Abstract
Key message
Host resistance in WI7120 cucumber to prevailing downy mildew pathogen field populations is conferred by two major-effect, one moderate-effect and two minor-effect QTL.
Abstract
Downy mildew (DM) caused by the obligate oomycete Pseudoperonospora cubensis is the most devastating fungal disease of cucumber worldwide. The molecular mechanism of DM resistance in cucumber is poorly understood, and use of marker-assisted breeding for DM resistance is not widely available. Here, we reported QTL mapping results for DM resistance with 243 F2:3 families from the cross between DM-resistant inbred line WI7120 (PI 330628) and susceptible ‘9930’. A linkage map was developed with 348 SSR and SNP markers. Phenotyping of DM inoculation responses were conducted in four field trails in 2 years at three locations. Four QTL, dm2.1, dm4.1, dm5.1, and dm6.1 were consistently and reliably detected across at least three of the four environments which together could explain 62–76 % phenotypic variations (R 2). Among them, dm4.1 and dm5.1 were major-effect QTL (R 2 = 15–30 %) with only additive effects; dm2.1 (R 2 = 5–15 %) and dm6.1 (R 2 = 4–8 %) had moderate and minor effects, respectively. Epistatic effects were detected for dm2.1 and dm6.1 with both dm4.1 and dm5.1. One additional minor-effect QTL, dm6.2 (R 2 = 3–5 %) was only detectable with the chlorosis rating criterion. All alleles contributing to DM resistance were from WI7120. This study revealed two novel QTL for DM resistance and the unique genetic architecture of DM resistance in WI7120 conferring high level resistance to prevailing DM populations in multiple countries. The effects of disease rating scales, rating time and criteria, population size in phenotyping DM resistance on the power of QTL detection, and the use of DM resistance in WI7120 in cucumber breeding were discussed.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Downy mildew (DM) caused by the obligate biotrophic oomycete Pseudoperonospora cubensis [(Berkeley & M. A. Curtis) Rostoyzev] is an important foliage disease of all cucurbit crops worldwide. DM is especially devastating in cucumber. Disease symptoms on susceptible cucumbers are characterized by the appearance of small and water-soaked lesions on the underside of leaves. Adaxial lesions are often angular and bound by leaf veins, eventually turning chlorotic and necrotic (Oerke et al. 2006). Heavy sporulation can be observed within the lesions on the abaxial leaf surface. A general yellowing of affected leaves typically occurs as the lesions coalesce into one large lesion, eventually causing the leaf to wilt and die. Under favorable conditions, the disease can progress quite rapidly defoliating a cucumber field in a matter of days rendering limited flexibility of fungicide spray (Savory et al. 2011). Pseudoperonospora cubensis spores spread rapidly by wind, splashing rain and/or irrigation water. Disease management and prevention requires destruction of all plants from infected nurseries and disinfection of the facilities (Ojiambo et al. 2015). Seed transmission of Ps. cubensis has been found in cucurbits, and it was suspected that international trade of fruits or seeds of cucurbits might be associated with the recent global change in the population structure of Ps. cubensis (Cohen et al. 2014).
Among integrated pest management practises for disease control, deployment of resistant cultivars is clearly the most economic and environmentally sound method. The Unites States has a long history of breeding cucumber for DM resistance (DMR, hereinafter). Jenkins (1942) was the first to conduct systematic screening of DMR in cucumber who found that, among 12 varieties examined, Puerto Rico 37 (PR37) and a line from China (Chinese Long) had high resistance, and the progeny from the cross between them did not segregate for DMR. Jenkins (1946) then studied the inheritance of DMR in PR37 and found normal distribution of disease ratings in F2 and F3 families; he also found consistent performance of resistance at both seedling (in the greenhouse) and adult plant (in the field) stages. From the Chinese Long resistance source, Barnes (1948) developed the cucumber cultivar ‘Palmetto’; its resistance, however, was lost only 2 years after its release probably due to the appearance of new DM strain in the field (Epps and Barnes 1952). Following the defeat of resistance in Palmetto, Barnes and Epps (1954) identified a new source of resistance, PI 197087 from India. The resistance responses in PI 197087 were characterized by sparse pathogen sporulation, small necrotic lesions, tissue browning, and rapid cell death, indicative of the classical hypersensitive response (HR)-type resistance (Barnes and Epps 1954). Since then, DMR conferred by the recessive gene dm-1 in PI 197087 has been widely used in commercial cultivar development in the US. Representative cultivars carrying dm-1 include Polaris, Gy14, M21, Clinton, Chipper, and the Poinsett or Marketmore series (Wehner and Shetty 1997; Call et al. 2012a). The resistance conferred by dm-1 was sufficient to prevent losses caused by DM without the use of fungicides for nearly 50 years until the emergence of a new DM strain in 2004 (Holmes et al. 2004, 2006). Fortunately, multi-year, multi-location, large scale screening tests identified several plant introduction (PI) lines with high resistance to the new DM strain including PI 197085, PI 197088, PI 330628, and PI 605996 (Call et al. 2012b).
In the post-2004 cucumber fields in the US, resistance tracing back to Puerto Rico 40 (PR40) provides only a slight advantage over lines that lack any resistance in their pedigree: cultivars containing the dm-1 gene show moderate resistance whereas susceptible ones without any DM resistance gene become infected earlier in the season, and exhibit more severe damages than was observed previously (Holmes et al. 2004; Call et al. 2012a, b). Cucumber lines possibly carrying both dm-1 and PR40 DM resistances (for example, SC50 or PI 234517, WI2757, and M21), in general, perform much better than dm-1 only lines suggesting residual resistances of the two defeated genes (Call et al. 2012b; Kozik et al. 2013). Meanwhile, from the progeny of a cross between Marketmore 97 and Ivory Queen with intermediate DM resistance, Holdsworth et al. (2014) selected a breeding line, MRNY264, with a high level of resistance to the new DM strain.
The inheritance of DM resistance in cucumber has been investigated in a number of studies through either classical quantitative genetic analysis or QTL mapping but with varying results. Vliet and Meysing (1974) suggested that a single recessive gene, dm-1 was underlying the DM resistance in Poinsett, which was later located in cucumber chromosome 5 (Fanourakis and Simon 1987; Kennard et al. 1994; Horejsi et al. 2000). Petrov et al. (2000) proposed that the resistance in WI2843 derived from PI 197087 was controlled by incomplete dominant genes. Genetic variance studies for Ames 2354 (a selection from SC50) revealed a small number of genes controlling DM resistance in this line, and one of them seems to be allelic to dm-1 (Kozik et al. 2013). Several QTL mapping studies identified five QTL in four cucumber chromosomes (Chr 1, 4, 5, and 6) for DM resistance in north China fresh market type (Chinese Long) cucumber lines ‘129’ (Ding et al. 2007), ‘S94’ (Bai et al. 2008), and ‘K8’ (Zhang et al. 2013). Pang et al. (2013) detected 3 QTLs for DM resistance (1 on Chr6 and 2 on Chr5) that were believed to be from a C. hystrix introgression line IL52. Since the F2 population used for mapping in Pang et al. (2013) was derived from a cross between IL52 (in the background of a North China type cucumber ‘Beijingjietou’) and a susceptible line ‘Changcunmici’, and the DM resistance in IL52 was comparable to that in the moderately resistant cucumber line M21, it is not known whether the resistance was contributed by IL52 or ‘Beijingjietou’ or both.
Although the four PI lines, PI 197085, PI 197088, PI 330628, and PI 605996 all show high resistance against the post-2004 DM strain in the US, it is not known whether they share a similar genetic basis for DM resistance. This knowledge is important in efficient use of these resistance sources in cucumber breeding. Angelov (1982) suggested that DM resistance in PI 197088 is under the control of two recessive genes whereas that in Poinsett is due to one recessive gene. Yoshioka et al. (2014) conducted QTL mapping for DM resistance in the cucumber line CS-PMR1 (DM resistance from PI 197088) and identified 10 QTLs in 5 chromosomes (1, 3, 5, 6 and 7) including 3 QTLs from the parental line Santou and the major-effect QTL contributed by PI 107088 being located in Chr5. More recently, Szczechura et al. (2015) identified three DM resistance QTLs in Chr5 from PI 197085. In field screening tests, VandenLangenberg (2015) observed a significant number of susceptible offspring in F2 populations derived from crosses among PI 197088, PI 330628, and PI 605996 suggesting possible genetic differences in these lines. VandenLangenberg (2015) also found that these PI lines responded differently to the DM disease over plant developmental stages, and suggested the preference of PI 330628 over PI 197088 due to its ability to maintain resistance for longer time. Therefore, the objective of the present study was to conduct QTL mapping to understand the genetic basis of DM resistance in WI7120 (PI 330628). Using F3 families derived from two inbred lines, the DM-resistant WI7120 and susceptible 9930, we collected phenotypic data from multiple environments in three countries over 2 years. QTL analysis identified QTL for DM resistance conferred by WI7120 in four cucumber chromosomes.
Materials and methods
Plant materials
Two inbred lines, WI7120 and 9930 were used as the parental lines to develop an F2 and F2-derive F3 populations for QTL mapping in the present study. WI7120 was the advanced self-pollinated inbred line from PI 330628 which was originally collected from Pakistan and is highly resistant to DM (Wehner and Shetty 1997; Call et al. 2012a). The North China type line 9930 was susceptible to DM. Ninety-one F2 plants and F2:3 families from 9930 × WI7120 mating were used for developing a linkage map and initial QTL mapping. Phenotypic data collection and refined QTL mapping in target chromosomal regions were based on 243 F2:3 families of the same population.
Phenotypic data collection and statistical analysis
Phenotypic data of inoculation responses of Ps. cubensis for 243 F2:3 families were collected in four environments over 2 years (2013, 2014) in three countries, which were designated as US2013, US2014, NL2013, and IT2013, respectively. US2013 and US2014 were conducted at the Clinton Horticultural Crop Research Station of North Carolina State University at Raleigh, North Carolina in 2013 and 2014, respectively. NL2013 and IT2013 were performed in the field of Bayer Vegetable Seeds in the Netherlands, and in the greenhouses of Magnum Seeds in Fondi, Italy, respectively. The two parental lines and their F1 were included in all screening tests. Some historical DM-resistant cultivars carrying the PI 197087-derived dm-1 resistance gene like Poinsett 87 and WI 2757, as well as the susceptible cultivar Straight 8 were also included as controls in the US2013 or US2014 trials.
The US2013 and US2014 experiments were a randomized complete block design with three replications. There were five plants per F3 family per replication. Plots were exposed to natural epidemics encouraged by overhead irrigation during the growing season at least three times per week, or as needed. All entries were hand planted on June 27, 2013 and June 30, 2014. Disease severity of individual plants was evaluated weekly for three consecutive weeks based on percentage of symptomatic leaf area using disease rating scale 1–9, where 1 = 0–10 %, 2 = 11–20 %, 3 = 21–30 %, 4 = 31–40 %, 5 = 41–50 %, 6 = 51–60 %, 7 = 61–70 %, 8 = 71–80 %, 9 = 81–100 %, or dead (supplemental Figure S1). Since one of the major block effects was the genetic variations within F3 families and there was no significant effect among replications in each environment, the family means from 15 plants of each family were used for statistical analysis and QTL mapping.
Field layout of the NL2013 experiment was similar to US2013 or US2014 but had two replications and seven plants per family per replication (plot). The DM symptoms of each plot were measured twice with 1 week apart with three criteria: general impression (GI), necrotic lesion size (Nec), and chlorotic lesion size (Chl). Leaf chlorosis or yellowing was a symptom of the DM fungal infection, and depends on both infection spread and plant growth rate. Leaf necrosis was the appearance of necrotic (dead) spots on the leaf, either as a symptom of infection or as a hypersensitive response from the plant. The GI score was based on the general health and symptom expression in the plant (Pang et al. 2013). For each trait, the disease severity was rated on the plant breeder’s 1–9 scale, where scale 1 = maximal disease symptoms and 9 = no visible symptom.
The IT2013 experiment was conducted in plastic tunnels with natural infection. There were ten plants per F3 family and no replications. To assure optimal conditions for DM symptom development, after transplanting, high humidity inside the tunnels was maintained by spraying with water twice a day when necessary. Rating of disease symptoms on each plant was based on a 1–5 scale where 1 = absence of symptoms and 5 = maximal disease symptoms.
Statistical analysis of phenotypic data was all performed in R (Version 3.1.1, http://www.r-project.org/). Means of disease scores of each family were calculated by rating time, replication, trait, and experiment, and used for statistical analysis. Quantile–Quantile (Q–Q) plot was performed for each location to confirm the normal distribution of the phenotypic data. The ordinal rating data for each environment were standardized to standard score (z score) for the same scale independently. Analysis of variance (ANOVA) was performed with the R/lme4 package to estimate the genetic and environment effects with the following model: \(R_{{ij}} = \mu + G_{{i}} + E_{{j}} + \varepsilon_{{ij}}\), where R = observed value for a given trait, μ = grand mean, G = genotype, E = environment, and ε = residuals. Best linear unbiased predictors (BLUPs) were also extracted from the model and used for QTL analysis. Broad-sense heritability estimates were calculated from variance components using \({{h^{ 2} = \sigma_{\text{G}}^{ 2} } \mathord{\left/ {\vphantom {{h^{ 2} = \sigma_{\text{G}}^{ 2} } {\left( {\sigma_{\text{G}}^{ 2} + \sigma_{\varepsilon }^{ 2} } \right)}}} \right. \kern-0pt} {\left( {\sigma_{\text{G}}^{ 2} + \sigma_{\varepsilon }^{ 2} } \right)}}\), where \(\sigma_{\text{G}}^{ 2}\) was the genotypic variance and \(\sigma_{\varepsilon }^{ 2}\) was the residual variance. Spearman’s rank correlation coefficient (r s) among different environments was calculated before and after the scale standardization.
Marker development and linkage map construction
The linkage map was developed with both SSR and SNP markers. Cucumber SSR markers described by Ren et al. (2009), Cavagnaro et al. (2010), and Yang et al. (2012, 2013) were used for polymorphism screening between WI7120 and 9930. SNP genotyping was performed in the Bayer Vegetable Seeds, the Netherlands genotyping facility using the KBiosciences Competitive Allele-Specific PCR SNP genotyping system (KASPar). Polymorphic markers were used to genotype a subset of 91 F2 plants. To refine QTL locations in four target regions, these markers mapped in 91 F2 plants and 27 new SSRs were also applied to the larger population including all 243 F2 plants. All markers were tested against the expected segregation ratio of 1:2:1 or 3:1 using Chi-squared tests (χ 2, P < 0.05). Linkage analysis was carried out with JoinMap 4.0. Genetic map was developed with the regression mapping method and Kosambi mapping function. The physical locations of all mapped SSR markers in the Gy14 (V1.0) (Yang et al. 2012) and 9930 (V2.0) (Li et al. 2011; http://www.icugi.org/) scaffold and draft genome assemblies were used to verify their genetic map positions.
DNA extraction, PCR amplification of molecular markers, and gel electrophoreses were conducted as described in Li et al. (2013).
QTL analysis
QTL analysis was performed using the R/qtl package with the multiple-QTL model (MQM) (Broman et al. 2003; Arends et al. 2010). QTL detection started with “scanone” for preliminary QTL identification, and followed with “addqtl” and “addpair” to scan additional QTLs and QTL pairs. Then, the function ‘refineqtl’ was used to refine the position of QTLs. The significance of each QTL interval was tested by a likelihood-ratio statistic (LOD). The LOD threshold for declaring significant QTLs was established separately for each trait and each environment using 1000 permutations at significance threshold of 0.05. The refined significant QTLs were assessed for the additive and dominant effects, and percentage of phenotypic variations (R 2) explained. The support intervals for these QTLs were calculated using a 1.5 LOD drop interval. QTL naming conventions followed He et al. (2013) except that each QTL was preceded with dm (for downy mildew).
Results
Phenotyping DM inoculation responses among F2:3 families
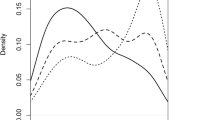
Phenotypic data of DM inoculation responses among 243 F2:3 families of 9930 × WI7120 were collected from four environments (US2013, US2014, NL2013, and IT2013) over 2 years (2013, 2014) in three countries. While disease rating in all experiments was based on a GI of foliage symptom development, two additional criteria, Nec and Chl were used in rating of DM disease symptoms in NL2013. The grand means and standard deviation (SD) of DM disease scores for the F2:3 families, the two parental lines, F1 and controls across the four environments are presented in Table 1. Frequency distribution of F3 family means of the four experiments is illustrated in Fig. 1a–c. In all experiments, the mean disease scores of the two parents (WI7120 and 9930) were located at the extreme ends of the largely normally distributed family means, and those for the F1 and the F3 families were close to mid-parent values suggesting the quantitative nature of DM resistance in WI7120. The normal distribution of residual phenotypic variations could also be reflected from Q-Q plots of the four experiments (supplemental Fig. S2A). As expected, the susceptible control Straight 8 remained highly susceptible as 9930, whereas the pre-2004 DM-resistant Poinsett 76 and WI2757 showed intermediate resistance although the latter performed a bit better (Table 1). These observations indicated satisfactory control of environmental factors in each experiment.
Different rating scales were employed in the three locations (1–9 in US2013, US2014; 9–1 in NL2013; 1–5 in IT2013). To compare environmental effects on DM resistance, the phenotypic data for each experiment were standardized using the z score in which the IT2013 and NL2013 datasets were converted to the same scale as the US experiments. The frequency distribution of the standardized data from the four environments is shown in Fig. 1d, which again was largely normal (Fig. S2B). A one-way ANOVA was performed using the standardized data, and the results are presented in Table 2 which indicated no significant environmental effects on DM resistance. The broad-sense heritability estimate of mean disease scores (H2) based on ANOVA was as high as 0.89 (Table 2) further indicating effective control of environments in these trials.
We calculated the Spearman’s rank correlation coefficient (r s) of different traits from the four environments using both the raw and standardized data. The results are provided in Table 3. The data among the four experiments were highly and significantly correlated with r s ranging from 0.592 to 0.796 (P < 0.001) implying the consistence and reliability in DM ratings among environments. When the raw data were used in calculation of r s, the mean disease scores of NL2013 (GI, Nec, and Chl) were negatively correlated to those from other three experiments. This is reasonable because NL2013 experiment used a reversed rating scale (plants rated 1 were the most susceptible).
DM infection may result in chlorosis and necrosis of the plants. In NL2013, phenotypic data of DM inoculation responses were collected using three criteria: Chl, Nec, and Gl. No significant differences in mean disease scores were observed among the three criteria in both parents and segregating populations (Table 1). All three parameters were positively and highly correlated with each other and with those in US2013, US2014, and IT2013 (Table 3) suggesting that these symptoms may share similar underlying genetic mechanism in WI7120.
In US2013, US2014, and NL2013 trials, rating of disease symptoms was conducted multiple times with 1 week apart. The boxplots of mean disease scores at each time point are shown in supplemental Fig. S3A (online material). In US2013 and US2014, it was clear that, with the progress of the disease, the overall mean disease score of the entire population shifted toward susceptibility with increasing variations within the population. On the other hand, this trend was not obvious in the NL2013 trial. The population dynamics had obvious consequences in QTL detection (see below).
To summarize, despite the different environments, scoring scales, and methods used in the four phenotyping experiments, data collected from these trials were highly correlated, consistent, and of good quality, which provided a solid foundation for subsequent QTL analysis.
Linkage map construction
Among 1440 cucumber SSR markers screened, 441 (30.6 %) were polymorphic between 9930 and WI7120, 271 of which were genotyped in 91 F2 individuals. These plants were also subjected to KASPar SNP assay and 76 polymorphic SNPs were successfully mapped. On the resulting map, there was a large gap (~16 cM) at the top of Chr4. This F2:3 population was segregating for mature fruit skin color and fruit spine color: WI7120 set netted brown fruits with black spines and 9930 had light yellow fruits with white spines. We phenotyped spine color in this population, and the underlying gene for black spine happened to be mapped in the 16-cM gap region. The location of the gene was consistent with previous work by Li et al. (2013) who identified a candidate gene for the black spine color gene B in the distal region of the short arm of cucumber chromosome 4. Therefore, it is reasonable to conclude that the black spine color in WI7120 was controlled by the same B locus as identified in Li et al. (2013).
The resulting genetic map is illustrated in supplemental Fig. S4, and the main statistics of the map are presented in Table S1. Detailed information of 348 mapped loci is provided in supplemental Table S2 (online materials). While the majority of the marker loci fitted the expected 1:2:1 or 3:1 segregation ratio, 23 markers (~7 %) showed distorted segregation (loci with asterisks in Table S2) which were mainly in two clusters in Chr5 and Chr7. In both clusters, the 9930 alleles showed preferred transmission over those of WI7120, which is consistent with observations in two previous studies (Miao et al. 2011; Rubinstein et al. 2015).
The genetic map covered 674.7 cM with seven linkage groups (chromosomes) with an average interval of 2.0 cM (Table S1). According to the 9930 and Gy14 draft genome scaffolds associated with these markers, this map seemed to physically cover the majority of the cucumber genome. The marker orders were also highly consistent with their physical locations.
Detection of DM resistance QTLs
We conducted QTL analysis using the MQM approach with data for each location, time point, and disease rating criterion. The LOD threshold to declare significance of QTL for each trait was determined with 1000 permutations (P = 0.05), which varied from 3.4 to 3.6. We first investigated the effects of scoring time on the ability of QTL detection, and the results are shown in supplemental Fig. S3B. We found that for US2013 and US2014, QTL on chromosomes 4 and 5 could be detected with data across all rating times whereas, the additional QTL on chromosome 2 and 6 could only be detected in the later scorings (2nd and 3rd). On the other hand, both scoring times detected three QTLs on chromosome 2, 4, and 5 for GI (general impression) and Nec (necrotic lesion size) in the NL2013 experiment (Fig. S3B). This result was consistent with the phenotypic variations of mean disease scores at these time points. That is, larger variations within the population allowed detection of more QTL (Fig. S3A–B). Since the last scoring time detected the most DMR QTL in all experiments, we used the datasets at this time point in subsequent discussions.
We examined the relationships of QTL detected with three different scoring criteria (GI, Nec, Chl) in NL2013 using MQM. The results are presented in supplemental Table S3 and Fig. S5. All three datasets detected QTL in Chr2, Chr4, and Chr5. The 1.5-LOD intervals and peak locations of QTL in Chr4 and Chr5 were largely overlapped suggesting that they probably belonged to the same QTL in each chromosome although the LOD support value and the effects were somewhat different (Table S3). For the QTL in Chr2, the Nec QTL peak location was shifted from that for GI and Chl (Fig. S5), which was probably a sampling error, because its peak location was much closer to that of the other two QTLs when a large population (243 F2:3 families) was used in QTL analysis (see below). In chromosome 6, two QTLs were detected, one with Chl (chlorotic lesion size) designated as dm6.2-Chl, which might be associated with chlorosis symptom development in response to DM infection. Another QTL, dm6.1, was detected with Nec, which had a LOD support score of 2.1 (Table S3) but was likely true since it was also detected with US2013, US2014 data, and BLUPs (see below). These two QTL, dm6.1 and dm6.2-Chl were clearly different, because their peak locations in chromosome 6 were far away from each other (Fig. S5, Table S3). Overall, the results from NL2013 suggest that GI, Nec, and Chl shared similar QTL in chromosomes 2, 4, and 5 whereas, the QTL in Chr6 may be unique to responses to chlorosis. Since GI represented integrated information of both necrotic and chlorotic responses to DM infection, the GI data from NL2013 were used for subsequent analysis with data from other environments.
QTL analysis was performed using the MQM approach with mean disease scores (raw data) from the four environments and BLUPs of the entire study. Details of each detected QTL including map location, LOD support value, percentages of total phenotypic variances explained (R 2), additive and dominant effects, and 1.5-LOD support interval are provided in supplemental Table S3 and graphically presented in supplemental Fig. S6 (online material). Note that the graph in Fig. S6 was generated with the ‘mqmscan’ utility in R/qtl, thus, the positions of QTLs in Fig. S6 might be somewhat different from data presented in Table S3. Comparing results among these experiments, it was clear that three DMR QTLs, dm2.1, dm4.1 and dm5.1 located in chromosomes 2, 4, and 5, respectively, were consistently and reproducibly detected in all four environments and BLUPs. These QTLs could explain 56.2–75.5 % phenotypic variations. A fourth QTL, dm6.1 in chromosome 6 could be detected in US2013, US2014, NL2013-Chl, as well as with BLUPs (Table S3).
Among the four DMR QTLs, dm4.1 on Chr4 had the largest effect accounting for 20.9–50.7 % phenotypic variations followed by dm5.1 (R 2 = 9.5–22.4 %). The QTL on Chr2, dm2.1 showed moderate effects with R2 ranging from 5.4 % in US2013 to 15.7 % in IT2013. The fourth one, dm6.1 in Chr6 was a minor-effect QTL that was detected in two environments, and the BLUPs explained only 3.8–5.5 % of the phenotypic variations (Table S3).
In US2013, US2014, and IT2013, all resistance contributing alleles were from WI7120 QTL which could be reflected from the negative additive effects of each QTL (reduction of disease score). Since the reverse DM rating scales were used in NL2013, the positive additive effects of all QTLs indicated increase of the score (higher DM resistance). Dominance effects of all QTLs were, in general, weak (Table S3).
Refinement of QTL locations
Our above-described initial QTL analysis with 91 F2:3 families identified four DM-resistant QTLs in WI7120 including two major-effect (dm4.1 and dm5.1), one moderate-effect (dm2.1) and two minor-effect (dm6.1 and dm6.2-Chl) QTLs (Table S3). While the 1.5-LOD interval and peak locations of dm4.1 and dm5.1 were relatively consistent across different environments, those for dm2.1 and dm6.1 showed more variations (Fig. S6), which may be due to the small population size and their relatively smaller contributions to overall phenotypic variations. Thus, we expanded the population size to include all 243 F2:3 families that we phenotyped in four environments aiming to refine the QTL locations on the genetic map. We also added more markers in the four target QTL regions. Information on the resulting genetic map (99 loci with 27 new SSR or indel markers) is provided in supplemental Table S4 (online material). LOD profiles of dm2.1, dm4.1, dm5.1, and dm6.1 based on data of four environments and BLUPs are presented in Fig. 2; detailed information for each QTL is summarized in Table 4.
The increased population size (from 91 to 243) allowed detection of new QTL. First, dm6.1 was identified with the NL2013-GI data which failed to detect in the 91-F3 population (Table S3). Second, the dm6.2-Chl locus was supported by IT2013 dataset which did not identify dm6.1 though. This was probably due to the fact that the scoring of disease symptoms in IT2013 relied more on chlorotic lesion sizes. This large population size not only confirmed the four QTLs (dm2.1, dm4.1, dm5.1, and dm6.1), but also significantly improved the precision of the location of each QTL. The peak location and 1.5-LOD support interval of each QTL from each environment as well as BLUPs were highly consistent. The QTL detected with BLUPs had the highest LOD support (Fig. 2) suggesting that BLUP is more powerful in detecting the small-effects QTL in this population. The size of the 1.5-LOD interval was also significantly narrowed down as compared with that based on the small population size (Fig. 2). For example, the physical size of the 1.5-LOD interval for the major-effect QTL dm4.1 in Chr4 was reduced from 4.2 Mbp (Table S3) to 1.2 Mbp in the 9930 V2.0 draft genome assembly that was delimited by two flanking markers, UW083868 and UW004293 (Table S4).
We investigated possible interactions among these QTLs. Line interaction plots of dm2.1 (CHXSSR00134 at 37.6 cM), dm4.1 (UW059163 at 22.9 cM), dm5.1 (SSR15321 at 51.6 cM), and dm6.1 (SSR18651 at 46.9 cM) (Table S4) are illustrated in Fig. 3. Typical parallels across all genotypes were observed indicating that there was no strong epistatic interaction between two major-effect QTL dm4.1 and dm5.1 (Fig. 3b). However, it seems that both dm2.1 and dm6.1 had epistatic interactions with either dm4.1 or dm5.1, and with each other (Fig. 3a, c–f). The dominant effect of dm2.1 in either dm4.1 (Fig. 3a) or dm5.1(Fig. 3c) background was evident from the fact that the mean disease score of the heterozygotes (ab) was the same as the homozygotes (bb) in both interaction plots.
Effect plots showing interactions among loci near dm2.1 (CHXSSR00134, 37.6 cM), dm4.1 (UW059163, 22.9 cM), dm5.1 (SSR15321, 51.6 cM), and dm6.1 (SSR18651, 46.9 cM) on DM resistance in 243 F2:3 families from 9930 × WI 7120. Line interaction plots demonstrate that dm2.1 has a completely dominant effect on DM resistance and strong epistatic interactions with dm4.1 and dm5.1. There are no significant interactions between dm4.1 and dm5.1. Note that “a” is allele from susceptible parent line 9930, while “b” is allele from resistant parent WI7120
Discussion
Rating scales and times for phenotyping DM inoculation responses in cucumber: more is better?
In this study, we performed QTL analysis using phenotypic data collected in four environments (US2013, US2014, IT2013, and NL2013) and identified four DMR QTLs in four chromosomes (dm2.1, dm4.1, dm5.1, and dm6.1). Despite the fact that the four experiments were different in experimental design, rating criteria and scales, environmental conditions, and population dynamics of the DM pathogen (see below for discussion), all four QTLs were consistently identified in at least three of the four environments with high LOD support (Fig. 2; Table 4; Table S4) suggesting that these DM resistance QTLs are authentic and the phenotypic data are of high quality. Many factors affect the QTL detection power, some of which, such as multiple-location and multi-year trials, or large segregating population sizes for data collection, are well known from the statistical point of view. Our work also revealed some other important factors such as disease rating scale, scoring time or summary statistics that may affect the detection power for DM resistance QTL in cucumber.
In QTL mapping studies for DM resistance in cucumber, the rating scales of disease severity varied from 0 to 5 (Zhang et al. 2013), 1–9 (Kozik et al. 2013), 9–1 (Pang et al. 2013), and 0–9 (Call et al. 2012a) (from most resistant to most susceptible). It is a challenge to compare the data from different studies. In the present study, three different rating scales were used including 1–9 in US2013 and US2014 (Fig. S1), 9–1 in NL2013, and 1–5 in IT2013, which provided a good opportunity to assess the efficiency of different rating scales on QTL detection. Since the phenotypic data showed largely normal distribution in all four environments (Fig. 1; Fig. S2), we were able to standardize the ordinal rating data for each environment using the same scale as US2013 and US2014, which made it possible to evaluate the inheritance of DM resistance in WI7120 across multiple environments and extract BLUPs from linear models.
The different ordinal rating scales seemed to contribute to the varying power for detection of DM resistance QTL. As compared with other three experiments using nine rating scales, IT2013 data (with five rating scales) failed to detect dm6.1 in both small and large F3 populations, and the LOD support score for QTL detected with this dataset was relatively lower (Table 4; S3). Since there were only five categories (1, 2, 3, 4, and 5), each interval includes a larger range of disease severities, which causes the deficiency of additivity and reduces the power in QTL detection (Xie et al. 2012). Increasing the categories in ordinal rating allows addition of more distinguishable units, thus reduce bias of the class midpoints and residual variances. In general, the closer the scale of collected data is to a ratio scale with normal distribution, the more powerful methods are available for analysis (Hartung and Piepho 2007; Poland and Nelson 2011). Therefore, for phenotyping of DM inoculation responses, a 9-scale rating system may be preferable to the 5-scale method. However, considering that other factors (phenotyping environment, inoculation method, pathogenicity of DM pathogen inoculum etc.,) may also influence the QTL detection power in the IT2013 experiment, more rigorous experiments may be needed to make a solid conclusion. Nevertheless, while the 9-scale rating system can increase the QTL detection power, it also offers advantage for comparison with data deposited in public databases such as the USDA Germplasm Resources Information Network (GRIN) (Postman et al. 2010).
Natural inoculation of the DM pathogen was used in all trials, and phenotypic data were collected from the adult plants. The performance of the plants was the interaction of intrinsic genetics and environmental factors. In the field, the distribution of DM inoculum was not uniform across the plots, which depends largely on the direction of the airflow (wind). This was evident from the heat maps of disease scores in the field (data not shown) over time. Therefore, to reveal the true QTL underlying host resistance, it is important to conduct the scoring multiple times especially for evaluating adult plants. Indeed, in US2013 and US2014 trials, among the three datasets collected at three time points, the last dataset could detect the most DM-resistant QTL, whereas, no additional QTL information was obtained in the second scoring time in NL2013 (Fig. S3, Table S3) suggesting that the third time of US2013 and US2014 or either time of NL2013 was the preferred time points for phenotypic data collection.
In reality, it is difficult to predict the best time for scoring. Due to the associated cost in time and labor, the number of times for scoring is also limited. Often, DM-resistant QTL detected in different environments were not overlapped (e.g., Zhang et al. 2013; Yoshioka et al. 2014), or the QTL detected were in the same region but with different peak locations, large LOD support interval or insignificant LOD support (e.g. Fig. S6). In the present study, we found that use of BLUPs was able to help mitigate the effects of growth stages or scoring time on disease scores and increase QTL detection power (Fig. 2). As compared to using the grand means to estimate the true value, BLUP allows information to be borrowed across trials, depending on the size of the genetic correlations, which can improve the prediction at each particular trial, especially when within-site replication is low (Gilmour et al. 2009). This was especially true in the large population (243 F2:3 families) (Fig. 2). In a small population, the environment variations could be overestimated and the BLUP would deviate too much to the “true value” while the estimation of the environment and population variances is more accurate in a larger population. This can explain why the BLUP-based DM resistance QTLs had the strongest LOD support and were highly consistent in location and 1.5-LOD interval with other QTLs (Fig. 2).
Chlorosis, necrosis and general impression: different criteria under the same mechanisms?
Cucumber DM is a foliar disease. Its symptoms are easily recognizable by the formation of chlorotic lesions on leaf surface, which become necrotic with the progress of the disease (Oerke et al. 2006; Savory et al. 2011). The speed of necrosis depends on the environment condition which occurs more quickly in hot and dry weather (Cohen and Rotem 1971). Criswell et al. (2008) and Call et al. (2012b) found that Chl and Nec of DM symptoms are highly correlated and suggested that they are likely the same trait. Pang et al. (2013) used Chl (yellowing), Nec as well as GI to evaluate DM inoculation responses, and identified two QTLs for both Chl (YL-5.1, YL-5.2) and GI (DM-5.1 and DM-5.2); YL-5.1 and DM-5.1, as well as YL-5.2 and DM-5.2 were, respectively co-localized in chromosome 5 suggesting that the two traits may be under the control of the same genetic mechanism(s). Of the two Nec QTLs identified, one (Nec-5.1) was co-localized with YL-5.1/GI-5.1, and the other was mapped in chromosome 6 (Nec-6.1) (Pang et al. 2013) which seemed to be consistent with the dm6.1 location in the present study (Fig. 2). In this study, we found that QTLs detected by Chl, Nec, and GI in NL2013 were co-localized in chromosomes 2, 4, 5, and 6. These locations were also consistent with corresponding QTLs detected with datasets from other three environments (Fig. 2; Table 4). On the other hand, the minor-effect QTL dm6.2-Chl detected by NL2013-Chl data was mapped in a different location from dm6.1 in chromosome 6 that was also detected with IT2013 data in the 243-F3 population (Fig. S5; Fig. 2), which may be uniquely associated with chlorosis development upon DM pathogen infection. Despite this, since the two studies (Pang et al. 2013; this study) employed very different sets of plant materials, these results suggest that DM resistance for chlorosis, necrosis or general impression in cucumber may share the common mechanism(s). Therefore, it is probably not necessary to use all the three criteria in future QTL mapping studies for DM resistance.
WI7120 (PI 330628): a genetically unique source of resistance for downy mildew pathogen?
In this study, the alleles of the four DMR QTLs contributing to increased disease resistance were all from WI7120 (Table 4; Fig. 2). We compared QTL mapping results from the present study and early studies, and the approximate locations of previously identified QTL are aligned on the genetic map developed herein, which is shown in supplemental Fig. S4. This map only contains QTLs detected from PI lines that were reported to confer high resistance to the post-2004 strain in the US (Call et al. 2012a, b) including PI 197085 (Szczechura et al. 2015), PI 197088 (Yoshioka et al. 2014), and PI 330628 (this study). QTL mapping studies with lines from North China cucumber types were also included (Ding et al. 2007; Bai et al. 2008; Zhang et al. 2013).
DMR QTLs have been detected in all seven cucumber chromosomes. Yoshioka et al. (2014) detected two QTLs in Chr3 (from CS-PMR1), one in Chr7 (from Santou). Chr2 harbored the moderate-effect QTL dm2.1 detected only in the present study. Using 77 F2:3 families and the BSA (bulked segregant analysis) approach, Ding et al. (2007) identified a single recessive locus that was responsible for the DM resistance in a north China type cucumber line ‘129’, which seemed to be close to dm4.1, the major-effect QTL identified in WI7120 from our study (Fig. S4). Bai et al. (2008); Zhang et al. (2013), and Yoshioka et al. (2014) all detected a DM resistance QTL at the distal end of the short arm of chromosome 1 with the resistance from north China or Japanese cucumber lines ‘S94’, ‘K8’, and ‘Santou’, respectively. Since Santou also had north China cucumber in its pedigree (Yoshioka et al. 2014), it seems that the north China fresh market cucumber carries the DM resistance QTL in chromosome 1 (Fig. S4), which was not detected in any PI lines of India or Pakistan origin (PI 197085, PI 197088 or PI 330628). On the other hand, the minor-effect QTL dm6.1 detected in the present study shared the similar location with a minor-effect QTL detected in the north China type cucumber line K8 by Zhang et al. (2013).
Cucumber chromosome 5 seems to harbor major-effect QTL for DM resistance from different sources (Fig. S4) including dm-1, the single recessive DM resistance locus originated from PI 197087 which was loosely linked with the dull fruit skin gene D (Vliet and Meysing 1974; Fanourakis and Simon 1987; Kennard et al. 1994; Horejsi et al. 2000). Zhang et al. (2013), Yoshioka et al. (2014), and Szczechura et al. (2015) each identified three QTLs in chromosome 5 for DM resistance in K8, PI 197088, and PI 197085, respectively, although the exact locations of these QTLs in each line and their relationships are not well characterized. Bai et al. (2008) identified one QTL from S94 in chromosome 5 which was in the distal end of the short arm of Chr5, whereas, the only QTL, dm5.1 we identified in WI7120 seemed to correspond to the QTL region detected by Zhang et al. (2013) and Yoshioka et al. (2014) (Fig. S4). These results suggest that, although PI 197085, PI 197088, and PI 330628 were all highly resistant to the post-2004 DM pathogen strain in the US cucumber field, the underlying genetic mechanisms may not be the same. Regardless, our study revealed the unique genetic architecture of DM resistance in WI7120. The three QTLs, dm2.1, dm4.1, and dm5.1 together could explain up to 75 % phenotypic variations with both dm4.1 and dm5.1 major-effect QTLs showing additive effects (Table 4). Such information is important for efficient use of this resistance source in cucumber breeding for DM resistance.
DM resistance in WI7120 versus Ps. cubensis races: one-fit-all?
The population of the DM pathogen Ps. cubensis in the field is highly heterogeneous and dynamic, which may consist of many isolates, pathotypes or races with varying degree of pathogenicity or virulence (Lebeda and Urban 2007; Quesada-Ocampo et al. 2012; Lebeda et al. 2013; reviewed by Cohen et al. 2015). Molecular fingerprinting studies have revealed that, while most Ps. cubensis isolate groups were present in all continents examined, the dominant ones were structured by geographic origin across continental scales (Quesada-Ocampo et al. 2012), which may result in differential inoculation responses on the same cucumber genotypes. For example, the Ps. cubensis population structure in the post-2004 cucumber field in the US was different from that in Europe; cucumber cultivars carrying dm-1 from PI 197087 have only moderate resistance in post-2004 fields in the US (Call et al. 2012a, b, which, however, still performs quite well in European countries (e.g. Lebeda 1999; Kozik et al. 2013). The pathogen population structure may change over time, and the virulence of Ps. cubensis population in the field may also shift (Quesada-Ocampo et al. 2012; Lebeda et al. 2013) thus rendering host resistance ineffective. This may be the reason for the defeat of host resistance genes in cucumber to the DM pathogen in the US in 1950s and early 2000s (Epps and Barnes 1952; Holmes et al. 2004, 2006).
In this study, the responses of the WI7120 × 9930 F3 mapping population to natural inoculation of the DM pathogen in all experiments were highly consistent which could be reflected from the significantly high and positive correlations of mean disease scores across these trials (Table 3). QTL analysis with data from these environments all supported the presence of four DM resistance QTLs in WI7120 (Fig. 2; Table 4). It is known that the population structures of Ps. cubensis in these fields are different (Quesada-Ocampo et al. 2012). Such results may suggest that host resistance in WI7120 is highly effective to current dominant isolate groups in the fields of the three countries (Italy, the Netherlands and the US), which is consistent with screening results in which PI 197088 and PI 330628 showed high resistance to DM inoculation in different countries in Asia, Europe, and the US (Call et al. 2012b; Chen and Cohen 2013). However, this does not mean that WI7120 is resistant to all isolates in the field pathogen populations. Innark et al. (2014) conducted field screening of cucumbers for DM resistance in Thailand and found that cucumber accessions from East Asia or Southeast Asia (e.g., PI 489752 from China) exhibited better DM resistance than those from India or surrounding areas; for example, PI 330628 from Pakistan only had moderate resistance. Chen and Cohen (2013) found that PI 330628 (WI7120) was highly resistant against multiple isolates (A, B, and C) of Ps. cubensis under laboratory, greenhouse, and field conditions in Israel. However, the F1 and F2 plants of PI 330628 × SMR18 (susceptible) showed differential inoculation responses to the three isolates. Therefore, more investigations are needed to understand the interactions of DM resistance QTL in WI7120 with isolates in different environments, which is important for efficient deployment of this resistance especially in the context of population dynamics of the DM pathogen in the fields.
Author contribution statement
YHW performed the research and conducted data analysis. YQW designed this experiment, participated in data analysis. YHW, KV, and TCW collected field data for US2013 and US2014 field trials. PAG and JS collected data for NL2013 experiment. XYZ and KO conducted IT2013 experiment. YHW and YQW wrote the manuscript with input from other coauthors. All authors reviewed and approved this submission.
References
Angelov D (1982) Relation of various samples of cucumbers to the mildew disease, Pseudoperonospora cubensis (Berk. & Curt.) Rostow; in Rep 2nd. Natl Symp Plant Immun (Plovdiv) 3:99–105
Arends D, Prins P, Jansen RC, Broman KW (2010) R/qtl: high-throughput multiple QTL mapping. Bioinformatics 26:2990–2992
Bai Z, Yuan X, Cai R, Liu L, He H, Zhou H, Pan J (2008) QTL analysis of downy mildew resistance in cucumber. Prog Nat Sci 18:706–710 (in Chinese)
Barnes WC (1948) The performance of Palmetto, a new downy mildew-resistant cucumber variety. Proc Amer Soc Hort Sci 51:437–444
Barnes WC, Epps WM (1954) An unreported type of resistance to cucumber downy mildew. Plant Dis Report 38:409–415
Broman KW, Wu H, Sen S, Churchill GA (2003) R/qtl: QTL mapping in experimental crosses. Bioinformatics 19:889–890
Call AD, Criswell AD, Wehner TC et al (2012a) Resistance of cucumber cultivars to a new strain of cucurbit downy mildew. HortSci 47:171–178
Call AD, Criswell AD, Wehner TC et al (2012b) Screening cucumber for resistance to downy mildew caused by Pseudoperonospora cubensis (Berk. and Curt.) Rostov. Crop Sci 52:577–592
Cavagnaro PF, Senalik DA, Yang LM, Simon PW, Harkins TT, Kodira CD, Huang SW, Weng Y (2010) Genome-wide characterization of simple sequence repeats in cucumber (Cucumis sativus L.). BMC Genom 11:569
Chen T, Cohen Y (2013) Isolate-dependent inheritance of resistance to downy mildew in cucumber (Abstracts of Presentations at the 34th Congress of the Israeli Phytopathological Society). Phytoparasitica 41:462
Cohen Y, Rotem J (1971) Rate of lesion development in relation to sporulating potential of Pseudoperonospora cubensis in cucumbers. Phytopathol 61:265–268
Cohen Y, Rubin AE, Galperin M, Ploch S, Runge F, Thines M (2014) Seed transmission of Pseudoperonospora cubensis. PLoS One 9:e109766
Cohen Y, Van den Langenberg KM, Wehner TC, Ojiambo PS, Hausbeck M, Quesada-Ocampo LM, Lebeda A, Sierotzki H, Gisi U (2015) Resurgence of Pseudoperonospora cubensis: the causal agent of cucurbit downy mildew. Phytopathol 105:998–1012
Criswell AD, Wehner TC, Klosinska U, Kozik E (2008) Use of sporulation and other leaf and vine traits for evaluation of resistance to downy mildew in cucumber. In: Pitrat M (ed) Proc Cucurbitaceae. INRA, Avignon, May 21–24th, 2008
Ding G, Qin Z, Zhou X, Fan J (2007) RAPD and SCAR markers linked to downy mildew resistance genes in cucumber. Acta Botanica Boreali-Occidentalia Sinica 27:1747–1751 (in Chinese)
Epps WM, Barnes WC (1952) The increased susceptibility of the palmetto cucumber to downy mildew in South Carolina. Plant Dis Rept 36:14–15
Fanourakis NE, Simon PW (1987) Analysis of genetic linkage in the cucumber. J Hered 78:238–242
Gilmour AR, Gogel BJ, Cullis BR, Thompson R (2009) ASReml user guide release 3.0
Hartung K, Piepho HP (2007) Are ordinal rating scales better than percent ratings? a statistical and “psychological” view. Euphytica 155:15–26
He X, Li YH, Pandey S, Yandell BS, Pathak M, Weng Y (2013) QTL mapping of powdery mildew resistance in WI 2757 cucumber (Cucumis sativus L.). Theor Appl Genet 126:2149–2161
Holdsworth WL, Summers CF, Glos M et al (2014) Development of downy mildew-resistant cucumbers for late-season production in the northeastern United States. HortSci 49:10–17
Holmes GJ, Main CE, Zeever ZT (2004) Cucurbit downy mildew: a unique pathosystem for disease forecasting. In: Spencer-Phillips PTN, Jeger M (eds) Advances in Downy Mildew Research. Kluwer Academic Publishers, Dordercht, pp 69–80
Holmes GJ, Wehner TC, Thornton A (2006) An old enemy re-emerges. American Vegetable Grower, pp 14–15
Horejsi T, Staub JE, Thomas C (2000) Linkage of random amplified polymorphic DNA markers to downy mildew resistance in cucumber (Cucumis sativus L.). Euphytica 115:105–113
Innark P, Ratanachan T, Khanobdee C, Samipak S, Jantasuriyarat C (2014) Downy mildew resistant/susceptible cucumber germplasm (Cucumis sativus L.) genetic diversity assessment using ISSR markers. Crop Prot 60:56–61
Jenkins SF Jr (1942) Downy mildew resistances in cucumber. J Hered 33:35–39
Jenkins SF Jr (1946) Studies on the inheritance of downy mildew resistance and of other characters in cucumbers. J Hered 37:261–271
Kennard WC, Poetter K, Dijkhuizen A, Meglic V, Staub JE, Havey MJ (1994) Linkages among RFLP, RAPD, isozyme, disease-resistance, and morphological markers in narrow and wide crosses of cucumber. Theor Appl Genet 89:42–48
Kozik EU, Klosińska U, Call AD, Wehner TC (2013) Heritability and genetic variance estimates for resistance to downy mildew in cucumber accession Ames 2354. Crop Sci 53:177–182
Lebeda A (1999) Pseudoperonospora cubensis on Cucumis and Cucurbita spp: resistance breeding aspects. Proc 1st Intl Symp Cucurbits Acta Hort 492:363–370
Lebeda A, Urban J (2007) Temporal changes in pathogenicity and fungicide resistance in Pseudoperonospora cubensis populations. Acta Hortic 731:327–336
Lebeda A, Pavelkova J, Sedlakova B, Urban J (2013) Structure and temporal shifts in virulence of Pseudoperonospora cubensis populations in the Czech Republic. Plant Pathol 62:336–345
Li Z, Zhang Z, Yan P, Huang S, Fei Z, Lin K (2011) RNA-Seq improves annotation of protein-coding genes in the cucumber genome. BMC Genom 12:540
Li Y, Wen C, Weng Y (2013) Fine mapping of the pleiotropic locus B for black spine and orange mature fruit color in cucumber identifies a 50 kb region containing a R2R3-MYB transcription factor. Theor Appl Genet 126:2187–2196
Miao H, Zhang S, Wang X et al (2011) A linkage map of cultivated cucumber (Cucumis sativus L.) with 248 microsatellite marker loci and seven genes for horticulturally important traits. Euphytica 182:167–176
Oerke E, Steiner U, Dehne H, Lindenthal M (2006) Thermal imaging of cucumber leaves affected by downy mildew and environmental conditions. J Exp Bot 57:2121–2132
Ojiambo PS, Gent DH, Quesada-Ocampo LM et al (2015) Epidemiology and population biology of Pseudoperonospora cubensis : a model system for management of downy mildews. Annu Rev Phytopathol 53:223–246
Pang X, Zhou X, Wan H, Chen J (2013) QTL mapping of downy mildew resistance in an introgression line derived from interspecific hybridization between cucumber and Cucumis hystrix. J Phytopathol 161:536–543
Petrov L, Boogert K, Sheck L, Baider A, Rubin E, Cohen Y (2000) Resistance to downy mildew, Pseudoperonospora cubensis, in cucumbers. Acta Hort (ISHS) 510:203–210
Poland J, Nelson RJ (2011) In the eye of the beholder: the effect of rater variability and different rating scales on QTL mapping. Phytopathology 101:290–298
Postman J, Volk G, Aldwinckle H (2010) Standardized plant disease evaluations will enhance resistance gene discovery. HortSci 45:1317–1320
Quesada-Ocampo LM, Granke LL, Olsen J, Gutting HC, Runge F, Thines M, Lebeda A, Hausbeck MK (2012) The genetic structure of Pseudoperonospora cubensis populations. Plant Dis 96:1459–1470
Ren Y, Zhang Z, Liu J et al (2009) An integrated genetic and cytogenetic map of the cucumber genome. PLoS One 4:e5795
Rubinstein M, Katzenellenbogen M, Eshed R, Rozen A, Katzir N, Colle M, Yang L, Grumet R, Weng Y, Sherman A, Ophir R (2015) Ultrahigh-density linkage map for cultivated cucumber (Cucumis sativus L.) using a single-nucleotide polymorphism genotyping array. PLoS One 10:e0124101
Savory EA, Granke LL, Quesada-Ocampo LM, Varbanova M, Hausbeck MK, Day B (2011) The cucurbit downy mildew pathogen Pseudoperonospora cubensis. Mol Plant Pathol 12:217–226
Szczechura W, Staniaszek M, Klosinska U, Kozik EU (2015) Molecular analysis of new sources of resistance to Pseudoperonospora cubensis (Berk. et Curt.) Rostov. in cucumber. Russian J Genet 51:974–979
van Vliet GJA, Meysing W (1974) Inheritance of resistance to Pseudoperonospora Cubensis Rost in cucumber (Cucumis Sativus L.). Euphytica 23:251–255
VandenLangenberg KM (2015) Studies on downy mildew resistance in cucumber (Cucumis sativus L.). North Carolina State University, Raleigh
Wehner TC, Shetty NV (1997) Downy mildew resistance of the cucumber germplasm collection in North Carolina field tests. Crop Sci 37:1331–1340
Xie W, Yu K, Pauls KP, Navabi A (2012) Application of image analysis in studies of quantitative disease resistance exemplified using common bacterial blight–common bean pathosystem. Phytopathology 102:434–442
Yang L, Koo DH, Li YH, Zhang XJ, Luan FS, Havey MJ, Jiang JM, Weng Y (2012) Chromosome rearrangements during domestication of cucumber as revealed by high-density genetic mapping and draft genome assembly. Plant J 71:895–906
Yang L, Li DW, Li YH, Gu XF, Huang SW, Garcia-Mas J, Weng Y (2013) A 1,681-locus consensus genetic map of cultivated cucumber including 67 NB-LRR resistance gene homolog and ten gene loci. BMC Plant Biol 13:53
Yoshioka Y, Sakata Y, Sugiyama M, Fukino N (2014) Identification of quantitative trait loci for downy mildew resistance in cucumber (Cucumis sativus L.). Euphytica 198:265–276
Zhang S, Liu MM, Miao H et al (2013) Chromosomal mapping and QTL analysis of resistance to downy mildew in Cucumis sativus. Plant Dis 97:245–251
Acknowledgments
The authors thank Kristin Haider for technical help. We are also indebted to Vittorio Stravato, and Giuseppe Carannante for help phenotyping of DM resistance in the IT2013 experiment. This research was supported by the US Department of Agriculture (USDA)-Specialty Crop Research Initiative Grant (SCRI, project # 2011-51181-30661) to YW. Names are necessary to report factually on available data; however, the USDA neither guarantees nor warrants the standard of the product, and the use of the name by USDA implies no approval of the product to the exclusion of others that may also be suitable. USDA is an equal opportunity provider and employer.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declares no conflict of interest.
Additional information
Communicated by S. Huang.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Wang, Y., VandenLangenberg, K., Wehner, T.C. et al. QTL mapping for downy mildew resistance in cucumber inbred line WI7120 (PI 330628). Theor Appl Genet 129, 1493–1505 (2016). https://doi.org/10.1007/s00122-016-2719-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00122-016-2719-x