Abstract
Key message
Diachronic analysis showed no significant changes in the level of genetic diversity occurred over the past 27 years’ domestication, which indicated genetic diversity was successfully maintained under on-farm conservation.
Abstract
Rice (Oryza sativa L.) is one of the earliest domesticated crop species. Its genetic diversity has been declining as a result of natural and artificial selection. In this study, we performed the first analysis of the levels and patterns of nucleotide variation in rice genomes under on-farm conservation in Yunnan during a 27-year period of domestication. We performed large-scale sequencing of 600 rice accessions with high diversity, which were collected in 1980 and 2007, using ten unlinked nuclear loci. Diachronic analysis showed no significant changes in the level of genetic diversity occurring over the past 27 years’ domestication, which indicated genetic diversity was successfully maintained under on-farm conservation. Population structure revealed that the rice landraces could be grouped into two subpopulations, namely the indica and japonica groups. Interestingly, the alternate distribution of indica and japonica rice landraces could be found in each ecological zone. The results of AMOVA showed that on-farm conservation provides opportunities for continued differentiation and variation of landraces. Therefore, dynamic conservation measures such as on-farm conservation (which is a backup, complementary strategy to ex situ conservation) should be encouraged and enhanced, especially in crop genetic diversity centers. The results of this study offered accurate insights into short-term evolutionary processes and provided a scientific basis for on-farm management practices.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Rice (Oryza sativa L.) is one of the earliest domesticated crop species. Its genetic diversity declined during domestication and artificial selection processes (Londo et al. 2006). The most extreme loss of diversity occurred in modern, high yield rice cultivars, which are often invariant for many genetic makers (Pusadee et al. 2009). By contrast, traditional rice landraces that have been preserved by indigenous farmers have retained rich genetic variation. This variation can be used in breeding for improved yield and increased resistance to pests, diseases, alkali, and salt; therefore, these germplasms are extremely important for enriching the gene pool. Rice landraces were successfully employed in many rice-breeding projects in China (Dai et al. 1999; Zhu et al. 2003; Liu et al. 2007). However, with the development of modern agriculture, local rice landraces were largely replaced by genetically uniform modern varieties, which led to an erosion of the rice gene pool. Rice landraces are no longer planted in most provinces in China, with the exception of some ethnic minority regions in areas such as Yunnan and Guizhou.
Yunnan Province in southwest China is characterized by an alternate distribution of mountains, plateaus, and basins. These features provide diverse geographic and climatic types, which help make this region one of the largest centers of genetic diversity for rice in China and indeed the world (Zeng et al. 2001, 2007; Zhang et al. 2006). Rice landraces in Yunnan are widely distributed in a region from 21°8′32″N to 29°11′18″N and 97°31′39″E to 106°11′47″E and are planted at different altitudes and under diverse climate conditions (Zeng et al. 2007). Such wide geographic distribution and diverse growing conditions contribute to the high genetic diversity of rice landraces in Yunnan. A remarkably diverse set of rice landraces, including upland and lowland, glutinous and non-glutinous, and indica and japonica varieties are all found in Yunnan (Zeng et al. 2003). Therefore, the strategic conservation and use of rice landraces in Yunnan are of great importance.
Since the 1970s, large numbers of landraces have been sampled and stored in ex situ gene banks. However, the storage of genetic resources in collections as backup seed stocks for ex situ conservation does not substitute for the evolution of crop plants in the field. Another form of conservation, on-farm (in situ), is best viewed as a complementary strategy to ex situ conservation. Plant genetic resources grown using on-farm conservation strategies continue to evolve in the agro-ecosystem and are influenced by natural selection in addition to selection pressures imposed by farmers, thus providing opportunities for continuous differentiation and variation (Bellon et al. 1997). Plant populations on farms have the capacity to support a greater number of rare alleles and different genotypes compared to accessions maintained in gene banks (Brown 2000). Therefore, on-farm conservation received a great deal of attention (Altieri and Merrick 1987; Brush 1991; Brush and Meng 1998; Maxted et al. 2002; Hammera et al. 2003), although farmers have been using this technique for centuries. It is essential to develop a dynamic view of the diversity that exists on farms, which would enable us to accurately pinpoint suitable areas for conservation and to design effective, targeted intervention strategies. Several studies examined the dynamics of genetic diversity over time in rice landraces under on-farm conservation in Yunnan; these studies were based on morphological traits or SSR markers (Xu et al. 2010a, 2011; Yan et al. 2012; Sun et al. 2012). Unfortunately, the results of these studies are controversial, which may be due to the limited number of accessions/locations sampled or the different marker systems used.
Recently, the development of DNA sequencing technology allowed multi-locus DNA sequences to be successfully used to address the genetic diversity and domestication history of crop species (Wright and Gaut 2005; Zhu et al. 2007; Zheng and Ge 2010; Li et al. 2011a, b; Wei et al. 2012). Differences in DNA sequences can directly reflect genetic differences, making direct sequencing an ideal method for revealing genetic diversity.
In this study, we performed the first analysis of the levels and patterns of nucleotide variation in Yunnan rice landraces under on-farm conservation conditions; this analysis was based on the sequences of ten unlinked nuclear loci. Furthermore, we performed large-scale sequencing of 600 rice accessions with high diversity, which were collected in 1980 and 2007. The aim of this study was to explore the dynamics of genetic diversity over a 27-year period in rice landraces under on-farm conservation in Yunnan through multi-locus analysis of nucleotide variation. The results of this study provide a scientific basis for the conservation and use of rice landrace germplasms, particularly for on-farm management practices.
Materials and methods
Plant materials
This study was performed at a large scale using 600 on-farm conserved rice landraces from Yunnan Province, China. These included 332 accessions collected in 1980, and 268 accessions collected in 2007 (Supplementary Table 1). According to documenting Ting’s taxonomic system of rice, the catalogue identified the counties, in which the accessions were collected; on the basis of this, accessions were grouped into five ecological zones (EZI, EZII, EZIII, EZIV, and EZV; Zhou 1988) (Supplementary Table 2), covering a wide geographic distribution and diverse growing conditions; these represent most of the diversity of rice landraces in Yunnan. The geographic localities of the rice landraces sampled in this study are shown in Fig. 1.
Geographical distribution of five rice ecological zones and the localities of the rice landraces sampled in this study. I, Indica/Japonica rice ecological zone at the middle of Yunnan; II, Single/double-season rice ecological zone of southern Yunnan; III, Lowland/upland-rice ecological zone at the edge of southern Yunnan; IV, Japonica-rice ecological zone on the plateau of northeast Yunnan; V, Cold Japonica-rice ecological zone of northwest Yunnan. Solid circles and triangles represent the rice landraces in 1980 and 2007, respectively
DNA extraction, SSR genotyping, and sequencing
Total genomic DNA was extracted from fresh young leaves using a modified CTAB procedure (Doyle and Dickson 1987). A set of 48 SSRs (Supplementary Table 3), evenly distributed throughout the rice genome, was used to analyze population structure. SSRs were amplified using a polymerase chain reaction (PCR) with fluorescently labeled primers in a 10-μL reaction volume containing 20 ng genomic DNA, 10 × PCR reaction buffer, 10 mM Mixture dNTP, 2 μM primers, and 0.5 unit of Taq polymerase. The PCR profile was as follows: pre-denature at 94 °C for 5 min; 36 cycles of denaturation at 94 °C for 30 s; annealing at 55–60 °C (dependent on primers) for 30 s; and extension at 72 °C for 40 s, followed by a final extension at 72 °C for 10 min. PCR products were size separated on a 3730XL DNA Sequencer equipped with GENESCAN software (ABI, USA). Fragment size was recorded using Gene Marker V1.6 (SoftGene, State College, PA, USA) and manually re-checked.
Genes used in this study represent 10 unlinked nuclear loci across the rice chromosomes. Five of these genes (CatA, GBSSII, Os1977, STS22, and STS90) were used in a previous study on the nucleotide diversity of rice (Caicedo et al. 2007; Li et al. 2011a). In addition, five genes (Ehd1, S5, Pid3, GS3, and GS5) associated with important agronomic traits of rice were also chosen (Doi et al. 2004; Chen et al. 2008; Shang et al. 2009; Mao et al. 2010; Li et al. 2011b). The schematic diagrams of all ten nuclear loci are shown in Supplementary Fig. 1. Information on the genomic location and putative gene functions of these genes and the primer sequences of their amplified regions is given in Supplementary Table 4.
For the detection of genes, PCR was performed in a 25-μl volume consisting of 0.2 μM of each primer, 200 μM of each dNTP, 10 mM Tris–HCl (pH 8.3), 50 mM KCl, 1.5 mM MgCl2, 0.5 U HiFi DNA polymerase (Transgen, China), and 30 ng genomic DNA. For GC-rich sequences, 10 % DMSO was added to the reaction mixture to facilitate denaturation during the PCR. The PCR profile was as follows: pre-denature at 94 °C for 5 min; 36 cycles of run, each followed by denaturation at 94 °C for 30 s, annealing at 55–60 °C (dependent on primers) for 30 s, extension at 72 °C for 1.5 min; and a final extension at 72 °C for 10 min. The PCR products were electrophoresed in 1 % agarose gels, and then DNA fragments were cut from the gel and purified using a Tiangen Gel Extraction kit (Tiangen, China).
Sequencing reactions were performed using an ABI 3730 automated sequencer (Applied Biosystems, USA). Initially, all samples were directly sequenced. However, if haplotypes could not be readily inferred because of heterozygosity, the PCR product was ligated into an EASY vector (Transgen, China), and at least four clones sequenced. When singletons (polymorphisms that occurred in only one sequence relative to all remainder sequences) were found, repeated PCR amplification and sequencing was conducted (to exclude singletons resulting from Taq polymerase errors).
Sequence analysis and neutrality test
DNA sequences were aligned using the ClustalX 1.83 program (Thompson et al. 1997) and edited using BioEdit 7.0.9.0 (Hall 1999). Indels (insertions/deletions) were not included in the analysis. Statistical analysis was conducted using DnaSP version 5.0 (Rozas 2009); items analyzed included the number of segregating sites (S), the number of haplotypes (h), haplotype diversity (Hd), and two parameters of nucleotide diversity: mean pairwise differences (π) (Nei 1987), and Watterson’s estimator based on the number of segregating sites (θw) (Watterson 1975). The minimum number of recombination events (Rm) was estimated using the four-gamete test (Hudson and Kaplan 1985). Considering the effects of sample size on estimating genetic diversity, haplotype richness was estimated by a rarefaction method implemented in HP-RARE software (Kalinowski 2005).
Two neutrality tests, Tajima’s D value (Tajima 1989), and Fu and Li’s D*/F* (Fu and Li 1993), were calculated for all loci, to test the neutral mutation hypothesis. Tajima’s D value is based on the discrepancy between π and θw, whereas Fu and Li’s D*/F* relies on differences between the number of polymorphic sites in external and internal groups. In these two tests, negative values indicate an excess of low-frequency polymorphisms, whereas positive values indicate an excess of intermediate variants.
Phylogene network analysis
Haplotype networks were constructed by mutational steps using Network 4.5 (Bandelt et al. 1999). These networks represent the genetic distance of DNA sequences or alleles and are mainly composed of circles of different size and color, and lines linking the circles. The circle size is proportional to the number of samples within a given haplotype, while the lines between haplotypes represent mutational steps between the alleles. The numbers next to the circle represent the haplotype number. Circle color represents different collection times of rice landraces. If more than one nucleotide difference exists between linked haplotypes, it is indicated by numbers next to the lines. Because many haplotypes were obtained for the nuclear loci, only major haplotypes, containing more than three individuals each, were selected for construction of the networks.
Population structure and differentiation
The model-based program STRUCTURE 2.2 (Pritchard et al. 2000; Falush et al. 2003) was used to determine population structure based on the 48 SSRs. Five independent runs were performed for each k value (from 1 to 12), using a burn-in length of 100,000, a run length of 100,000, and a model for admixture and correlated allele frequencies. The k value was determined by LnP(D) in STRUCTURE output and an ad hoc statistical Δk (Evanno et al. 2005).
The overall distribution of nucleotide diversity among periods was investigated using analysis of molecular variance (AMOVA), as implemented in the Arlequin 3.01 program (Excoffier et al. 1992). Sequence variation was hierarchically partitioned between the two periods, among ecological zones within periods, and within ecological zones. The significance of all estimated fixation indices was tested using 10,000 permutations, as described by Excoffier et al. (1992). Pairwise F ST, generally expressed as the proportion of genetic diversity as a result of allele frequency differences among populations (Holsinger and Weir 2009), was used to measure population differentiation within and between periods, as implemented in the Arlequin 3.01 program (Excoffier et al. 1992).
Results
Nucleotide diversity and neutrality test
Ten unlinked gene regions, including CatA, GBSSII, Os1977, STS22, STS90, S5, Pid3, Ehd1, GS3, and GS5, were sequenced in 332 accessions of Yunnan rice landraces collected in 1980, as well as 268 accessions collected in 2007. The length of aligned sequence for each locus ranged from 420 to 627 bp, with a total length of 4994 bp. Ten indel polymorphisms, ranging from 1 to 5 bp in length, were identified; these were not considered in the data analyses. The standard statistics of sequence polymorphisms for each locus are summarized in Table 1. The term θw indicates the diversity of segregating sites, which was calculated for each locus. This term ranged from 0.0011 (STS22) to 0.0079 (CatA) in accessions from 1980, and from 0.0012 (STS22) to 0.0082 (CatA) in accessions from 2007. The average silent nucleotide variation across loci was not significantly different between rice landraces from 1980 (π = 0.0061, θw = 0.0032) versus 2007 (π = 0.0058, θw = 0.0033; P > 0.05 for both π and θw). Furthermore, there were no fixed polymorphisms; however, a much larger number of shared polymorphisms were detected between rice landraces from 1980 versus 2007 (Table 2).
Four-gamete testing revealed that the Rm ranged from 0 to 6 in landraces from 1980, with an average of 2.8, and from 0 to 4 in landraces from 2007, with an average of 2.0, indicating that there was low heterozygosity in the rice landraces due to high levels of inbreeding. Neutrality test results for each locus, including Tajima’s D and Fu and Li’s D* and F* values, are presented in Table 1. Neutrality test values for most gene regions were not significant. In accessions from 1980, significantly positive Tajima’s D values were found for CatA (D = 2.131), Os1977 (D = 2.668), Ehd1 (D = 2.626), and GS5 (D = 2.412), while in accessions from 2007, GBSSII (D = 2.168) and Os1977 (D = 2.549) were significantly positive. It is noteworthy that these loci had significantly positive Tajima’s D values. Such values indicate the occurrence of numerous moderate-frequency polymorphisms in the rice landraces, which may be a consequence of positive or balancing selection during domestication.
Phylogenetic and geographic analyses with haplotypes
Figure 2 shows haplotype networks constructed based on the major haplotypes for all ten loci. The total number of haplotypes for CatA, GBSSII, Os1977, STS22, STS90, S5, Pid3, Ehd1, GS3, and GS5 was 7, 4, 3, 6, 9, 5, 7, 18, 10, and 11, respectively. The haplotype numbers of rice landraces from 1980 and 2007 in these networks were 7, 7; 4, 4; 3, 3; 6, 5; 9, 9; 5, 4; 6, 7; 16, 14; 10, 10; and 9, 10, respectively. In total, rice landraces from 2007 had maintained 90.7 % of the haplotypes found in 1980. But seven haplotypes were lost (H_6 of STS22; H_4 of S5; H_10, 12, 14 and H_16 of Ehd1; H_6 of GS5), with an average frequency of 1.38 %, ranging from 0.90 to 3.59 %; five haplotypes (H_7 of Pid3; H_17 and H_18 of Ehd1; H_10 and H_11 of GS5) were new, with an average frequency of 1.90 %, ranging from 1.16 to 4.25 % (data not shown). Therefore, with the exception of a few lost and new haplotypes, rice landraces grown under on-farm conservation conditions in 2007 maintained almost all of the haplotypes found in 1980 after 27 years of domestication. In addition, there were some distinct differences in haplotype frequency between rice landraces from 1980 versus 2007. Six haplotypes (H_5 of STS22; H_2 of S5; H_3 of Pid3; H_5 of Ehd1; and H_2 and H_7 of CatA) were common in rice landraces from 1980, with an average frequency of 8.02 %, ranging from 5.31 to 12.65 %, but rare in those from 2007, with an average frequency of 3.85 %, ranging from 1.54 to 4.92 %, whereas two haplotypes (H_5 of S5 and H_5 of STS90) were rare in rice from 1980, with an average frequency of 1.88 %, ranging from 0.92 to 2.84 %, but more frequent in those from 2007, with an average frequency of 9.59 %, ranging from 5.93 to 13.26 % (data not shown).
Haplotype networks of the ten loci. The circle size is proportional to the quantity of the samples within a given haplotype, and the numbers next to the circle represent the haplotype number. Lines between haplotypes represent mutational steps between alleles. When more than one nucleotide difference existed between linked haplotypes, it is indicated by the numbers next to the lines. Colours for rice landraces collected from different periods: yellow, from 1980; blue, from 2007 (color figure online)
To compare the geographic distribution of haplotypes between the two periods, we employed the Ehd1 (Early heading date 1) gene, which carried the most polymorphisms, for geographic analysis. Figure 3 shows that haplotypes H_1 and H_3 were widely distributed in Yunnan in both 1980 and 2007, which indicates that these two haplotypes were favored in local planting systems. However, with the exception of Yuanyang (EZII), Jiangcheng (EZIII), Lancang (EZIII), Menghai (EZIII), and Mengla (EZIII), the number of haplotypes in rice landraces in each county within various ecological zones was sharply lower in 2007 than in 1980, with the average number decreasing from 5.1 in 1980 to 2.8 in 2007 (data not shown). This result indicates that the geographic distribution of haplotypes have been changed by natural or artificial selection, during the 27 years of domestication.
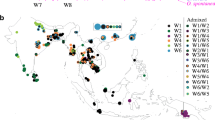
A map showing the sampled populations of rice landraces and the distribution of haplotypes. a, b Show rice landraces in 1980 and 2007, respectively. Phylogenetic relationship of the haplotype based on the NJ analysis is indicated below the map. Pie charts show the proportions of the haplotypes within each county. Haplotypes are indicated by different colors (color figure online)
Genetic variations in various ecological zones
We calculated haplotype richness for rice landraces grown in each ecological zone from 1980 and 2007 (Supplementary Table 5). Haplotype richness represents the genetic diversity of a sample. Therefore, rice landraces from EZIII had the richest diversity in both 1980 and 2007, with an average haplotype richness of 6.29 and 6.39, ranging from 2.84 to 13.30 and 3.00 to 12.99, respectively. In fact, EZIII not only contains a larger number of rice landraces, but also harbors three species of wild rice (O. granulata, O. rufipogon, and O. officinalis) (Zeng et al. 2007). Additionally, regression analysis indicated that the genetic diversity of rice landraces in Yunnan exhibited a geographic decline from south to north (Supplementary Fig. 2). This result provides clear-cut molecular evidence that southwest Yunnan is the center of genetic diversity of rice landraces in Yunnan, which is in agreement with the results of previous studies (Zeng et al. 2001, 2007; Zhang et al. 2006).
Table 3 summarizes the private haplotype richness of rice landraces in each ecological zone from 1980 and 2007, respectively. The average private haplotype richness of rice landraces in all ecological zones was 1.25 and 0.95, ranging from 0.67 (EZIV) to 1.68 (EZI) in 1980 and 0.52 (EZIV) to 1.26 (EZIII) in 2007. Among all ecological zones, rice landraces from EZI in 1980 had the highest percentage of private haplotype richness (33.1 %) and the lowest percentage of private haplotype richness (16.5 %) in 2007, whereas rice landraces in EZIII had the lowest percentage of private haplotype richness (18.3 %) in 1980. This result revealed that rice landraces grown in the ecological zone with the richest genetic diversity (EZIII) lost less haplotype richness than those grown in the other ecological zones, and they better maintained the genetic diversity during 27 years of domestication.
Genetic structure and differentiation
To infer population structure, we performed model-based simulations for rice from both 1980 and 2007 using 48 SSRs. The LnP(D) value increased with increasing k, but there was a sharp peak of Δk at k = 2 (Supplementary Fig. 3) in both 1980 and 2007. These results suggest that rice landraces from both 1980 and 2007 could be grouped into two subpopulations, including P1 and P2, respectively (Fig. 4). For rice from 1980, P1 includes 177 accessions; all are indica or indica-like. P2 contains 155 accessions; all are japonica or japonica-like. Similarly, for rice from 2007, most accessions in P1 and P2 are indica or indica-like and japonica or japonica-like, respectively (Supplementary Table 1). These results demonstrate that rice landraces from Yunnan strongly differentiated into indica and japonica groups. Figure 4c and d shows the distribution of rice accessions from the two subpopulations in each ecological zone. Interestingly, both indica and japonica were present in each ecological zone, which may be the result of the unique vertical three-dimensional climate in Yunnan.
Hierarchical AMOVA results are presented in Table 4. When considering only the two periods, variance components attributed between the periods were significant for CatA, STS22, S5, Ehd1, and GS3; however, variance components between periods comprised less than 3 % of the total variation for each locus. Genetic differentiation within periods was significant at all ten loci, with an average value of 99.22 %. When considering both ecological zones and periods, variance components attributed to between periods were not significant for any loci. Interestingly, genetic differentiation among ecological zones within periods and within ecological zones was significant at all ten loci. Of the total variation, components among ecological zones within periods ranged from 1.96 % (S5) to 7.51 % (GBSSII), while within ecological zones, the variation ranged from 95.34 % (GBSSII) to 98.46 % (Pid3). For all gene loci, the average genetic variation among ecological zones within periods (3.88 %), and within ecological zones within periods (96.65 %), was greater than that found between periods (−0.53 %).
The F ST results reveal that genetic differentiation between periods (−0.0139 to 0.0537) was lower than that among ecological zones within periods (−0.0021 to 0.0644 for rice accessions from 1980, and −0.0138 to 0.1728 for rice accessions from 2007; Table 5). This result, which is fairly consistent with the results of AMOVA analysis, reveals that there was little differentiation between periods. The average F ST value among ecological zones in 2007 (0.0664) was higher than in 1980 (0.0310), which demonstrates that the effects of different selection events are revealed by genetic differentiation among ecological zones and that genetic differentiation became stronger among different ecological zones during 27 years of domestication.
Discussion
Diachronic analysis of genetic diversity
As Yunnan is a known diversity center for rice in China, much research focused on genetic diversity and recent dynamics in Yunnan rice landraces (Zhang et al. 2006; Zeng et al. 2007; Xu et al. 2011; Yan et al. 2012). However, to date, such research focused exclusively on morphological traits or SSR markers in a limited number of varieties (Xu et al. 2010, 2011; Yan et al. 2012; Sun et al. 2012). The DNA sequences used in the current study included ten unlinked nuclear genes, which could directly be used to reflect genetic variation (Zheng and Ge 2010; Li et al. 2011a; Wei et al. 2012). Furthermore, we employed samples covering a wide geographic distribution and diverse growing conditions in Yunnan, which represent most of the known diversity within Yunnan rice landraces. Therefore, this study represents a thorough survey of changes in genetic diversity in Yunnan rice landraces under on-farm conservation across two different periods.
In this study, the average silent nucleotide variation across loci was not significantly different between rice landraces grown under on-farm conservation in 1980 versus 2007. Furthermore, no fixed polymorphisms, but a large number of shared polymorphisms, were detected between the two periods. This result provides clear-cut molecular evidence that no significant temporal changes in the level of genetic diversity occurred over the past 27 years. This view is further supported by the results of haplotype analyses, which indicate that, with the exception of a few lost and new haplotypes, the rice landraces grown under on-farm conservation in 2007 maintained almost all of the haplotypes of rice landraces grown in 1980.
Globally, our results are in agreement with the results of a temporal survey employing SSR markers to examine genetic diversity of rice landraces from Hani’s terraced fields in Yuanyang county, Yunnan (Xu et al. 2011). Similar results were obtained in a study of agricultural areas linked to 71 villages in Niger by Deu et al. using spatio-temporal analysis of sorghum gene diversity (Deu et al. 2010). By contrast, Tsegaye and Berg identified strong genetic erosion (i.e., loss of landraces) in tetraploid wheat landraces in central Ethiopia (Tsegaye and Berg 2007). It is clear from our research that genetic diversity levels in rice landrace germplasms grown under on-farm conservation conditions remained largely unchanged during the period investigated and were thus successfully maintained by local farmers in Yunnan.
Dynamic changes in haplotype
Most haplotypes occurring in on-farm-conserved rice landraces from 1980 were also found in landraces from 2007. Only seven haplotypes were lost, while five were acquired. Interestingly, all of the lost haplotypes were rare haplotypes with frequencies of less than 0.05, indicating that rare haplotypes were lost more easily than frequently occurring ones. This phenomenon was observed in previous studies (Fu et al. 2005). In addition, distinct differences in haplotype frequency were observed between rice landraces grown under on-farm conservation in 1980 versus 2007. Six haplotypes were common in rice landraces in 1980, with an average of 8.02 %, but rare in 2007, with an average of 3.85 %, whereas two rare haplotypes in 1980 were more frequent in 2007, with an average of 1.88 and 9.59 %, respectively. Although this phenomenon may have been caused by the sampling locations and scale, and in fact we carefully employed samples from almost the same locations in two periods. In each location, we selected samples which can represent most of the diversity within Yunnan rice landraces in order to reduce sampling error as much as possible. These findings indicate that on-farm conservation enabled continued differentiation of conserved crop varieties in their original habitats, generating new genetic variations (haplotypes) through farmers’ cultivation and management; meanwhile, the haplotype frequency also dynamically changed. As a dynamic form of conservation, on-farm conservation, which is presently used as a backup, complementary strategy to ex situ conservation, should be encouraged and enhanced, especially in genetic centers of crop diversity.
The private haplotype richness of rice landraces grown under on-farm conservation conditions were detected in each ecological zone during the period examined. Among all ecological zones, rice landraces in EZI lost the most haplotype richness (33.1 %), while rice landraces in EZIII lost the least (18.3 %). This result indicates that EZIII, which has the richest diversity, could better maintain genetic diversity during the period of domestication. However, EZI has the most convenient transportation system and a more developed economy, making it the most agriculturally developed ecological zone in Yunnan. Therefore, local rice landraces in EZI were largely replaced by improved varieties, leading to the loss of genetic diversity in rice landraces in this zone.
Geographic analysis in this study revealed that the distribution of haplotypes changed during the 27-year period under investigation. For the Ehd1 gene (Fig. 3), with the exception of a few counties, the number of haplotypes of rice landraces in each county within ecological zones was sharply lower in 2007 (2.8) than in 1980 (5.1). This result indicates that the frequency of some haplotypes gradually decreased (or they were even eliminated) during a 27-year test period. Ehd1 is an important photoperiod sensitivity gene in rice encoding a B-type response regulator that confers short-day promotion of flowering and controls FT-like gene expression (Doi et al. 2004). Indeed, the average days to heading value of rice landraces in 2007 increased by 3 days compared with that in 1980, which may have resulted from changes in the cropping system. In the past, double-season rice landraces were widely grown in most regions of Yunnan, whereas single-season rice landraces are currently planted in most regions. When we compared the agronomic traits of rice landraces grown in 1980 versus 2007, we found that there was a significant decrease in plant height and an increase in grains per panicle. This indicated that under on-farm conversation, farmer selection in combination with natural selection results in landraces with high levels of adaptation to the environments and as well as for agronomic traits (Pusadee et al. 2009). These findings demonstrate that rice landraces under on-farm conservation may provide a dynamic gene pool for favorable traits, and on-farm, in situ conservation is an essential strategy for future crop breeding efforts.
Genetic differentiation between and within periods
Model-based structure analysis revealed the presence of two subpopulations of rice landraces in Yunnan in 1980 and 2007, which included indica and japonica groups, respectively. Cultivated rice is classified into two major subspecies of O. sativa (indica and japonica) and is further subdivided into genetically differentiated groups, including Glaszmann's six groups (I–VI) (Glaszmann 1987) and Garris et al.'s five groups (indica, aus, aromatic, temperate japonica, and tropical japonica) (Garris et al.2005). The results of this study are in agreement with those of Zeng et al. (2007), i.e., differences between indica and japonica subspecies are very apparent at the DNA level. Interestingly, both indica and japonica were found in each ecological zone (Fig. 4c, d). It is generally recognized that indica–japonica differentiation in O. sativa is largely attributable to adaptation to very different environments, including different temperature and water conditions (Second 1982; Glaszmann 1987). In Yunnan, indica rice germplasm is mainly distributed in areas below 1400 m, while japonica rice germplasm is mostly grown in areas above 1800 m. An interesting interaction distribution of indica and japonica rice occurs between 1400 and 1800 m (Gao 2003). Yunnan has a unique vertical three-dimensional climate, with wide-ranging altitudes in each ecological zone. For example, the altitudes of the counties examined in this study varied from 425 to 2041 m in EZII. Therefore, alternate distribution of indica and japonica rice landraces occurred in each ecological zone.
Notably, significant genetic differentiation between rice landraces between the two periods occurred in half of the loci sampled, with less than 3 % of total genetic variation occurring for each locus when considering only the two periods (Table 4). These results indicate that the domestication of rice landraces under on-farm conservation conditions occurred by natural and artificial selection in the agro-ecosystem, providing opportunities for continued differentiation and variation of landraces. When considering both ecological zones and periods, AMOVA analysis revealed significant gene differentiation among ecological zones within periods, as well as within ecological zones within periods, at all ten loci, with an average of 3.88 and 96.65 % of total genetic variation, respectively. The F ST values were fairly consistent with the AMOVA analysis, revealing little differentiation between periods. Furthermore, the average F ST value among ecological zones was higher in 2007 (0.0664) than in 1980 (0.0310). This result indicates that the adaptation/domestication of rice landraces continued in their original ecological zones, and genetic differentiation gradually became stronger among ecological zones.
An ideal location for on-farm conservation
As previously mentioned, genetic diversity in rice landraces under on-farm conservation conditions was successfully maintained by local farmers in Yunnan, and the domestication of rice landraces continues to occur by natural and artificial selection in the agro-ecosystem. Thus, what are the factors that maintain genetic diversity of rice landraces under on-farm conservation in Yunnan?
Many conditions in a genetic diversity center accelerate evolution and help maintain genetic variation in rice landraces (Harlan 1975), such as the following:
1. A long history of cultivation in subsistence systems. According to archaeological evidence, rice cultivation has a long history in Yunnan (over 5000 years). Some of the primitive, extensive cultivation approaches used by the ancestors of some ethnic minorities are still used in these areas today (Zhang et al. 2006).
2. Ecological diversity. The genetic diversity of rice landraces in Yunnan is closely associated with the diverse climatic ecotypes and environmental heterogeneity, which brought about a comparatively diverse selection of rice. The landscape and topography in Yunnan is extremely complicated, and alternative distribution of rice is obvious (the altitude difference is as high as 1500 m), especially in the Hengduan Mountains, Gaoligong Mountains, West Yunnan Great Gorge, and southern part of the Yun-Gui Plateau (Zeng et al. 2007). According to the different ecological environments, cropping systems, and cultivation habits, there are five ecological zones for rice planting in Yunnan. In this study, we analyzed the genetic differentiation of rice landraces among ecological zones under on-farm conservation and found that genetic differentiation gradually became stronger during 27 years of domestication. These findings indicate that the diverse demands and cultivation methods of rice landraces used by local farmers in different ecological zones contributed to much of the genetic variation and that continued on-farm conservation will improve the genetic diversity of rice landraces within agro-ecosystems.
3. Ethnological diversity. The abundant rice landraces in Yunnan are partly a result of land use by diverse indigenous minorities. For example, glutinous rice is the main food of the Dai people (Xu et al. 2010b) and is also used as a food for ritual sacrifice to ancestors and deities. Therefore, glutinous rice is one of the most well-protected rice landraces in Yunnan. Furthermore, numerous types of colored rice are preserved by local farmers due to the dietary customs of the Blang people. In this study, we found the genetic differentiation of landraces used among ethnic groups to be significant (data not shown), demonstrating that the genetic diversity of rice landraces at the DNA level is closely related to the cultural customs and cultivation methods of different ethnic groups.
4. Crop features. Rice landraces are defined as geographically or ecologically distinctive populations, displaying conspicuously diverse in their genetic composition both between landraces and within them (Brown 1978). To date, many traditional rice landraces are still planted in Yunnan despite the availability of newer varieties due to the beneficial characteristics of the traditional landraces, such as their better adaptability, built-in resistance, high quality, and relationship to the culture of ethnic minorities; many of these landraces have been planted for more than 50 years. In addition, the rice landraces found in Yunnan have genetic heterogeneity, displaying rich genetic variation within the same variety.
5. Introgression with wild relatives. Gene flow between landraces and their wild relatives greatly enhances the genetic variation of rice landraces in Yunnan (Zhang et al. 2006). Furthermore, gene flow can occur by seed exchanges between farmers, which may also help maintain variation both between landraces and within them (Parzies et al. 2004; Sirabanchongkran et al. 2004; Dong et al. 2010). Therefore, Yunnan, a genetic diversity center, is an ideal location for on-farm conservation.
Author contribution statement
D.C. performed statistical analysis and wrote the paper. L. Z. H. and D.C. conceived and designed the experiments; D.C., J. M. L. and X. D. M. performed the experiments; C. F. T., X. X. A., T. Q. Y., E. L. Z., G. L. C. and F. R. X. contributed reagents/materials/analysis tools. Y. L. Q., L. Y. D. and L. Z. H. supervised the thesis.
References
Altieri MA, Merrick LC (1987) In situ conservation of crop genetic resources maintenance of traditional farming systems. Econ Bot 41:86–96
Bellon MR, Pham JL, Jackson MT (1997) Genetic conservation: a role for rice farmers. In: Hawkes JG (ed) Plant conservation: the in situ approach. Chapman and Hall, IPGRI, London
Brown AHD (2000) The genetic structure of crop landraces and the challenge to conserve them in situ on farms. In: Brush SB (ed) Genes in the fields: on–farm conservation of crop diversity. International Plant Genetic Resources Institute (IPGRI), Rome, International Development Research Centre (IDRC), Ottawa and Lewis Publishers, Boca Raton, pp 29–48
Brush SB (1991) A farmer–based approach to conserving crop germplasm. Econ Bot 45:153–165
Brush SB, Meng E (1998) Farmers’ valuation and conservation of crop genetic resources. Genet Resour Crop Evol 45:139–150
Bandelt HJ, Forster P, Rohl A (1999) Median–joining networks for inferring intraspecific phylogenies. Mol Biol Evol 16:37–48
Brown AHD (1978) Isozymes, plant population genetics structure and genetic conservation. Theor Appl Genet 52:145–157
Caicedo AL, Williamson SH, Hernandez RD et al (2007) Genome–wide patterns of nucleotide polymorphism in domesticated rice. PLoS Genet 3:1745–1756
Chen JJ, Ding JH, Ouyang YD et al (2008) A triallelic system of S5 is a major regulator of the reproductive barrier and compatibility of indica–japonica hybrids in rice. Proc Natl Acad Sci 105:11436–11441
Dai LY, Ye CR, Xu FR et al (1999) Genetic analysis on cold tolerance characteristics of Yunnan rice landrace (Oryza sativa L.) Kunmingxiaobaigu. Chin J Rice Sci 13:73–76
Doyle JJ, Dickson EE (1987) Preservation of plant samples for DNA restriction endonuclease analysis. Taxon 36:715–722
Deu M, Sagnard F, Chantereau J et al (2010) Spatio–temporal dynamics of genetic diversity in Sorghum bicolor in Niger. Theor Appl Genet 120:1301–1313
Doi K, Izawa T, Fuse T et al (2004) Ehd1, a B–type response regulator in rice, confers short–day promotion of flowering and controls FT–like gene expression independently of Hd1. Genes Dev 18:926–936
Dong SB, Lu BR, Wang YY et al (2010) Preliminary studies on the within-varietal genetic diversity and its maintenance of traditional rice from Yunnan. J Yunnan Agric Univ 25:1–9
Evanno G, Regnaut S, Goudet J (2005) Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol 14:2611–2620
Excoffier L, Smouse PE, Quattro JM (1992) Analysis of molecular variance inferred from metric distances among DNA haplotypes: application to human mitochondrial DNA restriction data. Genetics 131:479–491
Fu YX, Li WH (1993) Statistical tests of neutrality of mutations. Genetics 133:693–709
Falush D, Stephens M, Pritchard JK (2003) Inference of population structure using multilocus genotype data: linked loci and correlated allele frequencies. Genetics 164:1567–1587
Fu YB, Peterson GW, Richards KW et al (2005) Allelic reduction and genetic shift in the Canadian hard red spring wheat germplasm released from 1845 to 2004. Theor Appl Genet 110:1505–1516
Glaszmann JC (1987) Isozymes and classification of Asian rice varieties. Theor Appl Genet 74:21–30
Garris AJ, Tai TH, Coburn J et al (2005) Genetic structure and diversity in Oryza sativa L. Genetics 169:1631–1638
Gao LZ (2003) The conservation of Chinese rice biodiversity: genetic erosion, ethnobotany and prospects. Genet Resour Crop Evol 50:17–32
Hammera K, Gladisa Th, Diederichsen A (2003) In situ and on-farm management of plant genetic resources. Eur J Agron 19:509–517
Hall TA (1999) Bioedit: a user–friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser 41:95–98
Hudson RR, Kaplan NL (1985) Statistical properties of the number of recombination events in the history of a sample of DNA sequences. Genetics 111:147–164
Holsinger KE, Weir BS (2009) Genetics in geographically structured populations: defining, estimating and interpreting F(ST). Nat Rev Genet 10:639–650
Harlan JR (1975) Crops and man. American Society of Agronomy, Madison
Londo JP, Chiang YC, Hung KH et al (2006) Phylogeography of Asian wild rice, Oryza rufipogon, reveals multiple independent domestications of cultivated rice, Oryza sativa. Proc Natl Acad Sci 103:9578–9583
Liu XZ, Zhao MF, He YQ et al (2007) Breeding and blast resistance identification of Lijiangxintuanheigu nearnisogenic pyramid lines. Acta Agron Sin 33:20–24
Li ZM, Zheng XM, Ge S (2011a) Genetic diversity and domestication history of African rice (Oryza glaberrima) as inferred from multiple gene sequences. Theor Appl Genet 123:21–31
Li YB, Fan CC, Xing YZ et al (2011b) Natural variation in GS5 plays an important role in regulating grain size and yield in rice. Nat Genet 43:1266–1269
Maxted N, Guarino L, Myer L et al (2002) Towards a methodology for on–farm conservation of plant genetic resources. Genet Resour Crop Evol 49:31–46
Mao HL, Sun SY, Yao JL et al (2010) Linking differential domain functions of the GS3 protein to natural variation of grain size in rice. Proc Natl Acad Sci 107:19579–19584
Nei M (1987) Molecular evolutionary genetics. Columbia University Press, New York
Pusadee T, Jamjod S, Chiang YC et al (2009) Genetic structure and isolation by distance in a landrace of Thai rice. Proc Natl Acad Sci 106:13880–13885
Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155:945–959
Parzies HK, Spoor W, Ennos RA (2004) Inferring seed exchange between farmers from population genetic structure of barley landrace Arabi Aswad from Northern Syria. Genet Resour Crop Evol 51:471–478
Rozas J (2009) DNA sequence polymorphism analysis using DnaSP. Methods Mol Biol 537:337–350
Second G (1982) Origin of the genetic diversity of cultivated rice (Oryza spp.): study of polymorphism scored at 40 isoezyme loci. Jpn J Genet 57:25–57
Sirabanchongkran A, Yimyam N, Boonma W et al (2004) Varietal turnover and seed exchange: implications for conservation of rice genetic diversity on-farm. Int Rice Res Notes 29(2):18–19
Shang JJ, Tao Y, Chen XW et al (2009) Identification of a new rice blast resistance gene, Pid3, by genomewide comparison of paired nucleotide-binding site–leucine-rich repeat genes and their pseudogene alleles between the two sequenced rice genomes. Genetics 182:1303–1311
Sun JC, Cao GL, Jing Ma et al (2012) Comparative genetic structure within single–origin pairs of rice (Oryza sativa L.) landraces from in situ and ex situ conservation programs in Yunnan of China using microsatellite markers. Genet Resour Crop Evol 59:1611–1623
Thompson JD, Gibson TJ, Plewniak F et al (1997) The CLUSTALX windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 25:4876–4882
Tajima F (1989) Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 123:585–595
Tsegaye B, Berg T (2007) Genetic erosion of Ethiopian tetraploid wheat landraces in Eastern Shewa, Central Ethiopia. Genet Resour Crop Evol 54:715–726
Wright SI, Gaut BS (2005) Molecular population genetics and the search for adaptive evolution in plants. Mol Biol Evol 22:506–519
Wei X, Qiao WH, Chen YT et al (2012) Domestication and geographic origin of Oryza sativa in China: insights from multilocus analysis of nucleotide variation of O. sativa and O. rufipogon. Mol Ecol 21:5073–5087
Watterson GA (1975) On the number of segregating sites in genetical models without recombination. Theor Popul Biol 7:256–276
Xu FR, Zhang EL, Dong C et al (2010a) Comparison of phenotypic traits of rice landraces, grown in two different periods in Hani’s terraced fields in Yuanyang County, Yunnan. Biodiversity Science 18:365–372
Xu FR, Tang CF, Yu TQ et al (2010b) Diversity of paddy rice varieties from Yuanyang Hani’s terraced fields in Yunnan, China. Acta Ecologica Sinica 30:3346–3357
Xu FR, Dong C, Yang WY et al (2011) Comparison of genetic diversity of rice landraces planted in two periods in Hani’s terraced fields in Yuanyang County, Yunnan province, China using microsatellite markers. Chin J Rice Sci 25:38l–386
Yan HM, Dong C, Zhang EL et al (2012) Analysis of genetic variation in rice paddy landraces across 30 years as revealed by microsatellite DNA markers. Hereditas 34:87–94
Zhou YH (1988) Regionalization of rice cropping in Yunnan. In: Min SK, Wu XZ (eds) Regionalization of rice cropping in China. Zhejiang Science, and Technology Pubublication, Hangzhou, pp 104–108
Zeng YW, Wang JJ, Yang ZY et al (2001) The diversity and sustainable devolopment of crop genetic resources in the Lancang River Valley. Genet Resour Crop Evol 48:297–306
Zhu YY, Chen HR, Fan JH et al (2003) The use of rice variety diversity for rice blast control. Sci Agric Sin 36:521–527
Zhang HL, Sun JL, Wang MX et al (2006) Genetic structure and phylogeography of rice landraces in Yunnan, China, revealed by SSR. Genome 50:72–83
Zeng YW, Zhang HL, Li ZC et al (2007) Evaluation of genetic diversity of rice landraces (Oryza sativa L.) in Yunnan, China. Breed Sci 57:91–99
Zeng YW, Shen SQ, Li ZC et al (2003) Ecogeographic and genetic diversity based on morphological characters of indigenous rice (Oryza sativa L.) in Yunnan, China. Genet Resour Crop Evol 50:567–577
Zhu QH, Zheng XM, Luo JC et al (2007) Multilocus analysis of nucleotide variation of Oryza sativa and its wild relatives: severe bottleneck during domestication of rice. Mol Biol Evol 24:875–888
Zheng XM, Ge S (2010) Ecological divergence in the presence of gene flow in two closely related Oryza species (Oryza rufipogon and O. nivara). Mol Ecol 19:2439–2454
Acknowledgments
We thank the Chinese National Germplasm Bank for providing the landrace rice seeds. This work was supported by the National Key Technology Research and Development Program of China (2013BAD01B02-2, 2013BAD01B0101-02), Science and Technology Innovation Program of CAAS, the platform of National Crop Germplasm Resources, the Project of 973 (2010CB125904-5), the Protective Program of Crop Germplasm of China (NB2013-2130135-25-01), International Cooperation Project from National Institute of Crop Science, RDA (PJ008685).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest statement
None.
Additional information
Communicated by M. Thomson.
Electronic supplementary material
Below is the link to the electronic supplementary material.
122_2015_2617_MOESM1_ESM.pdf
Supplementary Fig. 1 Schematic diagrams of ten nuclear loci and locations of the regions sequenced. Exons are shown as open boxes, and exon numbers are labeled with capital roman numbers. Thin lines between open boxes refer to introns. Locations of primers for each fragment are sketched above the diagrams. (PDF 151 kb)
122_2015_2617_MOESM4_ESM.pdf
Supplementary Table 1 List of the materials sampled in the study, origin and subpopulation. Code No.: Official numbers of the accessions conserved in the Chinese National Germplasm Bank. (PDF 639 kb)
122_2015_2617_MOESM8_ESM.pdf
Supplementary Table 5 Summary of haplotype richness for rice accessions in each ecological zone from 1980 and 2007. (PDF 103 kb)
Rights and permissions
About this article
Cite this article
Cui, D., Li, J., Tang, C. et al. Diachronic analysis of genetic diversity in rice landraces under on-farm conservation in Yunnan, China. Theor Appl Genet 129, 155–168 (2016). https://doi.org/10.1007/s00122-015-2617-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00122-015-2617-7