) Increased proportion of variance explained and prediction accuracy of survival of breast cancer patients with use of whole-genome multiomic profiles. Genetics 203:1425–1438
Wang S, Wong D, Forrest K, Allen A, Chao S, Huang BE, Maccaferri M, Salvi S, Milner SG, Cattivelli L, Mastrangelo AM, Whan A, Stephen S, Barker G, Wieseke R, Plieske J, International Wheat Genome Sequencing Consortium, Lillemo M, Mather D, Appels R, Dolferus R, Brown-Guedira G, Korol A, Akhunova AR, Feuillet C, Salse J, Morgante M, Pozniak C, Luo MC, Dvorak J, Morell M, Dubcovsky J, Ganal M, Tuberosa R, Lawley C, Mikoulitch I, Cavanagh C, Edwards KJ, Hayden M, Akhunov E (2014) Characterization of polyploid wheat genomic diversity using a high-density 90,000 single nucleotide polymorphism array. Plant Biotechnol J 12(6):787–796
Ward J, Rakszegi M, Bedo Z, Shewry PR, Mackay I (2015) Differentially penalized regression to predict agronomic traits from metabolites and markers in wheat. BMC Genet 16:19
Wiggans GR, Vanraden PM, Cooper TA (2011) The genomic evaluation system in the United States: past, present, future. J Dairy Sci 94:3202–3211
Xu SZ, Xu Y, Gong L, Zhang QF (2016) Metabolomic prediction of yield in hybrid rice. Plant J 88:219–227
Yang J, Benyamin B, McEvoy BP, Gordon S, Henders AK, Nyholt DR, Madden PA, Heath AC, Martin NG, Montgomery GW, Goddard ME, Visscher PM (2010) Common SNPs explain a large proportion of the heritability for human height. Nat Genet 42:565–569
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflicts of interest.
Funding
The authors are grateful for funding from Dow AgroSciences for some of the research in this study. The authors are grateful for funding from Department of Economic Development, Jobs, Transport and Resources for some of the research in this study.
Additional information
Communicated by Dr. Ian Mackay.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Hayes, B.J., Panozzo, J., Walker, C.K. et al. Accelerating wheat breeding for end-use quality with multi-trait genomic predictions incorporating near infrared and nuclear magnetic resonance-derived phenotypes. Theor Appl Genet 130, 2505–2519 (2017). https://doi.org/10.1007/s00122-017-2972-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00122-017-2972-7