Abstract
Key message
This first association analysis between plant architecture and BR candidate genes in sorghum suggests that natural allelic variation has significant and pleiotropic effects on plant architecture phenotypes.
Abstract
Sorghum bicolor (L) Moench is a self-pollinated species traditionally used as a staple crop for human consumption and as a forage crop for livestock feed. Recently, sorghum has received attention as a bioenergy crop due to its water use efficiency and biomass yield potential. Breeding for superior bioenergy-type lines requires knowledge of the genetic mechanisms controlling plant architecture. Brassinosteroids (BRs) are a group of hormones that determine plant growth, development, and architecture. Biochemical and genetic information on BRs are available from model species but the application of that knowledge to crop species has been very limited. A candidate gene association mapping approach and a diverse sorghum collection of 315 accessions were used to assess marker-trait associations between BR biosynthesis and signaling genes and six plant architecture traits. A total of 263 single nucleotide polymorphisms (SNPs) from 26 BR genes were tested, 73 SNPs were significantly associated with the phenotypes of interest and 18 of those were associated with more than one trait. An analysis of the phenotypic variation explained by each BR pathway revealed that the signaling pathway had a larger effect for most phenotypes (R 2 = 0.05–0.23). This study constitutes the first association analysis between plant architecture and BR genes in sorghum and the first LD mapping for leaf angle, stem circumference, panicle exsertion and panicle length. Markers on or close to BKI1 associated with all phenotypes and thus, they are the most important outcomes of this study and will be further validated for their future application in breeding programs.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
During the last decade there has been a considerable interest in identifying alternative energy sources that can help to replace or decrease the use of petroleum. Sorghum (Sorghum bicolor L. Moench), the fifth most widely grown cereal crop in the world, is considered a promising alternative crop for biofuel production (Rooney et al. 2007). Sorghum possesses characteristics that are attractive for this use, such as: high yield potential, high water use efficiency, drought tolerance, well-developed production systems, a C4 photosynthesis mechanism, and abundant genetic variation for its improvement as a biofuel crop (Rooney et al. 2007; Paterson et al. 2009). Currently, increasing biomass of bioenergy crops is a breeding goal as important as ensuring food security. This goal has induced a paradigm shift in breeding for plant architecture to obtain high biomass varieties (Salas Fernandez et al. 2009). Therefore, the new challenges are to understand the genetics responsible for variation in plant architecture and apply that knowledge in breeding programs for biofuel production.
Brassinosteroids (BRs) are a group of steroid hormones that regulate important processes and traits such as germination, cell elongation, fertility, stress tolerance, and plant architecture (Taiz and Zeiger 2010). The BR biosynthesis and signaling pathways have been extensively studied in the model species Arabidopsis thaliana (Clouse 2011; Hao et al. 2013; Jiang et al. 2013; Zhao and Li 2012; Zhu et al. 2013), and more recently, in rice (Oryza sativa) (Tong and Chu 2012), barley (Hordeum vulgare) (Gruszka et al. 2011) and maize (Zea maize) (Hartwig et al. 2011, 2012; Makarevitch et al. 2012). Those studies have identified not only the enzymes involved in the BR pathways but also the genes involved in BR signal transduction and their biological mechanisms.
The BR biosynthesis pathway is a complex metabolic pathway with multiple parallel routes that are conserved across plant species (Taiz and Zeiger 2010; Gruszka et al. 2011). In general, the biosynthesis pathway converts campesterol (CR) into brassinolide (BL) through the activity of several enzymes that generate numerous intermediate compounds as shown in Fig. 1 (modified from RIKEN 2010). CR can be converted to campestanol (CN) through the enzyme DEETIOLATED2 (DET2). Although DET2 is usually the first enzyme that participates in the modification of CR, sometimes enzymes like DWARF4 (DWF4) and CYTOCHROME P450 C-3 OXIDASE (CPD/P90A1) can act before DET2 in a route called early C-22 oxidation pathway (Fujioka and Yokota 2003; Taiz and Zeiger 2010). The early C-6 oxidation pathway and late C-6 oxidation pathway involve different enzymes such as DWF4, CYTOCHROME P450 (CP450/CYP90D1), 3-EPI-6-DEOXOCATHASTERONE 23-MONOOXYGENASE (ROT3/CYP90C1) and BRASSINOSTEROID-6-OXIDASE 1 or 2 (BR6ox1 or BR6ox2) (Shimada et al. 2001; Fujioka and Yokota 2003; Taiz and Zeiger 2010; RIKEN RIKEN 2010). The final step is the conversion of castasterone (CS) into BL, the most biologically active BR compound (Fujioka and Yokota 2003). The levels of BR are also regulated by PHYB ACTIVATION-TAGGED SUPPRESSOR 1 (BAS1), which is a key component in the inactivation pathway (Turk et al. 2005; Taiz and Zeiger 2010).
BR biosynthesis pathway (modified from RIKEN 2010). CR campesterol; CN campestanol; CS castasterone; BL brassinolide; DET2 DEETIOLATED2; DWF4 DWARF4; CP450/CYP90D1 CYTOCHROME P450; CPD/P90A1 CYTOCHROME P450 C-3 OXIDASE, CYP90C1, 3-EPI-6-DEOXOCATHASTERONE 23MONOOXYGENASE; BR6ox1 or BR6ox2, BRASSINOSTEROID-6-OXIDASE 1 or 2, BAS1 PHYB ACTIVATION-TAGGED SUPPRESSOR 1. In blue boxes: associated candidate genes with phenotypes of interest. LA leaf angle, FL flowering time, PH plant height, PE panicle exsertion, PL panicle length, SC stem circumference (color figure online)
The BR signaling pathway (Fig. 2) consists of a cascade of reactions that start with the interaction of BL with a cell membrane-localized receptor-like kinase BR INSENSITIVE 1 (BRI1) (Hothorn et al. 2011; Li and Chory 1997; She et al. 2011). In the absence of BRs, BRI KINASE INHIBITOR 1 (BKI1) binds and inhibits BRI1 function (Jaillais et al. 2011; Wang et al. 2011; Wang and Chory 2006). A leucine carboxyl methyltransferase (LCMT) SBI1 also inhibits BRI1 function through PP2A (Wu et al. 2011). Also, in the absence of BRs, BR INSENSITIVE 2 (BIN2) kinase phosphorylates and inhibits the function of bri1 EMS SUPPRESSOR 1 (BES1) and BRASSINAZOLE RESISTANT 1 (BZR1) family transcription factors (Yin et al. 2002, 2005; Ye et al. 2011; He et al. 2005; Wang et al. 2002). During the activation, BL binds to the extracellular domain of BRI1 and a cascade of auto-phosphorylation events occur at multiple intracellular domains of BRI1 (Clouse 2011). Active BRI1 phosphorylates and inhibits the suppressor BKI1. BRI1 then associates with its co-receptor BRI1-ASSOCIATED RECEPTOR KINASE 1 (BAK1) (Gou et al. 2012; Wang et al. 2008). This association induces the phosphorylation of other positive-acting substrates such as: BRI1 SIGNALING KINASES (BSK1, 2 and 3), CONSTITUTIVE DIFFERENTIAL GROWTH 1 (CDG1), TGF-β RECEPTOR-INTERACTING PROTEIN-1 (TRIP-1) and TRANSTHYRETIN-LIKE PROTEIN (TTL) (Kim et al. 2011, Ehsan et al. 2005; Nam and Li 2004; Tang et al. 2008). It is not completely known how TRIP-1 and TTL are involved in the pathway, but some studies provide evidence of their importance in the cascade of signaling reactions (Nam and Li 2004; Ehsan et al. 2005). BRI1 phosphorylation of BSKs leads to the phosphorylation and activation of BR1 SUPPRESSOR 1 (BSU1), which may be responsible for the BIN2 dephosphorylation, causing its inactivation and allowing accumulation of unphosphorylated BES1/BZR1 (Kim et al. 2009). Simultaneously, PROTEIN PHOSPHATASE 2A (PP2A) (Mora-Garcia et al. 2004; Tang et al. 2011) dephosphorylates BES1/BZR1, which moves to the nucleus to interact with other transcription factors and regulates gene expression (Guo et al. 2013; Li 2010) (Fig. 2).
BR deactivation and activation signaling pathways. Deactivation signaling pathway (shown with red lines and arrow) occurs in absence of brassinosteroids (BRs). Activation signaling pathway (shown with green lines and arrows) occurs in the presence of BRs. BRI1 BR INTENSIVE 1; BAK1, BRL2 BRI1 LIKE PROTEIN 2; BRI1-ASSOCIATED RECEPTOR KINASE 1; BKI1 BRI KINASE INHIBITOR 1; CDG CONSTITUTIVE DIFFERENTIAL GROWTH; BSK1,2,3 BRI1 SIGNALING KINASES 1, 2, or 3; TTL TRANSTHYRETIN-LIKE PROTEIN; TRIP1, TGF-β RECEPTOR-INTERACTING PROTEIN-1; BSU1 BR1 SUPPRESSOR 1; BIN2 BR INTENSIVE 2; BES1, bri1 EMS SUPPRESSOR 1; BZR1 BRASSINAZOLE RESISTANT 1. In the blue boxes: associated phenotypes with the indicated candidate gene. LA leaf angle, FL flowering time, PH plant height, PE panicle exsertion, PL panicle length, SC stem circumference (color figure online)
Mutations in BR biosynthesis and signaling genes in model species have demonstrated their effects on plant architecture phenotypes such as: plant height (Clouse 1996; Mandava 1988; Taiz and Zeiger 2010), leaf length and shape (Yin et al. 2002; Clouse 1996; Mandava 1988; Zhiponova et al. 2013; Divi and Krishna 2009; Taiz and Zeiger 2010), flowering time (Taiz and Zeiger 2010), and abiotic stress response (Divi and Krishna 2009). However, there is no knowledge on the effects of BR in sorghum and whether or not BR genes are responsible for natural variation in plant architecture phenotypes. Therefore, there is a critical need to utilize quantitative genetic methods to test the hypothesis that sorghum orthologous BR genes are responsible for variation in plant architecture traits. The primary goal of our study was to utilize a candidate gene linkage disequilibrium (LD) mapping approach to test associations between plant architecture phenotypes and allelic variations in BR candidate genes identified in a diverse sorghum association panel.
LD mapping, also known as association mapping, is an alternative to linkage analysis and is based on the structure of LD across the genome. LD mapping explores the genotype–phenotype correlations within populations of unrelated individuals, exploiting historical recombination events (Nordborg and Tavaré 2002; Zhu et al. 2008). Some of the advantages of LD mapping include its low cost and short time of implementation, that together with the advances in genome sequencing and large number of available markers, have made it one of the most frequently used methods for gene/marker discovery (Flint-Garcia et al. 2003; Myles et al. 2009). In sorghum, the extensive diversity of the species has been exploited in both linkage and association mapping studies. Some of those studies have shed light on the genes/genomic regions associated with specific plant characteristics (Brown et al. 2008; Shehzad et al. 2009; Zou et al. 2012; Morris et al. 2013), but a complete understanding of the hormonal control of plant architecture remains elusive. Our study focused on BR candidate genes and the complete characterization of their effect on plant architecture. The ultimate goal is to identify markers that can be utilized in sorghum breeding programs to efficiently develop high biomass germplasm with optimal plant architecture components.
Materials and methods
Germplasm
The diverse sorghum collection used in this study consists of 288 accessions from the sorghum conversion program (SCP) and 149 breeding lines with significant genetic and geographical diversity and widely used in breeding programs (Casa et al. 2008). This panel has been successfully used in previous LD mapping studies (Brown et al. 2008; Sukumaran et al. 2012; Morris et al. 2013).
Experimental design and phenotypic data collection
A final set of 315 accessions from the association panel were planted in a randomized complete block design (RCBD) with two replications per location during summer 2010 and 2012. In 2010, sorghum lines were planted in Ames, Crawfordsville and Lewis (IA, USA) to evaluate five plant architecture traits: flowering time, plant height, panicle length, panicle exsertion and stem circumference. In 2012, lines were planted in Ames, Crawfordsville and Greenfield (IA, USA) to collect leaf angle data. Three plants per replication were evaluated for all traits except for leaf angle, for which two plants per replication were measured.
Flowering time was expressed as the number of days from planting to 50 % anthesis (50 % of plants in 50 % anthesis). Plant height was measured from ground level to the top of the panicle, and panicle exsertion was taken as the distance between the flag leaf and the base of the panicle. Panicle length was measured as the panicle raquis length. Stem circumference was determined at the base of the stem, after removal of leaf sheaths, and leaf angle was recorded on the leaf below the flag leaf, using a paper protractor.
Genotypic data
Protein sequences of BR genes from model species were obtained from the National Center for Biotechnology Information (NCBI) databases and used to identify the DNA sequence of sorghum orthologous genes. TBLASTN was performed on the sorghum genome sequence from phytozome V1.4 (Paterson et al. 2009). Predictions of gene structure for BR genes of model species and sorghum were obtained using FGENESH from Softberry Inc. (USA) (Solovyev et al. 2006) (Supplementary Fig. S1, S2). Proteins from model species and sorghum-predicted proteins were aligned using CLUSTAL2W (Larkin et al. 2007) to determine the level of identity, and their common domains were predicted using Pfam (Punta et al. 2012). A total of 26 BR candidate genes were identified: eleven from the biosynthesis pathway and 15 from the signaling pathway (Supplementary Tables T1, T2). Single nucleotide polymorphisms (SNPs) in the sorghum orthologous BR genes were extracted from the imputed set of ~260,000 SNPs developed using genotyping by sequencing (GBS) technology (Elshire et al. 2011; Morris et al. 2013). The SNP dataset for the association panel used in this study and other sorghum accessions has been publicly released by the Institute of Genomic Diversity at Cornell University and it is publicly available online at: http://www.igd.cornell.edu/index.cfm/page/projects/bread/sorgdata.htm. SNPs within 5 kb upstream or downstream of the BR genes were also extracted and used in the analysis. Sequenom (SQNM) MassARRAY iPLEX platform (Gabriel et al. 2009; Oeth et al. 2009) was used to identify additional SNPs in the candidate genes for which limited GBS data were available. A summary of GBS and SQNM developed SNPs for each candidate gene is presented in Supplementary Tables T1 and T2.
Phenotypic data analysis
Phenotypic data analysis was performed by ANOVA using PROC mixed as implemented in SAS® 9.2 (SAS Institute Inc., Cary, NC, USA), with location considered fixed, while replication, genotype and genotype × location interaction were considered random effects. Best linear unbiased predictions (BLUPs) were calculated using the library lme4 on R statistical software (R Core Team 2013). The BLUP for each line was used as its observed phenotypic value for the association analysis.
Genotypic data analysis
Population structure was determined using 702 genome-wide SNPs distributed across the sorghum genome. Those SNPs were selected from the GBS set of 260,000 SNPs based on the following criteria: (i) SNPs highly informative with less than 15 % of missing data; (ii) SNPs that have a minimum distance of 350 kb considering the average LD (r 2 < 0.1) reported by Morris et al. (2013); and (iii) SNPs randomly distributed in every chromosome. The program STRUCTURE 2.2.3 (Pritchard et al. 2000) was used to detect subpopulations. An admixture model was used varying the number of subpopulations (k) from one to ten. The analysis was performed using ten iterations per k, a 20,000 length of burn-in period and 30,000 MCMC (Markov Chain Monte Carlo, France) replicates. The optimal number of subpopulations was determined using: (i) the posterior probability plot, (ii) Delta K (ΔK) method (Evanno et al. 2005), (iii) stability of grouping patterns across ten runs, and (iv) germplasm information from previous studies. Population structure was verified by Principal Component Analysis (PCA), which was calculated using the statistical program Rx64 3.0 (R Core Team 2013). Results of PCA1 vs PCA2 and PCA1 vs PC3 were plotted, analyzed and compared with STRUCTURE results.
Kinship matrix, also known as coefficient of relatedness or co-ancestry matrix, was calculated using the same 702 SNPs, the algorithm described by Loiselle et al. (1995), and the software SPAGeDi 1.4 (Hardy and Vekemans 2002).
Association analysis
A total of 263 SNPs with a minimum allele frequency >5 % were used to test marker-trait associations. Two models for association analysis were tested using TASSEL 3.0 (Bradbury et al. 2007): GLM (General Linear Model) and MLM (Mixed Linear Model). GLM tests the marker–phenotype associations including population structure as a covariate (Q-matrix). Both marker effect and population structure are considered fixed (Bradbury et al. 2007). MLM tests the marker–phenotype associations including population structure and kinship matrix. Markers and population structure are fixed effects, while kinship is included as a random effect (Bradbury et al. 2007). Quantile–quantile (Q–Q) plots of the expected cumulative distribution of p values were performed for each model and trait to assess the best model to control for type I errors (Yu et al. 2006). False discovery rate (FDR) was used to assign a measure of significance to each test and correct for multiple comparisons. The FDR method assigns a q value to each p value after analyzing the p value distribution. The q value threshold was determined on a trait by trait basis. q values were obtained using QVALUE software (Storey and Tibshirani 2003) and R software (R Core Team 2013).
Analysis of the overall phenotypic variation explained by the significantly associated genes in the BR signaling and biosynthesis pathways was performed using SAS 9.2 (SAS Institute Inc., Cary, NC, USA). The original SAS script of mixed linear model for association analysis (by Gael Pressoir, Institute for Genomic Diversity, Cornell University, http://www.maizegenetics.net/images/stories/interests/qk.txt) was modified to fit multiple markers in the model. Those markers were selected based on the individual SNP analysis. Only one or two significantly associated markers for each associated BR gene were included in the model and the complete list of SNPs for each pathway is presented in Supplementary Table T3. This criterion was used to avoid an overestimation of the phenotypic variation explained by multiple markers that can be in LD (r 2 > 0.2). The mixed model based on maximum likelihood (ML) was used and the R 2-likelihood ratio (R 2 LR) was calculated to explain the phenotypic variation as recommended by Sun et al. (2010)
where −2logL M is the −2 log-likelihood of the model of interest; −2logL 0 is the −2 log-likelihood of the minimal model; and n is the number of observations. The minimal model was defined as the model that only considered population structure Q as fixed effect and kinship K as random effect (Sun et al. 2010).
Results
Phenotypic analysis
Descriptive statistics confirmed the large variability of the phenotypes in the association panel (Table 1). Results from ANOVA showed that replication nested within location was not a significant source of variation (p < 0.05) for any of the traits except for stem circumference (Table 2). The analysis of variance also indicated that there was a significant effect of location, genotype, and location × genotype interaction (p < 0.05) for all traits except leaf angle. Location was not significant for leaf angle, however, its interaction with genotype was marginally significant. This result suggests that for leaf angle, the location effect cannot be considered separately from the genotype effect.
Correlations between phenotypes were calculated using BLUPs (Table 3). Stem circumference and plant height were the phenotypes correlated with most other traits at a significance level p < 0.001. An interesting positive correlation was observed between stem circumference and flowering time. According to these results, lines with late flowering tended to present thicker stems, an important correlation to consider when breeding for biomass production. A significant (p < 0.001) and positive correlation was observed between leaf angle and plant height but negative correlations were identified with flowering time (p < 0.001) and stem circumference (p < 0.01).
BR candidate genes
A total of 26 candidate genes in sorghum were identified based on TBLASTN scores, gene structure similarity, protein alignment score and protein common domains. In most cases, a single sorghum orthologous gene could be identified for each of the known genes in the biosynthesis (Supplementary Table T1; Supplementary Figure S1) and signaling pathway (Supplementary Table T2; Supplementary Figure S2). An exemption was Sb01g015040, a sorghum gene very similar to both biosynthetic genes BR6ox1 and BR6ox2 (Supplementary Table T1). In the signaling pathway, the same sorghum gene (Sb02g019470) was identified as the most likely orthologous of both BRL1 and BRL3; while another genomic region (Sb02g037500) was similar to both BES1 and BZR1genes (Supplementary Table T2). Finally, one region in sorghum chromosome 3 (Sb03g012070) and one in chromosome 4 (Sb04g008580) were equally predicted as orthologous of BIN2, since both sequences presented high identity, low E value, similar alignment score and same common domains as the BIN2 protein in model species (Supplementary Table T2).
Important similarities in gene structure were observed for most BR candidate genes with the exemption of CYP90C1 and CYP90D1. For these two genes, the structure of the Arabidopsis homolog was drastically different than the predicted sorghum gene structure. Therefore, a rice-predicted ortholog was used in each case to elucidate these differences. As expected based on the shorter evolutionary distance between rice and sorghum, gene structure corresponding to their orthologs had high similarities as summarized in Supplementary Figure S1.
Genotypic analysis
Population structure analysis determined that five is the optimal number of subpopulations in the association panel. The five subpopulations were classified as: Q1, intermediate races Guinea/Bicolor; Q2, Caudatum; Q3, intermediate races Guinea-Caudatum (West Africa); Q4, Kafir and Q5, Durra (Supplementary Fig. S3). Principal Component Analysis (PCA) results were consistent with STRUCTURE. PCA1 explained 8.02 % of the variation and separated the subpopulations Caudatum and Guinea/Caudatum. PCA2 explained 7.48 % of the variation and separated the subpopulation Durra. PCA3 explained 6.19 % of the variation and separated the Kafir subpopulation (data not shown).
Association analysis
The mixed linear model (MLM), which includes population structure and kinship, was identified as the best model for the six plant architecture traits analyzed in this study. If the model includes population structure and kinship, the observed p values have a better approximation to the expected p value distribution in comparison with the model that only accounts for population structure (GLM).
Association analysis per trait
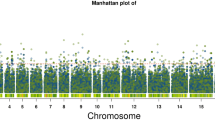
A total of 73 markers were found significantly associated with the six phenotypes of interest, 25 of them were in complete LD (r 2 = 1), and 18 markers were associated with more than one trait (Tables 4, 5). Traits with the highest number of associated markers were stem circumference, panicle length and flowering time with 26, 24 and 18 markers, respectively.
The FDR threshold (q value) was determined on a trait by trait basis (Tables 4, 5). The q value threshold was q < 0.26 for flowering time and panicle length, q < 0.23 for leaf angle, q < 0.28 for plant height and q < 0.17 for panicle exsertion and stem circumference. Therefore, the expected number of false associations varies from one to six depending on the trait.
Association analysis per candidate gene
Markers in 16 candidate genes were associated with the traits of interest; eight of those genes belong to the signaling pathway and eight to the biosynthesis pathway (Figs. 1, 2). The level of LD between markers in the same gene/genomic region varied depending on the number of markers, physical distance between them and LD structure in that genomic region (Tables 4, 5).
BKI1 was the BR signaling candidate gene associated with more traits (Table 4; Fig. 2). Although the phenotypic variation explained by each marker was small (R 2 < 0.055), this is a very interesting candidate gene since it was associated with all six phenotypes. Moreover, eight markers within BKI1 or in the nearby region were associated with more than one trait (Table 4) and these apparently pleiotropic effects are consistent with the known repressor function of BKI1 upstream in the BR signaling pathway (Fig. 2).
One of the most interesting results in this study was the identification of 18 markers from nine candidate genes that were associated with more than one trait (Figs. 1, 2). The effect of those markers on the phenotype was consistent with the phenotypic correlations previously described (Tables 3, 4, 5). For example, marker S2_61887636 in the candidate gene BKI1, was associated with flowering time, panicle length and stem circumference. The presence of the nucleotide G over the nucleotide C reduces the flowering time, panicle length and stem circumference. These results are consistent with phenotypic correlations because flowering time, panicle length and stem circumference were positively and significantly correlated.
The overall phenotypic variation explained by the associated markers/genes of both BR pathways ranged from 6 % for plant height to 26 % for panicle length (Table 6). When the phenotypic variation was analyzed independently per pathway, the signaling pathway explained a higher proportion of the phenotypic variation than the biosynthesis pathway for most of the plant architecture traits (Table 6).
Discussion
BR candidate genes
Multiple analyses were performed to identify the most likely sorghum BR orthologs. Gene structure was used to compare the number and size of introns and exons between species. According to Itoh et al. (2007), most of the rice and Arabidopsis genes share important similarities, however, there are some rice genes that may have diverged to such an extent that their homologs could not be detected by sequence similarity search, although the protein domains and apparent function are the same. It means that genome sequences drastically changed during evolution, but their functionality is the same. These findings can explain the difference in gene structure between Arabidopsis and predicted sorghum genes CYP90C1 and CYP90D1 (Supplementary Figure 1).
In a few cases, the same sorghum candidate gene was predicted as the most likely ortholog of two different genes in the model species. This observation corresponds to genes that have been reported to have redundant activity in Arabidopsis. According to Shimada et al. (2003), BR6ox1 and BR6ox2 genes have similar DNA sequence, similar protein sequence and have the same substrate specificity after yeast expression analysis was performed, so the authors concluded that these two genes arose by gene duplication in Arabidopsis and have redundant functions. Likewise, BRL1 and BRL3 proteins have also been reported with redundant functions in Arabidopsis and their corresponding genes have similar structure (Zhou et al. 2004; Morillo and Tax 2006). In the case of BES1 and BZR1, both genes encode closely related proteins that have approximately 90 % sequence identity, the same consensus sites (Clouse 2002) and overlapping functions (Wang et al. 2002; Clouse 2002). Even though we have not performed any studies to demonstrate functionality of the predicted BR sorghum genes and proteins, we have exhaustively compared them with genes from model species using multiple bioinformatic tools to determine sequence similarity, protein–protein identity levels, gene structure comparisons and functional domains.
Genetic diversity in the panel
The large morphological differentiation in the subspecies bicolor has been classified by Harlan and De Wet (1972) into five major races: Bicolor, Guinea, Kafir, Durra and Caudatum. This sorghum diversity panel has been previously characterized by different groups using molecular markers; however, the results have not been completely consistent between studies (Casa et al. 2008; Sukumaran et al. 2012; Brown et al. 2011; Morris et al. 2013). The population structure of k = 5 reported here is consistent with Sukumaran et al. (2012). Three major races (Durra, Kafir and Caudatum) were clearly distinguished as previously reported (Sukumaran et al. 2012; Brown et al. 2011; Casa et al. 2008) and the intermediate races, Guinea/Bicolor and Guinea/Caudatum (West Africa), were identified as separate groups, in agreement with Casa et al. (2008). In spite of the small representation of individuals of the intermediate race Guinea/Bicolor, they constituted a separate subpopulation as reported by Morris et al. (2013) at k = 5.
Association analysis
LD mapping studies have been extensively used in humans, animals and plants to assess marker-trait associations. Although the methodology per se can lead to spurious associations, the control for population structure and co-ancestry relationships has minimized the number of false positive associations (Yu et al. 2006; Sun et al. 2010). In our study, the total number of significant markers for each trait was reduced when the coefficient of co-ancestry was included in the model because it corrects for spurious associations that can be produced by multiple levels of relatedness among lines (Yu et al. 2006). These results are consistent with similar studies performed using the same diversity panel (Morris et al. 2013; Sukumaran et al. 2012). A total of 73 markers were associated with all phenotypes and each marker explained less than 7 % of the phenotypic variation, an expected result considering the highly quantitative nature of plant architecture phenotypes.
Panicle length was the trait associated with more candidate genes: three from the BR biosynthesis pathway and seven from the signaling pathway localized on chromosomes 1, 2, 3, 4, 5 and 8 (Figs. 1, 2). Morris et al. (2013) identified markers associated with inflorescence branch length, a panicle length-related trait, on the same chromosomes but not physically close to the markers reported in this study. On chromosome 1, Morris et al. (2013) identified five SNPs located 900–3,100 kb from BSK1 markers and 1,400–6,000 kb from BSK2 markers. Similar results were obtained for markers on chromosome 2 (gene BKI1) and chromosome 3 (BAS1) which are at a distance of 7,000 and 700 kb from associated SNPs in Morris et al. (2013), respectively. A QTL study for panicle length performed using RFLPs on recombinant inbred lines (RILs) identified a QTL on chromosome 2 between markers UMC04 and BNL12.06, which are 7,000 and 10,000 kb from the associated markers in BKI1, respectively (Rami et al. 1998). Another study also identified two QTL on chromosome 8: Bin2742 and Bin2753 (Zou et al. 2012), but markers on BSU1 are outside of those QTL regions at approximately 4,500 and 3,500 kb, respectively. Genotypic information of the parental lines used by Rami et al. (1998) and Zou et al. (2012) is not publicly available for comparative haplotype analysis.
Plant height has been extensively studied in sorghum and previous reports have identified QTL on chromosomes 1, 3, 6, 7, 9 and 10 (Pereira and Lee 1995; Salas Fernandez et al. 2009; Zou et al. 2012), but not on chromosomes 2 and 5, as reported in this study. It is important to emphasize that the gene DW3 (ortholog to DWF3 in Arabidopsis) has been cloned and identified on chromosome 7, while DW1 has been mapped on chromosome 9 (Brown et al. 2008; Morris et al. 2013; Thurber et al. 2013). Preliminary evidence suggests that DW2 is on chromosome 6 (Thurber et al. 2013). The fact that these three genes (DW1, DW2 and DW3) have major effects on plant height could explain that other studies were not able to identify genomic regions with small effects on the trait, as we have for markers located on chromosomes 2 and 5.
Significant markers for flowering time were localized on chromosomes 1, 2, 3 and 5. El Mannai et al. (2011) reported SSR markers associated with flowering time on chromosomes 1 and 2 that are distant from our associated markers. The SSRs Xtxp61, Xtxp75 and Xtxp279 on chromosome 1 are at approximately 8,000–10,000 kb from the associated markers on the BSK1 gene. On chromosome 2, markers for BKI1 are approximately 6,000 kb away from the SSR Xtxp100 (El Mannai et al. 2011). All markers with a significant effect on flowering time identified in our study explained a small proportion of the phenotypic variation (R 2 < 0.06) and these results are expected considering that Ma 1, mapped to chromosome 6, has been reported as a major gene explaining approximately 45 % of the variation (Murphy et al. 2011; Li et al. 2013).
Limited information is available about the genetic mechanisms controlling panicle exsertion and stem circumference in sorghum (Klein et al. 2001; Zou et al. 2012). According to Klein et al. (2001), there is a QTL on chromosome 1 that explains 11–13 % of variation in panicle exsertion. In our study, markers on BSK1 were associated with this trait, and these markers were located in the QTL region delimited by Xtxp37 and Xpxt61 (Klein et al. 2001). The RILs used by Klein et al. (2001) were developed by crossing the parental lines Sureno (PI 561472) and RTx436 (PI 561071). Both lines have different haplotypes in the genomic region corresponding to the candidate gene BSK1 and also have different alleles for the associated BSK1 SNPs. These results provide additional independent support to the findings reported in our study. The two QTL for panicle exsertion reported by Zou et al. (2012) on chromosome 1 did not correspond to the same genomic region of our significant markers on BSK1 gene (3,400–17,000 kb from Bin377 and Bin469). A similar result was obtained when the QTL on chromosome 3 was compared with our significant markers on DET2 (7,000 kb away from Bin1234) (Zou et al. 2012). Only one QTL (Bin253) for stem circumference previously identified (Zou et al. 2012) was on the same chromosome as our BRL2 and TTL markers. However, this previously reported that QTL is 750 kb from BRL2 and 1,800 kb from TTL. Genotypic information for the Chinese lines used in Zou et al. (2012) was not publicly available (personal communication with the authors) and thus, a comparative haplotype analysis in the genomic regions of the BR candidate genes was not possible.
Leaf angle is an important plant architecture phenotype for which very limited information is available. The only study for this trait in sorghum identified a major QTL on chromosome 7 that explained 45 % of the variation (Hart et al., 2001). In our study, markers on chromosomes 1, 2 and 3 were associated with leaf angle but none of them are in common with genomic regions reported by Hart et al. (2001). The RILs used by Hart et al. (2001) were obtained from the cross between BTx623 (PI 659985) and IS3620C (PI 533839). Both inbred lines have the same haplotype for the candidate genes BSK1, BES1, DWF7, and BAS1 but contrasting haplotypes for BKI1. Multiple experimental factors could have affected the capacity to identify a QTL in the BKI1 region by Hart et al. (2001) in spite of the contrasting parental haplotypes for the gene, particularly the masking effect of the major QTL on chromosome 7 that could have hindered the identification of QTL with small effects.
In summary, most of the significantly associated markers were localized in novel sorghum genomic regions not previously correlated with phenotypic variation in plant architecture. The new knowledge contributed by this study is of importance considering that most previous sorghum studies were conducted using biparental populations and thus, have sampled a very limited representation of the allelic variation available in the species. Additionally, in those studies, the identification of QTL/markers with small effects, as the ones reported here, was likely hindered by genes/QTL with large effects.
Multiple phenotypes associated with the same marker
The identification of 18 markers associated with more than one trait suggests that BR candidate genes in sorghum have pleiotropic effects. Pleiotropy has been reported for many BR Arabidopsis and rice mutants (Fujioka et al. 1997; Choe et al. 1999a, b; Yamamuro et al. 2000; Choe et al. 2002; Yin et al. 2002; Sakamoto et al. 2006; Wu et al. 2008).
In our study, markers on and nearby BKI1 were associated with all traits of interest: flowering time, leaf angle, panicle length, panicle exsertion, plant height and stem circumference. This gene is a negative regulator of the BR signaling pathway and Arabidopsis mutants in BKI1 have shown phenotypes affected in rosette radius, leaf angle, plant height and petiole length (Jaillais et al. 2011; Wang et al. 2011; Wang and Chory 2006). There is no additional knowledge on the effect of BKI1 mutants in other species and its effect on plant phenotypes. This is the first study suggesting that natural allelic variation in BKI1 is associated with changes in plant architecture characteristics.
BKI1 has been characterized as an unstructured protein that functions through short linear motifs involved in protein–protein interactions and/or modifications (Diella et al. 2008; Jaillais et al. 2011). The N-terminal of BKI1 is localized to the plasma membrane and has three conserved motifs. Motif-3 contains tandem repeats of basic residues [lysine (K)/arginine(R)] and the Y211 has been proposed as the phosphorylation site (Jaillais et al. 2011). The C-terminal is the most conserved region of BKI1 in several species (residues 253–337 in A. thaliana) and it was both necessary and sufficient to bind the BRI1-kinase domain. BKI1 associated specifically with the kinase domain of BRI1 but not with TTL, BIN2 or the kinase domain of other receptor like kinases such as BAK1, BRL1 and BRL3 (Wang and Chory 2006). The sorghum-predicted BKI1 protein has the tandem repeats of basic residues in motif-3 and a highly conserved C-terminal that is 62.3 and 91.3 % similar to the A. thaliana and rice C-terminal regions, respectively (data not shown). The markers on BKI1 significantly associated with flowering time, panicle length and stem circumference correspond to the amino acid residues 58, 158, and 186 (data not shown). Due to the nature of the genotyping technology used in this study (GBS), the gene was not saturated with markers and thus further investigation will be conducted to fully identify polymorphisms in BKI1 and relate them to known protein motifs and important amino acid residues.
BSK1 was another candidate gene associated with more than one phenotype (panicle exsertion, panicle length, leaf angle and flowering time). BSK1 protein mediates signal transduction from receptor kinase BRI1. In Arabidopsis, only BSK1 and BSK3 proteins interact with BRI1 (Tang et al. 2008) and their mutants presented reduced rosette size, leaf curling and higher leaf angle (Sreeramulu et al. 2013). No mutants have been reported for BKI1 and BSK1 in rice but, proteins from both genes interact with BRI1, and BRI1 mutants have been extensively characterized in this species. Sakamoto et al. (2013) identified three OsBRI1 mutants (d61–1 N, d61–11, and d61–12) which showed variation in plant height, leaf angle, internode elongation, and seed shape. Mutant phenotypes included shorter plants, reduced leaf angle, abnormal and twisted leaves, reduced internode elongation and plants without flowers (Sakamoto et al. 2013). Since proteins translated from BSK1 and BKI1 genes interact directly with BRI1, we can speculate that similar effects on the phenotype could be observed by allelic variations in BSK1 and BKI1, as shown in our study.
Arabidopsis mutants in the BR signaling pathway gene BES1 have been characterized by constitutive BR responses including longer petioles and curly leaves (Yin et al. 2002). Rice mutants of BZR1, the closest ortholog of BES1, resulted in dwarf plants with erect leaves (reduced leaf angle) and low yield (Bai et al. 2007; Zhang et al. 2012). In our study, three markers close to BES1 were associated with changes in leaf angle in agreement with the mutant phenotypes reported in rice for this gene. Additionally, novel phenotypes have been associated with BES1 in this study, since stem circumference and panicle length have not been previously characterized in BES1 mutants of other species.
Overall phenotypic variation explained by BR pathways
Mutant studies on models species have demonstrated the importance of individual BR biosynthesis and signaling genes on plant architecture phenotypes. We have reported that natural allelic variations on individual BR candidate genes in sorghum have relatively small effects on plant architecture characteristics. However, it is important to determine the total variation that all significant BR genes could have on the phenotypes of interest. Based on the R 2 LR statistics, all associated genes from both BR pathways explained more than 15 % of the overall variation of each phenotype, with the exception of plant height. In general, natural allelic variations of signaling genes accounted for a higher proportion of the variation in all traits. Panicle length and stem circumference were the plant architecture characteristics more strongly affected by the allelic variations in the signaling pathway (R 2 = 0.23 and 0.20, respectively) (Table 6). Studies of natural allelic variations in BR genes have not been conducted in model or crop species and the overall proportion of the phenotypic variation determined by multiple BR genes has not been reported before. Therefore, our study provides the first evidence of the importance of both individual BR genes and entire pathways on the control of plant architecture in sorghum.
Final remarks
One nucleotide change could significantly alter the transcribed RNA and translated protein, causing new variation in a phenotype. Therefore, allelic variation associated with specific traits in LD mapping studies is proposed to be either the causal polymorphism or in LD with the causal polymorphism. If one of the associated markers is the real cause of variation and the level of LD is high, it was expected to obtain results in which other markers on the same genomic region were associated with the phenotype as well. Likewise, if the real cause of variation was not on the candidate gene but on the nearby genomic region, it was possible to identify markers on BR genes significantly associated with the phenotype of interest. These two alternative conclusions depend on the extent of LD for the particular species, genomic region and population under investigation. A high average LD has been previously reported for this diversity panel (27–44 kb, r 2 < 0.2) (Morris et al. 2013) but low and medium range LD have also been reported in sorghum (5–15 kb) (Hamblin et al. 2005), demonstrating the variability between genomic regions. In our study, the extent of LD between markers located on or nearby a BR gene was also variable (Tables 4, 5). Additional specific investigation of an extended region around the BKI1 gene demonstrated that there is a rapid decay of LD upstream and downstream of the gene (Supplementary Figure S4). This observation and the apparent highly pleiotropic nature of associated markers deepened our interest in this genomic region and we have thus initiated experiments to further investigate BKI1 and its effect on plant architecture.
Understanding the genetic control of plant architecture traits is a key element for the improvement of sorghum varieties for biofuel production and in general, for the efficient manipulation of plant characteristics determining yield. BRs are a group of hormones with important and pleiotropic effects on plant characteristics that have not been exploited in breeding programs to genetically improve crop species. Our study is the first LD mapping analysis of BR genes and their effect on plant architecture and thus, it contributes new knowledge about the natural genetic variation in BR genes and their potential effect on agronomically important traits. This is also the first sorghum study to investigate natural allelic variation affecting leaf angle, stem circumference, panicle exsertion and panicle length in a diverse sorghum set of germplasm. The overall phenotypic variation explained by each BR pathway, the 18 pleiotropically associated markers, and all significant associations with BKI1 are the most important outcomes of this project and will be further characterized and validated in other populations. If our results are confirmed, those markers could be utilized in breeding programs to efficiently improve plant architecture traits, to transfer the knowledge from sorghum to other important grass species such as rice, maize, wheat and sugarcane, and can be specifically exploited in sorghum breeding programs to design the best ideotype for each environment.
Author contributions
M.G.S.F., M.B.M.P., and J.Z. designed the experiment; M.B.M.P. and J.Z. performed phenotypic data collection and analysis; Y.Y. supervised BR candidate gene identification; M.B.M.P., J.Z., and J.H. collected and analyzed genotypic data; M.B.M.P. performed LD mapping analysis; M.G.S.F., M.B.M.P., J.Z., J.H., and Y.Y. contributed to writing and editing of the manuscript; M.G.S.F. directed the project.
References
Asami T, Min YK, Nagata N, Yamagishi K, Takatsuto S, Fujioka S, Murofushi N, Yamaguchi I, Yoshida S (2000) Characterization of brassinazole, a triazole-type brassinosteroid biosynthesis inhibitor. Plant Physiol 123:93–100
Bai MY, Zhang LY, Gampala SS, Zhu SW, Song WY, Chong K, Wang ZY (2007) Functions of OsBZR1 and 14-3-3 proteins in brassinosteroid signaling in rice. Proc Natl Acad Sci USA 104:13839–13844
Bradbury PJ, Zhang Z, Kroon DE, Casstevens TM, Randoss Y, Buckler ES (2007) TASSEL: software for association mapping of complex traits in diverse samples. Bioinformatics 23:2633–2635
Brown PJ, Rooney WL, Franks C, Kresovich S (2008) Efficient mapping of plant height quantitative trait loci in a sorghum association population with introgressed dwarfing genes. Genetics 180:629–637
Brown PJ, Myles S, Kresovich S (2011) Genetic support for phenotype-based racial classification in sorghum. Crop Sci 51:224–230
Casa AM, Pressoir G, Brown PJ, Mitchell SE, Rooney WL, Tuinstra MR, Franks CD, Kresovich S (2008) Community resources and strategies for association mapping in sorghum. Crop Sci 48:30–40
Choe S, Dilkes BP, Gregory BD, Ross AS, Yuan H, Noguchi T, Fujioka S, Takatsuto S, Tanaka A, Yoshida S, Tax FE, Feldmann KA (1999a) The Arabidopsis dwarf1 mutant is defective in the conversion of 24-methylenecholesterol to campesterol in brassinosteroid biosynthesis. Plant Physiol 119:897–907
Choe S, Noguchi T, Fujioka S, Takatsuto S, Tissier CP, Gregory BD, Ross AS, Tanaka A, Yoshida S, Tax FE, Feldmann KA (1999b) The Arabidopsis dwf7/ste1 mutant is defective in the ∆7 sterol C-5 desaturation step leading to brassinosteroid biosynthesis. Plant Cell 2:207–221
Choe S, Schmitz RJ, Fujioka S, Takatsuto S, Lee MO, Yoshida S, Feldmann KA, Tax FE (2002) Arabidopsis brassinosteroid-insensitive dwarf12 mutants are semidominant and defective in a glycogen synthase kinase 3β-like kinase. Plant Physiol 130:1506–1515
Clouse SD (1996) Molecular genetic studies confirm the role of brassinosteroids in plant growth and development. Plant J 10:1–8
Clouse SD (2002) Brassinosteroid signaling: novel downstream components emerge. Curr Biol 12:R485–R487
Clouse SD (2011) Brassinosteroid signal transduction: from receptor kinase activation to transcriptional networks regulating plant development. Plant Cell 23:1219–1230
Diella F, Haslam N, Chica C, Budd A, Michael S, Brown NP, Trave G, Gibson TJ (2008) Understanding eukaryotic linear motifs and their role in cell signaling and regulation. Front Biosci 13:6580–6603
Divi UK, Krishna P (2009) Brassinosteroid: a biotechnological target for enhancing crop yield and stress tolerance. New Biotechnol 26:3–4
Ehsan H, Ray WK, Phinney B, Wang X, Huber SC, Clouse SD (2005) Interaction of Arabidopsis BRASSINOSTEROID-INSENSITIVE 1 receptor kinase with a homolog of mammalian TGF-β receptor interacting protein. Plant J 43:251–261
El Mannai Y, Shehzad T, Okuno K (2011) Variation in flowering time in sorghum core collection and mapping of QTLs controlling flowering time by association analysis. Genet Resour Crop Evol 58:983–989
Elshire RJ, Glaubitz JC, Sun Q, Poland JA, Kawamoto K, Buckler ES, Mitchell SE (2011) A robust, simple genotyping-by-sequencing (GBS) approach for high diversity species. PLoS ONE 6:e19379
Evanno G, Regnaut S, Goudet J (2005) Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol 14:2611–2620
Flint-Garcia SA, Thornberry JM, Buckler ES (2003) Structure of linkage disequilibrium in plants. Annu Rev Plant Biol 54:357–374
Fujioka S, Yokota T (2003) Biosynthesis and metabolism of brassinosteroids. Annu Rev Plant Biol 54:137–164
Fujioka S, Li J, Choi YH, Seto H, Takatsuto S, Noguchi T, Watanabe T, Kuriyama H, Yokota T, Chory J, Sakurai A (1997) The Arabidopsis deetiolated2 mutant is blocked early in brassinosteroid biosynthesis. Plant Cell 11:1951–1962
Gabriel S, Ziaugra L, Tabbaa D (2009) SNP genotyping using the Sequenom MassARRAY iPLEX platform. Curr Protoc Hum Genet 60:2.12.1–2.12.16
Gou X, Yin H, He K, Du J, Yi J, Xu S, Lin H, Clouse SD, Li J (2012) Genetic evidence for an indispensable role of somatic embryogenesis receptor kinases in brassinosteroid signaling. PLoS Genet 8:e1002452
Gruszka D, Szarejko I, Maluszynski M (2011) Identification of barley DWARF gene involved in brassinosteroid synthesis. Plant Growth Regul 65:343–358
Guo H, Li L, Aluru M, Aluru S, Yin Y (2013) Mechanisms and networks for brassinosteroid regulated gene expression. Curr Opin Plant Biol 16:545–553
Hamblin MT, Salas Fernandez MG, Casa AM, Mitchell SE, Paterson AH, Kresovich S (2005) Equilibrium processes cannot explain high levels of short- and medium-range linkage disequilibrium in the domesticated grass Sorghum bicolor. Genetics 171:1247–1256
Hao J, Yin Y, Fei SZ (2013) Brassinosteroid signaling network: implications on yield and stress tolerance. Plant Cell Rep 32:1017–1030
Hardy OJ, Vekemans X (2002) SPAGeDi: a versatile computer program to analyze spatial genetic structure at the individual or population levels. Mol Ecol Notes 2:618–620
Harlan JR, De Wet JMJ (1972) A simplified classification of cultivated sorghum. Crop Sci 12:172–176
Hart GE, Schertz KF, Peng Y, Syed NH (2001) Genetic mapping of Sorghum bicolor (L.) Moench QTL that control variation in tillering and other morphological characters. Theor Appl Genet 103:1232–1242
Hartwig T, Chuck GS, Fujioka S, Klempien A, Weizbauer R, Potluri DP, Choe S, Johal GS, Schulz B (2011) Brassinosteroid control of sex determination in maize. Proc Natl Acad Sci USA 108:19814–19819
Hartwig T, Corvalan C, Best NB, Budka JS, Zhu JY, Choe S, Schulz B (2012) Propiconazole is a specific and accessible brassinosteroid (BR) biosynthesis inhibitor for Arabidopsis and maize. PLoS ONE 7:e36625
He JX, Gendron JM, Sun Y, Gampala SS, Gendron N, Sun CQ, Wang ZY (2005) BZR1 is a transcriptional repressor with dual roles in brassinosteroid homeostasis and growth responses. Science 307:1634–1638
Hothorn M, Belkhadir Y, Dreux M, Dabi T, Noel JP, Wilson IA, Chory J (2011) Structural basis of steroid hormone perception by the receptor kinase BRI1. Nature 474:467–471
Itoh T, Tanaka T, Barrero R, Yamasaki C, Fujii Y, Hilton PB, Antonio BA, Aono H, Apweiler R, Bruskiewich R, Bureau T, Burr F, Costa de Oliveira A, Fuks G, Habara T, Haberer G, Han B, Harada E, Hiraki AT, Hirochika H, Hoen D, Hokari H, Hosokawa S, Hsing YI, Ikawa H, Ikeo K, Imanishi T, Ito Y, Jaiswal P, Kanno M, Kawahara Y, Kawamura T, Kawashima H, Khurana JP, Kikuchi S, Komatsu S, Koyanagi KO, Kubooka H, Lieberherr D, Lin YC, Lonsdale D, Matsumoto T, Matsuya A, McCombie WR, Messing J, Miyao A, Mulder N, Nagamura Y, Nam J, Namiki N, Numa H, Nurimoto S, O’Donovan C, Ohyanagi H, Okido T, Oota S, Osato N, Palmer LE, Quetier F, Raghuvanshi S, Saichi N, Sakai H, Sakai Y, Sakata K, Sakurai T, Sato F, Sato Y, Schoof H, Seki M, Shibata M, Shimizu Y, Shinozaki K, Shinso Y, Singh NK, Smith-White B, Takeda J, Tanino M, Tatusova T, Thongjuea S, Todokoro F, Tsugane M, Tyagi AK, Vanavichit A, Wang A, Wing RA, Yamaguchi K, Yamamoto M, Yamamoto N, Yu Y, Zhang H, Zhao Q, Higo K, Burr B, Gojobori T, Sasaki T (2007) Curated genome annotation of Oryza sativa ssp. japonica and comparative genome analysis with Arabidopsis thaliana. Genome Res 17:175–183
Jaillais Y, Horthorn M, Belhadir Y, Dabi T, Nimchik ZL, Meyerowitz EM, Chory J (2011) Tyrosine phosphorylation controls brassinosteroid receptor activation by triggering membrane release of its kinase inhibitor. Genes Dev 25:232–237
Jiang J, Zhang C, Wang X (2013) Ligand perception, activation, and early signaling of plant steroid receptor brassinosteroid insensitive 1. J Integr Plant Biol 55:1198–1211
Kim TW, Guan S, Sun Y, Deng Z, Tang W, Shang JX, Burlingame AL, Wang ZY (2009) Brassinosteroid signal transduction from cell-surface receptor kinases to nuclear transcription factors. Nat Cell Biol 11:1254–1260
Kim TW, Guan S, Burlingame AL, Wang ZY (2011) The CDG1 kinase mediates brassinosteroid signal transduction from BRI1 receptor kinase to BSU1 phosphatase and GSK3-like kinase BIN2. Mol Cell 43:561–571
Klein RR, Rodriguez-Herrera R, Schlueter JA, Klein PE, Yu ZH, Rooney WL (2001) Identification of genomic regions that affect grain-mould incidence and other traits of agronomic importance in sorghum. Theor Appl Genet 102:307–319
Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG (2007) ClustalW and ClustalX version 2. Bioinformatics 1:2947–2948
Li J (2010) Regulation of the nuclear activities of brassinosteroid signaling. Curr Opin Plant Biol 13:540–547
Li J, Chory J (1997) A putative leucine-rich repeat receptor kinase involved in brassinosteroid signal transduction. Cell 90:929–938
Li X, Tesso T, Yu J (2013) Genotype by environment interaction of sorghum flowering time. Kansas State University. https://pag.confex.com/pag/xxi/webprogram/Paper5338.html. Accessed 01 November 2013
Loiselle BA, Sork VL, Nason J, Graham C (1995) Spatial genetic structure of a tropical understory shrub, Psychotria officinalis (Rubiaceae). Am J Bot 82:1420–1425
Makarevitch I, Thompson A, Muehlbauer GJ, Springer NM (2012) Brd1 gene in maize encodes a brassinosteroid C-6 oxidase. PLoS ONE 7:e30798
Mandava N (1988) Plant growth-promoting brassinosteroids. Annu Rev Plant Physiol Plant Mol Biol 39:23–52
Mora-Garcia S, Vert G, Yin Y, Cano-Delgado A, Cheong H, Chory J (2004) Nuclear protein phosphatases with Kelch-repeat domains modulate the response to brassinosteroids in Arabidopsis. Genes Dev 18:448–460
Morillo S, Tax F (2006) Functional analysis of receptor-like kinases in monocots and dicots. Curr Opin Plant Biol 9:460–469
Morris GP, Ramu P, Deshpande SP, Hash CT, Shah T, Upadhyaya HD, Riera-Lizarazu O, Brown PJ, Acharya CB, Mitchell SE, Harriman J, Glaubitz JC, Buckler ES, Kresovich S (2013) Population genomic and genome-wide association studies of agroclimatic traits in sorghum. Proc Natl Acad Sci USA 2:453–548
Murphy RL, Klein RR, Morishige DT, Brady JA, Rooney WL, Miller FR, Dugas DV, Klein PE, Mullet JE (2011) Coincident light and clock regulation of pseudoresponse regulator protein 37 (PRR37) controls photoperiodic flowering in sorghum. Proc Natl Acad Sci USA 39:16469–16474
Myles S, Peiffer J, Brown P, Ersoz E, Zhang Z, Costich D, Buckler E (2009) Association mapping: critical considerations shift from genotyping to experimental design. Plant Cell 21:2194–2202
Nam KH, Li J (2004) The Arabidopsis transthyretin-like protein is a potential substrate of BRASSINOSTEROID-INSENSITIVE 1. Plant Cell 16:2406–2417
Nordborg M, Tavaré S (2002) Linkage disequilibrium: what history has to tell us. Trends Genet 18:83–90
Oeth P, Mistro G, Marnellos G, Shi T (2009) Qualitative and quantitative genotyping using single base primer extension coupled with matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MassARRAY). In: Komar AA (ed) Single nucleotide polymorphisms methods and protocols, 2nd edn. Humana Press, Cleveland, pp 307–343
Paterson AH, Bowers JE, Bruggmann R et al (2009) The Sorghum bicolor genome and the diversification of grasses. Nature 457:551–556
Pereira MG, Lee M (1995) Identification of genomic regions affecting plant height in sorghum and maize. Theor Appl Genet 90:380–388
Pritchard JK, Stephens M, Rosenberg NA, Donnelly P (2000) Association mapping in structured populations. Am J Hum Genet 67:170–181
Punta M, Coggill PC, Eberhardt RY, Mistry J, Tate J, Boursnell C, Pang N, Forslund K, Ceric G, Clements J, Heger A, Holm L, Sonnhammer EL, Eddy SR, Bateman A (2012) The Pfam protein families database. Nucleic Acids Res 40:D290–D301
Rami JF, Dufour P, Trouche G, Fliedel G, Mestres C, Davrieux F, Blanchard P, Hamon P (1998) Quantitative trait loci for grain quality, productivity, morphological and agronomical traits in sorghum (Sorghum bicolor L. Moench). Theor Appl Genet 97:605–616
RIKEN Plant Hormone Research Network—Plant Science Center (2010) Brassinosteroid metabolomic pathway in Arabidopsis. In: Fujioka S, Shimada Y (eds) http://hormones.psc.riken.jp/pathway_br.shtml. Accessed 02 February 2014
Rooney WL, Blumenthal J, Bean B, Mullet JE (2007) Designing sorghum as a dedicated bioenergy feedstock. Biofuel Bioprod Biorefin 1:147–157
Ryu H, Kim K, Cho H, Hwang I (2010) Predominant actions of cytosolic BSU1 and nuclear BIN2 regulate subcellular localization of BES1 in brassinosteroid signaling. Mol Cells 29:291–296
R Core Team (2013) R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. ISBN 3-900051-07-0. http://www.r-project.org/ Accessed 05 Nov 2013
Sakamoto T, Morinaka Y, Ohnishi T, Sunohara H, Fujioka S, Ueguchi-Tanaka M, Mizutani M, Sakata K, Takatsuto S, Yoshida S, Tanaka H, Kitano H, Matsuoka M (2006) Erect leaves caused by brassinosteroid deficiency increase biomass production and grain yield in rice. Nat Biotechnol 24:105–109
Sakamoto T, Morinaka Y, Fujioka S (2013) Genetic background influences brassinosteroid-related mutant phenotypes in rice. Am J Plant Sci 4:212–221
Salas Fernandez MG, Becraft PW, Yin Y, Lübberstedt T (2009) From dwarves to giants? Plant height manipulation for biomass yield. Trends Plant Sci 14:454–461
SAS Institute (2009) The SAS system for windows. Release 9.2. SAS Inst, Cary, NC
She J, Han Z, Kim TW, Wang J, Cheng W, Chang J, Shi S, Yang M, Wang ZY, Chai J (2011) Structural insight into brassinosteroid perception by BRI1. Nature 474:472–476
Shehzad T, Hiroyoshi I, Kazutoshi O (2009) Genome-wide association mapping of quantitative traits in sorghum [Sorghum bicolor (L.) Moench] by using multiple models. Breed Sci 59:217–227
Shimada Y, Fujioka S, Miyauchi N, Kushiro M, Takatsuto S, Nomura T, Yokota T, Kamiya Y, Bishop GJ, Yoshida S (2001) Brassinosteroid-6-oxidases from Arabidopsis and tomato catalyze multiple C-6 oxidations in brassinosteroid biosynthesis. Plant Physiol 126:770–779
Shimada Y, Goda H, Nakamura A, Takatsuto S, Fujioka S, Yoshida S (2003) Organ-specific expression of brassinosteorid-biosynthetic genes and distribution of endogenous brassinosteroids in Arabidopsis. Plant Physiol 131(287):297
Solovyev V, Kosarev P, Seledsov I, Vorobyev D (2006) Automatic annotation of eukaryotic genes, pseudogenes and promoters. Genome Biol 7(Suppl 1):10.1–10.12
Sreeramulu S, Mostizky Y, Sunitha S, Shani E, Nahum H, Salomon D, Hayun LB, Gruetter C, Rauh D, Ori N, Sessa G (2013) BSKs are partially redundant positive regulators of brassinosteroid signaling in Arabidopsis. The Plant J 74:905–919
Storey JD, Tibshirani R (2003) Statistical significance for genome-wide experiments. Proc Natl Acad Sci USA 100:9440–9445
Sukumaran S, Xiang W, Bean SR, Pedersen JF, Kresovich S, Tuinstra MR, Tesso TT, Hamblin MT, Yu J (2012) Association mapping for grain quality in a diverse sorghum collection. The Plant Genome 5:126–135
Sun G, Zhu C, Kramer MH, Yang SS, Song W, Piepho HP, Yu J (2010) Variation explained in mixed-model association mapping. Heredity 105:333–340
Taiz L, Zeiger E (2010) Plant Physiol, 5th edn. Sinauer Associates, Sunderland
Tang W, Kim TW, Oses-Prieto JA, Sun Y, Deng Z, Zhu S, Wang R, Burlingame AL, Wang ZY (2008) BSKs mediate signal transduction from the receptor kinase BRI1 in Arabidopsis. Science 321:557–560
Tang W, Yuan M, Wang R, Yang Y, Wang C, Oses-Prieto JA, Kim TW, Zhou HW, Deng Z, Gampala SS, Gendron JM, Jonassen EM, Lillo C, DeLong A, Burlingame AL, Sun Y, Wang ZY (2011) PP2A activates brassinosteroid-responsive gene expression and plant growth by dephosphorylating BZR1. Nat Cell Biol 13:124–131
Thurber CS, Ma JM, Higgins RH, Brown PJ (2013) Retrospective genomic analysis of sorghum adaptation to temperate-zone grain production. Gen Bio 14:R68
Tong H, Chu C (2012) Brassinosteroid signaling and application in rice. J Genet Genomics 39:3–9
Turk EM, Fujioka S, Seto H, Shimada Y, Takatsuto S, Yoshida S, Wang HC, Torres QI, Ward JM, Murthy G, Zhang JY, Walker JC, Neff MM (2005) BAS1 and SOB7 act redundantly to modulate Arabidopsis photomorphogenesis via unique brassinosteroid inactivation mechanisms. Plant J 42:23–34
Wang X, Chory J (2006) Brassinosteroids regulate dissociation of BKI1, a negative regulator of BRI1 signaling, from the plasma membrane. Science 313:1118–1122
Wang ZY, Nakano T, Gendron J, He J, Chen M, Vafeados D, Yang Y, Fujioka S, Yoshida S, Asami T, Chory J (2002) Nuclear-localized BZR1 mediates brassinosteroid-induced growth and feedback suppression of brassinosteroid biosynthesis. Dev Cell 2:505–513
Wang X, Kota U, He K, Blackburn K, Li J, Goshe MB, Huber SC, Clouse SD (2008) Sequential transphosphorylation of the BRI1/BAK1 receptor kinase complex impacts early events in brassinosteroid signaling. Dev Cell 15:220–235
Wang H, Yang C, Zhang C, Wang N, Lu D, Wang J, Zhang S, Wang ZX, Ma H, Wang X (2011) Dual role of BKI1 and 14-3-3 s in brassinosteroid signaling to link receptor with transcription factors. Dev Cell 21:825–834
Wu CY, Trieu A, Radhakrishnan P, Kwok SF, Harris S, Zhang K, Wang J, Wan J, Zhai H, Takatsuto S, Matsumoto S, Fujioka S, Feldmann KA, Pennell RI (2008) Brassinosteroids regulate grain filling in rice. Plant Cell 20:2130–2145
Wu G, Wang X, Li X, Kamiya Y, Otegui, Chory J (2011) Methylation of a phosphatase specifies dephosphorylation and degradation of activated brassinosteroid receptors. Sci Signal 4 172:ra29–ra29
Yamamuro C, Ihara Y, Wu X, Noguchi T, Fujioka S, Takatsuto S, Ashikari M, Kitano H, Matsuoka M (2000) Loss of function of a rice brassinosteroid insensitive1 homolog prevents internode elongation and bending of the lamina joint. Plant Cell 12:1591–1606
Ye H, Li L, Yin Y (2011) Recent advances in the regulation of brassinosteroid signaling and biosynthesis pathways. J Integr Plant Biol 53:455–468
Yin Y, Wang ZY, Mora-Garcia S, Li J, Yoshida S, Asami T, Chory J (2002) BES1 accumulates in the nucleus in response to brassinosteroids to regulate gene expression and promote stem elongation. Cell 109:181–191
Yin Y, Vafeados D, Tao Y, Yoshida S, Asami T, Chory J (2005) A new class of transcription factors mediates brassinosteroid-regulated gene expression in Arabidopsis. Cell 120:249–259
Yu J, Pressoir G, Briggs WH, Vroh-Bi I, Yamasaki M, Doebley JF, McMullen MD, Gaut BS, Nielsen DM, Holland JB, Kresovich S, Buckler ES (2006) A unified mixed-model method for association mapping that accounts for multiple levels of relatedness. Nat Genet 38:203–208
Zhang C, Xu Y, Guo S, Zhu J, Huan Q, Liu H, Wang L, Luo G, Wang X, Chong K (2012) Dynamics of brassinosteroid response modulated by negative regulator LIC in rice. PLoS Genet 8(4):e1002686
Zhao B, Li J (2012) Regulation of brassinosteroid biosynthesis and inactivation. J Integr Plant Biol 54:746–759
Zhiponova MK, Vanhoutte I, Boudolf V, Betti C, Dhondt S, Coppens F, Mylle E, Maes S, Gonzalez-Garcia MP, Cano-Delgado AI, Inze D, Beemster GT, De Veylder L, Russinova E (2013) Brassinosteroid production and signaling differentially control cell division and expansion in the leaf. New Phytol 197:490–502
Zhou A, Wang H, Walker JC, Li J (2004) BRL1, a leucine-rich repeat receptor-like protein kinase, is functionally redundant with BRI1 in regulating Arabidopsis brassinosteroid signaling. Plant J 40:399–409
Zhu C, Gore M, Buckler ES, Yu J (2008) Status and prospects of association mapping in plants. Plant Genome 1:5–20
Zhu JY, Sae-Seaw J, Wang ZY (2013) Brassinosteroid signaling. Development 140:1615–1620
Zou G, Zhai G, Feng Q, Yan S, Wang A, Zhao Q, Shao J, Zhipeng Z, Zou J, Han B, Tao Y (2012) Identification of QTLs for eight agronomically important traits using an ultra-high-density map based on SNPs generated from high-throughput sequencing in sorghum under contrasting photoperiods. J Exp Bot 63:5451–5462
Acknowledgments
The authors are grateful to Xiaochen Sun, for the development of R codes for the estimation of BLUPs; Dr. Jianming Yu, for his contribution to modify the SAS script to include multiple markers/genes in the mixed model and to RIKEN Plant Hormone Research Network—Plant Science Center, for the BR biosynthesis figure. This work was supported by the United States Department of Agriculture, National Institute of Food and Agriculture (Project #IOW05298), by the R.F. Baker Endowment, Iowa State University and by the National Secretary of Education, Science and Technology (SENESCYT), from the Republic of Ecuador.
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical standard
This experiment complies with the current laws of the US where the experiment was performed.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by Hai-Chun Jing.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Mantilla Perez, M.B., Zhao, J., Yin, Y. et al. Association mapping of brassinosteroid candidate genes and plant architecture in a diverse panel of Sorghum bicolor . Theor Appl Genet 127, 2645–2662 (2014). https://doi.org/10.1007/s00122-014-2405-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00122-014-2405-9