Abstract
Key message
Rootstock HKT1 genotype affected fruit [Na + ] and non-commercial fruit yield; QTL analysis of rootstock-mediated scion nutrition is a powerful forward genetic approach to identify wild genes for rootstock breeding.
Abstract
The present study approaches the QTL dissection of rootstock effects on a commercial hybrid variety grafted on a population of RILs derived from Solanum pimpinellifolium, genotyped for 4370 segregating SNPs from the SolCAP tomato panel and grown under moderate salinity. Results are compared to those previously obtained under high salinity. The most likely functional candidate genes controlling the scion [Na+] were rootstock HKT1;1 and HKT1;2 as it was previously reported for non-grafted genotypes. The higher fruit [Na+] found when rootstock genotype was homozygote for SpHKT1 supports the thesis that scion HKT1 is loading Na+ into the phloem sap in leaves and unloading it in sink organs. A significant increment of small, mostly seedless, fruits was found associated with SlHKT1 homozygous rootstocks. Just grafting increased the incidence of blossom end rot and delayed fruit maturation but there were rootstock RILs that increased commercial fruit yield under moderate salinity. The heritability and number of QTLs involved were lower and different than those found under high salinity. Four large contributing (>17 %) rootstock QTLs, controlling the leaf concentrations of B, K, Mg and Mo were detected whose 2 Mbp physical intervals contained B, K, Mg and Mo transporter-coding genes, respectively. Since a minimum of 3 QTLs (two of them coincident with leaf K and Ca QTLs) were also found governing rootstock-mediated soluble-solids content of the fruit under moderate salinity, grafting desirable crop varieties on stress-tolerant rootstocks tenders an opportunity to increase both salt tolerance and quality.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
More than 800 million hectares of land throughout the world are affected by salinity (FAO 2008), which can decrease yield and lead to increased poverty and reliance on imports (Witcombe et al. 2008). Tomato is one of the most important horticultural crops. In terms of human health, tomato fruit is a major component of daily meals in many countries and constitutes an important source of minerals, vitamins, and antioxidant compounds. However, the areas for tomato optimal growing conditions are becoming narrower around the world. Since salt tolerance, like tolerance to any abiotic stress, means adaptation, breeding for salt tolerance should take advantage of the evolution of Solanum species that occurred through adaptation to marginal environments. In this sense, S. pimpinellifolium L. has been frequently considered as possible donor of salt tolerance (Bolarin et al. 1991; Cuartero et al. 1992; Asins et al. 1993; Foolad and Lin 1997).
Grafting is a biotechnological tool used since ancient times to improve the amount and uniformity of crop yield, and currently most fruit crops and many horticultural species are grown as scion–rootstock combinations. Although this strategy triples the work required by breeders (selection for rootstock, scion and their combination), main problems to breeding food crops for biotic and abiotic resistance can be circumvented when using rootstocks bred or selected to confer resistance (King et al. 2010). Improved nutrient absorption, salt stress tolerance and fruit quality are rootstock breeding objectives that would also benefit soil-less grafted tomato production in climate-controlled greenhouses in Europe and the US where extending the harvest season is a major goal (King et al. 2010). Besides, the genetic study of rootstock effects on a given variety is a valuable strategy to understand root functions (particularly, nutrient uptake and transport) since they are regulated by shoot through materials cycling between roots and shoots (Wang et al. 2006).
Estañ et al. (2009) showed that an efficient and profitable utilization of wild germplasm can be carried out through the improvement of rootstocks that confer salt tolerance in terms of fruit yield to the grafted variety. These authors found that salt tolerance rootstock effect was a heritable trait (H 2 near 0.3), governed by at least 8 QTLs. However, different QTLs were detected depending on the RIL population and/or the salinity level because the S. cheesmaniae RIL population was tested under moderate salinity (75 mM NaCl, CE = 8.6 dS m−1) and the S. pimpinellifolium population, under high salinity (125 mM NaCl, 13.7 dS m−1). Since salinity is variable in time and space under field conditions in marginal areas, breeding tomato rootstocks for salt tolerance through marker-assisted selection should take into account the differences in the QTLs controlling rootstock-mediated fruit yield under different levels of salinities.
Nowadays, analysis of QTL can also reach further research objectives by contributing to fill the gap between agronomic performance and the DNA sequences involved. With the advent of the complete tomato genome sequence by The Tomato Genome Consortium (2012), and the availability of a large panel of SNPs (SolCAP panel, http://solgenomics.net/), it is feasible to narrow the QTL interval where the gene(s) lies without the need for generating a physical map. The genome assembly also allows the rapid identification of candidate genes around the physical position of the SNP(s) with observed maximum LOD score. Thus, in a previous paper (Asins et al. 2013) we tried this approach to identify the tomato Na+ transporters HKT1;1 and HKT1;2 as the most likely candidates for the major QTL controlling Na+/K+ homeostasis in tomato.
The objectives of this study were (1) to estimate the heritability of the rootstock effect on the fruit yield and quality of a commercial variety grafted on an S. pimpinellifollium RIL population grown under moderate salinity (2) to detect the QTLs involved, (3) to compare the results of this QTL analysis with those reported for the same population under high salinity (Estañ et al. 2009), (4) to investigate the genetic relationship of potential physiological components of salt tolerance conferred by the rootstock and searching for co-location of physiological and fruit yield QTLs, and (5) to test the efficiency of the present QTL analysis for the inference of candidate genes.
Materials and methods
Plant material, growth conditions and trait evaluation
A total of 130 F10 lines (P population) derived by single-seed descendent from the hybrid between a salt-sensitive genotype of Solanum lycopersicum var. Cerasiforme (formerly L. esculentum) and a salt-tolerant line from S. pimpinellifolium L. (formerly L. pimpinellifolium) (Monforte et al. 1997) were used for the study reported here.
The commercial tomato hybrid Solanum lycopersicum cv. Boludo (Bol) was used as scion, and plants from 124 lines of the P population were finally evaluated as rootstocks. Boludo (the scion) was also grafted onto roots derived from a different plant of the same genotype (Bol/Bol). Bol and Bol/Bol, non-grafted and self-grafted plants were included as controls. Bol/Bol plants were used to evaluate any physiological change that could be induced by the grafting process per se.
Grafting was performed when seedlings had developed 3–4 true leaves, seedlings were cut over the cotyledons, using the shoot as scion and the remaining plant part as rootstock. Grafts were made immediately after cutting the plants and grafting clips were used to adhere the graft union. After grafting, seedlings were grown as described in Estañ et al. (2009).
Five grafted plants per line and controls were transferred to a polycarbonate greenhouse, in Valencia, Spain (September 18th, 2012) at a density of 2.1 plants m−2 in an open soilless system using coco fiber as a substrate. The greenhouse had automatic roof ventilation and heating system (maintaining inside air temperature above 8 °C). The climate variables, in and out solar global radiation, inside photosynthetic active radiation, temperatures of the air, and air humidity were recorded by sensors connected to a data acquisition system. A latinised row–column design was proposed with 5 reps along the benches, 22 rows per rep and 6 columns (benches) across the reps. Extra repetitions were used to complete the 144 experimental units. A high-frequency fertirrigation system together with 4L/h drippers was used and handled to ensure homogeneity of the salinity of the roots of all plants in cultivation at the same time. To reach the salinity target level (75 mM NaCl) 10 days after the transplanting date, increasing amounts of NaCl were gradually added over a full-strength nutrient solution for tomato culture (in meq/L: NO3 − 11.1; H2PO4 − 1.3; SO4 2− 1; NH4 + 0.7; K+ 4.7; Ca2+ 3.5; Mg2+ 1; plus 0,025 g/L of an EDTA-microelements complex pH = 5.8, EC = 1.44 dS/m). The water for the nutrient solution was previously treated with reverse osmosis. Final electrical conductivity was 8.94 dS/m. The plants were cultivated to only one stem, eliminating all axillary buds.
The third (H3) and fifth (H5) Boludo leaves were harvested from 3 out of the 5 reps per RIL and controls for phenotyping after 46 days of salt treatment. Three evaluations of scion leaves at both H3 and H5 were carried out: leaf fresh weight (LFW, g); leaf dry weight (LDW, g) measured in samples dried at 80 °C for 3 days, and leaf water content (LWC, g) calculated as the difference between LFW and LDW. The difference between H5 and H3 for LDW and LWC was also considered and coded as dLDW and dLWC, respectively.
Each plant was evaluated during 8 weeks for fruit yield (number of ripe fruits, FN; their individual weight, FW and total fruit weight, TFW). If the weight of the fruit was <5 g (normally containing no seed), it was considered only for determining non-profitable fruit yield (FN < 5 and TFW < 5). Fruits larger than 5 g were used to estimate commercial fruit yield under moderate salinity (FN > 5, FW > 5 and TFW > 5). The variance of fruit weight per plant (varFW) was included as an additional trait. The number of fruits with blossom end rot (BER) and the number of days till harvesting the first ripe fruit (gDaysTFM) were also recorded.
Ripe fruits from the second and the third trusses of each plant (three plants per genotype) were evaluated for soluble-solids content (SSC2 and SSC3, respectively) and acidity. Fruit juice was obtained by squeezing of tomato through a cloth filter. This filtered juice was used to measure SSC, pH and citric acid content (Cit). Total soluble solids were measured as °Brix with a digital refractometer (PR-101α; Pallete, Atago). The citric content (%) and pH of fruit juice was measured in a 1:10 mL juice dilution with a digital pH meter (PH-Matic23; Crinson).
The type of scion/rootstock union was evaluated in all plants after 214 days of salt treatment using two classifications. Union1 corresponded to a three-type classification, where 1 means scion diameter smaller than rootstock diameter, 3 the opposite and 2, no diameter difference between rootstock and scion. Union2 concerned the presence, length and amount of roots arising from the union (from 0 to 4).
Two sets of 10 RILs each were selected by their HKT1 genotype (HKT1 sub-experiment). Within each HKT1 genotype, 10 RILs were chosen at random. One set was homozygous for the S. lycopersicum allele, EE; and the other set of 10, homozygous for the S. pimpinellifolium allele, PP (Asins et al. 2013). Three reps from each of these 20 RILs and from both controls (Bol and Bol/Bol) were further used for ionomic profile determination of the fruit juice (J) and the mesocarp (F), upper (TA) and lower stem (TB), and root (R) at the end of the experiment (20th May, 2013), after 234 days of saline treatment.
Tissue samples of F, H3, H5, TA, TB and R were fresh-weight determined, oven dried for 48 h at 80 °C, weighed (dry weight) and prepared for mineral analysis by digestion in a HNO3:HClO4 (2:1, v/v) solution. Inorganic solutes were determined in ppm by inductively coupled plasma spectrometry (ICP) (Ionomic Service; CEBAS–CSIC, Murcia, Spain). Cations in the fruit juice were determined by inductively coupled plasma spectrometry in ppm using a 1:10 mL dilution (Varian ICP 720-E, Scientific Instrumentation Service, Estación Experimental del Zaidín, CSIC, Granada, Spain). Third leaf Cl− concentration (mg/L) was measured as described by Gilliam (1971) using a Sherwood chloride analyser 926.
Statistical analysis
A general factorial mixed model with genotypes (fixed), benches and reps (random) was used to asses the significance of each source of variation following the latinised row–column design. The Bayesian Information Criteria (BIC) was used to select the best model. Given that for most traits the model with genotypes and benches was the most parsimonious with the lowest BIC, it was used to estimate the adjusted mean traits per rootstock genotype for the QTL analysis and to study the grafting effects by comparing Bol vs. Bol/Bol adjusted means.
Pearson and Spearman (for type of scion/rootstock union) correlation coefficients and principal component analysis based on the correlation matrix for the adjusted means were used to study the relations between the different traits.
Broad-sense heritability (H 2) was calculated for traits measured in both populations assuming that the individuals from the ninth self-pollinated generation were nearly homozygous for all loci. Heritability was calculated as reported previously by Villalta et al. (2007), using the formula: H 2 = V g/(V g + V e) where V g and V e are the estimates of genotype and environmental variance, respectively, by REML (Restricted Maximum Likelihood). These estimates were obtained by a model with the same sources of variation as above but considering genotypes as random effects.
Differences between HKT1 rootstock genotypes for the HKT1 sub-experiment were analyzed by a mixed model with benches (random), HKT1 genotype (fixed), and RILs (random) nested within HKT1 genotype as factors.
Molecular markers and QTL analysis
One hundred and thirty P-RILs at F10 were genotyped for 7720 SNPs from the SolCAP tomato panel (Illumina BeadXhip WG-401-1004) using an external genotyping service (Fundación Investigación Clínico, Valencia, Spain). Linkage groups were set at LOD ≥ 17 using Joinmap 4 software for Windows (Van Ooijen 2006). A first genotype file including good quality segregating SNPs was used to know their distribution among linkage groups and to find out groups of SNPs for which all RILs showed the same genotype (groups of redundant markers). A second genotype file including non-redundant, segregating SNPs was used for the map construction of each chromosome using the same software.
QTL analyses were carried out using Interval Mapping (IM) and Multiple QTL Mapping (MQM) procedures in MapQTL® 6 (Van Ooijen 2009). Kruskal–Wallis procedure was also used to genetically analyze Union1 and Union2. A 5 % experiment-wise significance level was assessed by permutation tests. These LOD critical values ranged from 2.1 to 2.3 depending on the trait and chromosome.
Results
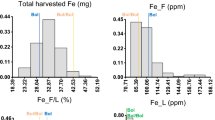
Significant differences between Bol and Bol/Bol, non-grafted and self-grafted plants were found for some traits (Table 1). Grafting delayed fruit maturation and increased BER, and fruits’ and leaves’ S concentrations. Mo concentration at third leaf also increased just by grafting (Fig. 1).
Heritabilities of rootstock effects (Table 2) were particularly large for H3_Na (0.86), H3_B (0.58), H3_Mg (0.49), H3_Mo (0.36), H3_Tl (0.35) and H3_Cd (0.33) but very low for total fruit yield. Nevertheless, some RILs conferred higher yield and fruit quality than the controls (Fig. 2).
Ordered adjusted means and standard errors for the fruit traits of Boludo: Total Fruit Weight (TFW) and Soluble-Solids Content (SSC) using the lines of the P population as rootstock in comparison to the controls, non-grafted and self-grafted Boludo plants (Bol and Bol/Bol). Vertical bars indicate the relative position of these controls
The relationships among traits are visualized through the representation of the principal component analysis by the first two axes (27.1 %) (Fig. 3). Evaluations at third and fifth leaves appeared almost orthogonal in this representation. Commercial fruit yield (TFW > 5) was highly significant and directly correlated to FN > 5 (0.78), FW > 5 (0.30), TFW < 5 (0.31), varFW (0.43), LFW5 (0.28) and LWC5 (0.28) and inversely related to quality traits SSC2 (−0.28) and SSC3 (−0.31) (Table 3). No significant correlation was obtained between fruit yield and H3_Na or H3_Cl.
A total of 4,370 out of 7,720 SNPs conforming the SolCAP tomato panel were segregating in the P population (130 RILs), and 2,059 of them were genetically non-redundant. The number of non-redundant SNPs that was finally included in the linkage map of each chromosome (from chromosome 1–12) was 207, 198, 171, 175, 178, 165, 144, 117, 138, 97, 192 and 117, respectively. Thus, the linkage map of the P population contained 1899 non-redundant SolCAP SNPs and covered 1,326.37 cM of genetic length. An acceptable relationship between the genetic and physical positions of SNPs was found (supplementary figure S1) to use them for QTL analysis.
A total of 54 significant QTLs were detected for 24 Boludo traits, including two related to the scion/rootstock union (Table 4). Chromosomes 3 and 7 were particularly QTL rich. The contribution of some of them was large: H3_Na in chromosome 7 (68.2 %), H3_Mo in 10 (37.3 %), H3_Mg in 3 (20.8 %) and H3_B in 6 (20.5 %). Fruit yield QTLs were detected in chromosome 1 for large (commercial) fruits, and in chromosomes 3 and 6 for small fruits. Interestingly, at least three QTLs for soluble-solid content of the fruit as influenced by the rootstock genotype were detected under moderate salinity in chromosomes 3, 6 and 7. Several QTLs for the type of rootstock–scion union were detected by interval mapping but the position of the SNPs showing maximum significance should be better estimated using the non-parametric methodology of Kruskal–Wallis (Table 5).
Results from the HKT1 sub-experiment, showed significant differences between HKT1 rootstock genotypes for Na+ concentrations of all tissues except for the root were K+ concentration was significant instead (Table 6). Na+ concentrations of all tissues of Boludo grafted on rootstocks homozygous for the S. pimpinellifolium HKT1 allele were larger than on rootstocks homozygous for the S. lycopersicum allele. Yield of fruits lighter than 5 g (TFW < 5) was significantly larger when the rootstock was homozygous for the S. lycopersicum allele than for the S. pimpinellifolium allele. Therefore, the HKT1 rootstock homozygote for the S. pimpinellifolium allele would be indirectly associated with agronomic salt tolerance given that it is associated with a lower non-commercial fruit yield.
Discussion
The effect of grafting
The first question regarding the benefits obtained using rootstocks is whether or not grafting (as a type of wounding) per se has any effect on the evaluated traits under salinity. This question has been addressed by comparing non-grafted Boludo (Bol) to Boludo grafted on Boludo (Bol/Bol). There are several indications suggesting that just grafting could benefit resistance to salinity regardless the rootstock. As shown in Table 1 and Fig. 1, just grafting translated into an increment of certain nutrients in leaves and fruits (Mo and, mostly, S). Sulfate and molybdate share a high degree of similarity and it has been suggested that molybdate import and distribution are facilitated by sulfate transporters or related systems. In addition, the sulfur and molybdenum metabolisms are interconnected at several other stages such as Moco (molybdenum cofactor) biosynthesis and sulfite detoxification, so that both nutrients appear to interact closely at various levels in a common metabolic network (Bittner 2014, and references therein). Therefore, it seems possible to hypothesize that grafting affects their uptake, transport and/or storage. Thus, the invigorating effect of rootstocks in general might be due, at least in part, to a higher Mo translocation easiness by the grafting process and not to the rootstock genotype itself.
We have found also an increment of Ca2+ concentration in root just by grafting. Early and late changes in gene expression in shoot induced by root wounding were studied in Arabidopsis (Hasegawa et al. 2011) and, among early up-regulated root-to-shoot-responsive genes, some of them related to signal transduction are Ca2+ binding proteins. Then, following Scrase-Field and Knight (2003), the root of any grafted plant would have the Ca2+ “switch” permanently on. Besides, many of the root-to-shoot-responsive genes were associated with systemic production of jasmonic acid, OPDA (12-oxo-phytodienoic acid) and possibly ethylene. Therefore, the fact of grafting seems to prepare the plant to resist physiologically. Drawbacks of grafting are delayed fruit maturation and a higher incidence of BER, possibly due to lower fruit Ca2+ availability from the root since BER is a well-known Ca2+ deficiency symptom (Simon 1978). Although the heritabilities of BER and gDaysTFM were very low (Table 2), there were RILs showing no BER incidence, and one QTL was detected for gDaysTFM (Table 4), suggesting that rootstocks can be improved for this trait through selection in the P population.
The rootstock genotype for HKT1 genes affects fruit [Na+] and non-commercial fruit yield
Asins et al. (2013) identified and mapped the location of two tomato HKT1 genes encoding Na+ transporters (HKT1;1 and HKT1;2) and studied their role as candidate genes for the major QTL involved in Na+/K+ homeostasis under high salinity. Now, we have shown that those HKT1 genes are also positional candidates (only 35 Kb from the same SNP showing maximum LOD score) for the major rootstock-mediated QTL (68 %) controlling leaf Na+ concentration (without any effect on K+ concentration) of the grafted variety under moderate salinity. Similarly to what was reported previously using non-grafted plants, the S. pimpinellifolium HKT1 allele, SpHKT1;2, coming from the wild salt-tolerant genotype in the rootstock, is associated with the highest Na+ concentration of the aerial parts of the grafted variety (Tables 4, 6). These results fit the observation that the wild ScHKT1;2 transcript level in roots was lower that SlHKT1;2 in the experiment with NILs reported by those authors, suggesting that reduced expression of ScHKT1;2 in roots translates into lower Na+ retrieval from the xylem in roots and consequently more Na+ is transported via the transpiration stream to the aerial part. We cannot discard that wild S. pimpinellifolium HKT1;2 had a lower affinity for Na+ in comparison to SlHKT1;2 as Almeida et al. (2014) have reported recently for S. pennellii HKT1;2.
The results from the HKT1-rootstock sub-experiment showed significant difference for K+ but not for Na+ concentration in root (Table 6) as it was observed previously when comparing near-isogenic lines differing for HKT1 genotypes (Asins et al. 2013). The maximum difference between the HKT1 rootstock genotypes for Boludo Na+ concentration corresponded to the oldest leaf, sampled after 46 days of salt treatment. Boludo fruits showed significant differences for Na+ concentration at the juice and the mesocarp accordingly to the rootstock genotype. Therefore, the scion HKT1;1 and/or HKT1;2 must be involved in Na+ loading into the phloem sap in leaves and unloading in sink organs, such as fruit and root, which would be in line with the hypothesis put forward by Berthomieu et al. (2003) that AtHKT1;1 plays an important role in Na+ phloem loading. Besides, this root unloading (Na+ transport from shoot to root) would explain the lack of significant differences between rootstock HKT1 genotypes for R_Na (Table 6). Nevertheless, the specific cellular/tissue location of tomato HKT1 transporters requires further study.
The concentrations of other elements appeared affected by the HKT1 rootstock genotype such as B in the fruit mesocarp. Regarding the relationship between rootstock HKT1 genotype and salt tolerance, no significant difference for vigor-related traits were detected but for yield of small fruits.
The tolerance to salinity depends on its level, moderate or high
Our results on fruit yield support the hypothesis that the genetic control of the rootstock-mediated salt tolerance depends on the level of salinity. First of all, a certain amount of small (seedless) fruits was yielded under moderate salinity. Second, only small-fruit QTLs detected in chromosomes 3 (98.4 cM) and 6 (48.0 cM) could be the same than those detected by Estañ et al. (2009) given that their position in the SNP map would be 95.5 and 39.9 cM, respectively. Besides, the increasing allele comes from the wild species in both cases. Correlations between fruit yield and vegetative traits also have shown several differences regarding the level of salinity. Under moderate salinity, fruit yield correlated with LWC5, while it correlated with both the LWC5 and LWC2 under high salinity (Asins et al. 2010). In fact, the physiology of the leaves at levels 3 and 5 under moderate salinity appeared as they were independent (see orthogonal positions of related traits in the PCA of Fig. 3). Contrary to the high-salinity experiment, we have not detected correlation with leaf Na+ concentration here. Third, the TFW QTL detected for commercial fruits under moderate salinity in chromosome 1 was not detected under high salinity. Since both experiments were similar and uniform (fruit yield was evaluated at the same scion variety) it cannot be argued that crop management would be the reason of these differences (Kromdijk et al. 2014). Instead, all those results together support the hypothesis that plants (grafted tomato plants) respond to different salinity levels through different receptors (Munnik and Meijer 2001) or different signaling systems that might depend on the stress-induced primary event (Kacperska 2004). Thus, the final agronomic performance would depend on different QTLs according to the level of salinity. Under moderate salinity, the rootstock effect on salt tolerance is more closely related to maintaining water content at the shoot than for ion homeostasis, although Na+ homeostasis might be involved in the production of non-commercial fruits. Non-commercial fruit yield was increased when rootstock genotypes for HKT1;1 and HKT1;2 were homozygotes for the lycopersicum allele, i.e., the allele responsible for avoiding Na+ accumulation in all scion tissues, including fruits (Table 6). Therefore, Na+ accumulation is not driving the reduction of fruit size through ovule abortion by salt stress (Sun et al. 2004). If it were so, it should happen for the S. pimpinellifolium HKT1 rootstocks. Besides, no significant QTL for small-fruit yield has been detected on chromosome 7 where HKT1 genes locate (Asins et al. 2013), discarding genetic linkage of QTLs as an explanation for the relationship between rootstock HKT1 genotype and TFW < 5. Alternatively, this relationship might be indirectly related to the effect of HKT1-rootstock genotype on the accumulation of other elements (Table 6) and would need future experiments to be tested.
Genetic effects of the rootstock on the tomato quality under moderate salinity
Numerous studies have shown that salinity increases the soluble-solid content of tomatoes (Cuartero and Fernández-Muñoz 1999; Dorais et al. 2001), even when using grafted plants (Fernandez-García et al. 2004; Estañ et al. 2008). However, Turhan et al. (2011) found this fruit quality trait was lower in the tomatoes of grafted plants than in the non-grafted ones. Here, we have shown the availability of genetic variability (H 2 = 0.17) in an RIL population derived from S. pimpinellifolium to increase the tomato total soluble solids of a grafted variety grown under moderate salinity. In fact, QTLs controlling this trait were detected in chromosomes 3, 6, 7 and possibly 9 (the only QTL where the wild allele would be increasing the trait). The small differences in the location of SNPs showing maximum LOD score for SSC2 and SSC3 (soluble-solid content of ripe fruits from the second and third trusses, respectively) could be explained by the sampling process itself (random errors). Full coincidence was obtained for SSC2 and SSC3 on chromosome 3. QTLs controlling soluble-solid and sugar contents of tomatoes from other non-grafted RIL populations were reported previously in chromosomes 9 (marker CT032) (Saliba-Colombani et al. 2001), 3 (marker TG214) (Saliba-Colombani et al. 2001, Ashrafi et al. 2012), 6 and 7 (marker TG252) (Ashrafi et al. 2012). Using the comparative map viewer tool (sol genomics network, http://solgenomics.net/) it was possible to compare the positions of the SNPs associated with the rootstock-mediated SSC QTLs (Tomato-Kazusa and SolCAP markers mapped to genome and TraitGenetics EXPEN2012) and the position of those reported previously for non-grafted plants (Tomato-EXPEN 2000) and no coincidence was found for those in chromosome 6 (43 cM apart), 9 (61 cM), 7 (12 cM), and 3 (88 cM). The QTL reported in chromosome 7 is the closest one but the increasing allele is different. Recently, Sauvage et al. (2014) have reported 9 peak SolCAP SNPs associated with SSC through GWA using a panel of 163 tomato accessions but none of them is coincident with the SSC peak SNPs of the present study. This fact suggests that rootstock-mediated SSC under moderate salinity is controlled by different genes from those governing SSC in non-grafted plants.
QTL analysis of rootstock effects as a powerful forward genetic approach to unveil wild genes with agronomic interest
Present QTL analysis, based on the SNPs from the SolCAP panel (http://solgenomics.net/), has been feasible without the need for generating a physical map for each parental line. In fact, the relationship between their physical and genetic positions (supplementary figure S1) was very similar to that reported by Sim et al. (2012) for populations EXPEN 2012 and, particularly, EXPIM 2012, derived from S. pennellii and S. pimpinellifolium, respectively. It also showed similarities with that reported by Pascual et al. (2014) for the MAGIC population whose total map length (2,156 cM) was the largest, followed by ours.
In general, there was good agreement between sensu lato-estimated heritabilities and number of QTLs that were detected in this experiment. There were some exceptions such as H3_Na and H3_Mo whose heritabilities were high (0.86 and 0.36, respectively) and only one QTL was detected although their contributions were large (68 and 37 %, respectively) (Table 4). More difficult to explain are the traits for which 1–2 QTLs were detected but their estimated heritabilities were <0.0001 (VarFW, TFW < 5) and should be considered with caution. In the case of qualitative traits, QTL location is better estimated by non-parametric methods. This seems the case of the QTLs for types of rootstock–scion union (Union1 on Table 4 through interval mapping, and through Kruskal–Wallis on Table 5). That one in chromosome 9 showed a peak at solcap_snp_sl_36845, precisely an SNP at gene Solyc09g089610.2.1 (ethylene receptor-like protein, ETR6). Ruzicka et al. (2007) proposed a model for ethylene-dependent root growth that accounts for auxin effects on root growth, and Aloni et al. (2008) proposed that the main cause for incompatibility is the occurrence of hormonal imbalance, primarily, of auxin and ethylene in the root system. Nevertheless, it is important to point out that none of the RILs resulted graft-incompatible with Boludo, so the undergrowth (Union1 class 1) and the overgrowth (Union1 class 3) of the scion cannot be considered here as an indication of graft incompatibility, but of an invigorating rootstock effect since Union1 was significantly correlated with most (commercial and non-commercial) fruit yield traits (Table 3).
Some examples of correlations found between fruit yield and other traits on Table 3, can be explained just by linkage of QTLs controlling both traits. These could be the cases of QTLs in chromosome 11 for FN < 5, LFW5 and Union1; on 3 for FN < 5, TFW < 5, SSC2, H3_Ca, H3_Mn and H3_Sr; on 6 for TFW < 5 and SSC2 (Table 4). The negative correlation between fruit yield and soluble-solid content of the fruit juice was also found in citrus within a similar experiment on rootstock effects (Raga et al. 2014). These results fit the hypothesis by Kromdijk et al. (2014) that fruit composition is heavily influenced by fruit load and go further because at least part of this relationship is genetically supported by linked QTLs in chromosomes 3 and 6 of the tomato rootstock under moderate salinity. From the three fruit compositional traits influenced by the fruit load: water and solute accumulation, metabolic interconversions and the incorporation of solute into structural material discussed by Kromdijk et al. (2014), it seems reasonable to assign to the rootstock an important role for the first one, water and nutrient availability.
Our results on the genetic analysis of leaf nutrients content mediated by rootstock (Table 4) prove the hypothesis that the efficiency of nutrient assimilation could be improved by rootstock breeding. This objective would enhance crop yield under organic production (Farias et al. 2013) and nutrient stress (Savvas et al. 2010; King et al. 2010), increasing agriculture sustainability. Noteworthy, Savvas et al. (2011) reported that grafting onto three commercial tomato rootstocks, significantly reduced the leaf Mg concentration resulting in clear Mg-deficiency symptoms of salinized tomato. Our results provide information on QTL positions at four chromosomes that could be used to improve the scion Mg content under moderate salinity through a proper rootstock selection.
The tomato genome assembly also allows the rapid identification of candidate genes within a 2 Mbp interval around the physical position of the SNP(s) with maximum observed LOD score. Since the QTL peak may accurately indicate the genes or gene clusters responsible (Price 2006), we have used this approach to search for candidate genes (mostly transporter-coding genes), responsible for the rootstock QTLs controlling leaf content of some nutrients such as B, Ca, Cl, K, Mg, Mn and Mo (supplementary table S2). There were only 2 QTLs (H3_B and H3_Ca in chromosomes 9 and 3, respectively) for which no candidate could be envisaged. Very likely functional candidates (such as the Boron and Molybdate transporters for H3_B and H3_Mo, respectively) were found for 50 % (9 out of 18) of QTLs using the available information of annotated genes (http://solgenomics.net/) within each 2 Mbp interval and performing blast analysis to refine the results. Four of them corresponded to highly contributing rootstock QTLs for leaf Mo (37.3 %), B (20.5 %), Mg (20.8 %) and K (17.5 %). Candidate genes were also identified for minor QTLs like for leaf [Cl−] in chromosome 6.
Noteworthy, two SSC QTLs co-located with Ca and K leaf concentration rootstock-mediated QTLs in chromosomes 3 (CL015369-0414) and 7 (Solcap_snp_sl_38698), respectively, providing a genetic support to phenotypic correlations. Is there any physiological connection behind it? It was surprising to find out that 2 Mbp around both SNPs, genes coding for proteins involved in fruit maturation (Steele et al. 1997; Rose et al. 1997) were also found: a glucan endo-1 3-beta-glucosidase (Solyc03g115200.2.1) and 5 expansins (Solyc03g115300.1.1, Solyc03g115310.1.1, Solyc03g115320.1.1, Solyc03g115340.1.1 and Solyc03g115350.1.1) for the former SNP, and a glucan endo-1 3-beta-glucosidase (Solyc03g017730.2.1) for the latter. Is there any joint root-to-shoot regulation at this genomic region?
In conclusion, a total of 37 QTLs have been mapped controlling important rootstock-mediated scion traits such as leaf concentration of nutrients, fruit yield, soluble-solids content of fruits and harvest time under moderate salinity that has allowed the search for candidate genes presumably involved. This will open the door to their validation by functional analysis and will facilitate marker-assisted selection in tomato rootstock breeding to improve fruit quality and the efficiency of nutrient uptake under salinity.
Author contribution statement
Conceived: MJA; Experimental design: EAC, MJA, DR; Performed experiment: VR, DR, MJA; Analyzed data: EAC, MJA, AB; Wrote paper: MJA, AB, EAC.
References
Almeida P, de Boer G-J, de Boer A-H (2014) Differences in shoot Na+ accumulation between two tomato species are due to differences in ion affinity of HKT1;2. J Plant Physiol 171:438–447
Aloni B, Kami L, Deveturero G, Levin Z, Cohen R, Kazir N, Lotan-Pompan M, Edelstein M, Aktas H, Turhan E, Joel DM, Horey C, Kapulnic Y (2008) Physiological and biochemical changes at the rootstock scion interface in graft combinations between Cucurbita rootstocks and a melon scion. J Hortic Sci Biotechnol 83:777–783
Ashrafi H, Kinkade MP, Merk HL, Foolad MR (2012) Identification of novel quantitative trait loci for increased lycopene content and other fruit quality traits in a tomato recombinant inbred line population. Mol Breed 30:549–567
Asins MJ, Breto MP, Carbonell EA (1993) Salt tolerance in Lycopersicon species. II. Genetic effects and search of associated traits. Theor Appl Genet 86:769–774
Asins MJ, Bolarín MC, Pérez-Alfocea F, Estañ MT, Martinez-Andújar C, Albacete A, Villalta I, Bernet GP, Dodd I, Carbonell EA (2010) Genetic analysis of physiological components of salt tolerance conferred by Solanum rootstocks. What is the rootstock doing for the scion? Theor Appl Genet 121:105–115
Asins MJ, Villalta I, Aly MM, Olías R, Álvarez De Morales P, Huertas R, Li J, Jaime-Pérez N, Haro R, Raga V, Carbonell EA, Belver A (2013) Two closely linked tomato HKT coding genes are positional candidates for the major tomato QTL involved in Na+/K+ homeostasis. Plant Cell Environ 36:1171–1191
Berthomieu P, Conejero G, Nublat A, Brackenbury WJ, Lambert C, Savio C, Uozumi N, Oiki S, Yamada K, Cellier F, Gosti F, Simonneau T, Essah PA, Tester M, Vèry A-A, Sentenac H, Casse F (2003) Functional analysis of AtHKT1 in Arabidopsis shows that Na+ recirculation by the phloem is crucial for salt tolerance. The EMBO J 22:2004–2014
Bittner F (2014) Molybdenum metabolism in plants and crosstalk to iron. Front Plant Sci 5:28. doi:10.3389/fpls.2014.00028
Bolarin MC, Fernandez FG, Cruz V, Cuartero J (1991) Salinity tolerance in 4 wild tomato species using vegetative yield salinity response curves. J Am Soc Hort Sci 116:286–290
Cuartero JM, Fernández-Muñoz R (1999) Tomato and salinity. Sci Hortic 78:83–125
Cuartero J, Yeo AR, Flowers TJ (1992) Selection of donors for salt-tolerance in tomato using physiological traits. New Phytol 121:63–69
Dorais M, Papadopoulos AP, Gosselin A (2001) Influence of electric conductivity management on greenhouse tomato yield and fruit quality. Agronomie 21:367–383
Estañ MT, Medina S, Morales B, Moyano E, Bolarín MC, Asins MJ (2008) Utilización como portainjertos de líneas RILs derivadas de S. lycopersicum x S. chesmaniae y S. lycopersicum x S. pimpinellifolium para mejorar el cultivo de tomate bajo condiciones salinas. In: Monreal LR, Ruiz JM, Blasco B, Rubio MM, Sánchez E, Ríos JJ, Cervilla LM (eds) Presente y Futuro de la Nutrición Mineral de Plantas, Héctor Santillán CP 18183, Granada, pp 225–235. ISBN: 978-84-89780-10-7
Estañ MT, Villalta I, Bolarín MC, Carbonell EA, Asins MJ (2009) Identification of fruit yield loci controlling the salt tolerance conferred by solanum rootstocks. Theor Appl Genet 118:305–312
FAO (2008) FAO land and plant nutrition management service. http://www.fao.org/ag/agl/agll/spush/
Farias EAD, Ferreira RLF, Neto SED, Costa FC, Nascimento DS (2013) Organic production of tomatoes in the Amazon Region by plants grafted on wild Solanum rootstocks. Cienc Agrotecnol 37:323–329
Fernandez-García N, Martinez V, Cerdá A, Carvajal M (2004) Fruit quality of grafted tomato plants grown under saline conditions. J Hortic Sci Biotech 79:995–1001
Foolad MR, Lin GY (1997) Genetic potential for salt tolerance during germination in Lycopersicon species. HortSci 32:296–300
Gilliam JW (1971) Rapid measurement of chlorine in plant materials. Soil Sci Soc Am Proc 35:512–513
Hasegawa S, Sogabe Y, Asano T, Nakagawa T, Nakamura H, Kodama H, Ohta H, Yamaguchi MK, Mueller MJ, Nishiuchi T (2011) Gene expression analysis of wounding-induced roots-to-shoot communication in Arabidopsis thaliana. Plant Cell Environ 34:705–716
Kacperska A (2004) Sensor types in signal transduction pathways in plant cells responding to abiotic stressors: do they depend on stress intensity? Physiol Plant 122:159–168
King SR, Davis AR, Zhang X, Crosby K (2010) Genetics, breeding and selection of rootstocks for Solanaceae and Cucurbitaceae. Scentia Horticulturae 127:106–111
Kromdijk J, Bertin N, Heuvelink E, Molenaar J, de Visser PHB, Marcelis LFM, Struik PC (2014) Crop management impacts the efficiency of quantitative trait loci (QTL) detection and use: case study of fruit loadxQTL interactions. J Exp Bot 65:11–22
Monforte AJ, Asins MJ, Carbonell EA (1997) Salt tolerance in Lycopersicon species. 5. Does genetic variability at quantitative trait loci affect their analysis? Theor Appl Genet 95:284–293
Munnik T, Meijer HJG (2001) Osmotic stress activates distinct lipid and MAPK signalling pathways in plants. FEBS Lett 498:72–178
Pascual L, Desplat N, Huang BE, Desgroux A, Bruguier L, Bouchet J-P, Le Q, Chauchard B, Verschave P, Causse M (2014) Potential of a tomato MAGIC population to decipher the genetic control of quantitative traits and detect causal variants in the resequencing era. Plant Biotechnol J. doi:10.1111/pbi.12282
Price AH (2006) Believe it or not, QTLs are accurate! Trends Plant Sci 11:213–216
Raga V, Bernet GP, Carbonell EA, Asins MJ (2014) Inheritance of rootstock effects and their association with salt-tolerance candidate genes in a progeny derived from ‘Volkamer’ lemon. J Am Soc Hort Sci 139:518–528
Rose JKC, Lee HH, Bennett AB (1997) Expression of a divergent expansin gene is fruit-specific and ripening-regulated. Proc Natl Acad Sci USA 94:5955–5960
Ruzicka K, Ljung K, Vanneste S, Podhorska R, Beeckman T, Friml J, Benkova E (2007) Ethylene regulates root growth through effects on auxin biosynthesis and transport–dependent auxin distribution. Plant Cell 19:2197–2212
Saliba-Colombani V, Causse M, Langlois D, Philouze J, Buret M (2001) Genetic analysis of organoleptic quality in fresh market tomato. 1. Mapping QTLs for physical and chemical traits. Theor Appl Genet 102:259–272
Sauvage C, Segura V, Bauchet G, Stevens R, Do PT, Nikoloski Z, Fernie A, Causse M (2014) Genome wide association in tomato reveals 44 candidate loci for fruit metabolic traits. Plant Physiol. doi:10.1104//pp.114.241521
Savvas D, Colla G, Rouphael Y, Scharz D (2010) Amelioration of heavy metal and nutrient stress in vegetables by grafting. Sci Hortic 127:156–161
Savvas D, Savva A, Ntatsi G, Ropokis A, Karapanos I, Krumbein A, Olympios C (2011) Effects of three commercial rootstocks on mineral nutrition, fruit yield, and quality of salinized tomato. J Plant Nutr Soil Sci 1:154–162
Scrase-Field AMG, Knight MR (2003) Calcium: just a chemical switch. Curr Opin Plant Biol 6:500–506
Sim S-C, Durstewitz G, Plieske J, Wieseke R, Ganal MW et al (2012) Development of a large SNP genotyping array and generation of high-density genetic maps in tomato. PLoS ONE 7(7):e40563. doi:10.1371/journal.pone.0040563
Simon EW (1978) The symptoms of calcium deficiency in plants. New Phytol 80:1–15
Steele NM, Mccann MC, Roberts K (1997) Pectin modification in cell walls of ripening tomatoes occurs in distinct domains. Plant Physiol 114:373–381
Sun K, Hunt K, Hauser BA (2004) Ovule abortion in Arabidopsis triggered by stress. Plant Physiol 135:2358–2367
The Tomato Genome Consortium (2012) The tomato genome sequence provides insights into fleshy fruit evolution. Nature 485:635–641
Turhan A, Ozmen N, Serbeci MS, Seniz V (2011) Effects of grafting on different rootstocks on tomato fruit yield and quality. Hortic Sci 38:42–1497
Van Ooijen JW (2006) JoinMap 4. Software for the calculation of genetic linkage maps in experimental populations. Kyazma BV, Wageningen, Netherlands
Van Ooijen JW (2009) MapQTL 6. Software for the mapping of quantitative trait loci in experimental populations of diploid species. Kyazma BV, Wageningen, Netherlands
Villalta I, Bernet GP, Carbonell EA, Asins MJ (2007) Comparative QTL analysis of salinity tolerance in terms of fruit yield using two Solanum populations of F7 lines. Theor Appl Genet 114:1001–1017
Wang H, Inukaia Y, Yamauchia A (2006) Root development and nutrient uptake. Crit Rev Plant Sci 25:279–301
Witcombe JR, Hollington PA, Howarth CJ, Reader S, Steele KA (2008) Breeding for abiotic stresses for sustainable agriculture. Phil Trans R Soc B 363:703–716
Acknowledgments
This work was supported in part by grants from the Spanish Government (AGL2008-00197/AGR, RTA2011-00132-C02) and the European Union (FP7-KBBE-2011-5), contract # 289365 (ROOTOPOWER). Authors thank UNIGENIA BIOSCIENCE SLU for the grafting labor, Dr. A.J. Monforte (IBMCP, Valencia, Spain) for comprehensive access to information on SolCAP SNPs, and Dr. G.P. Bernet (IBMCP, Valencia, Spain) for technical assistance.
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical standard
The authors declare that the experiment complies with the current laws of Spain.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by Richard G.F. Visser.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Asins, M.J., Raga, V., Roca, D. et al. Genetic dissection of tomato rootstock effects on scion traits under moderate salinity. Theor Appl Genet 128, 667–679 (2015). https://doi.org/10.1007/s00122-015-2462-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00122-015-2462-8