Abstract
Key message
Seven sharp eyespot resistance QTL were detected consistently across five environments and delimited to seven DNA marker intervals, respectively, six of which were independent of plant height and heading time.
Abstract
Sharp eyespot, caused mainly by the soil-borne fungus Rhizoctonia cerealis, is one of the important diseases of bread wheat (Triticum aestivum L.). This disease has escalated into a major threat to wheat production in some regions of the world. Wheat resistance to sharp eyespot can be a potential means to reduce the needs for application of fungicides and agricultural inputs. In the present study, the winter wheat lines, Luke and AQ24788-83, both of which possess quantitative resistance to sharp eyespot, were crossed and a population consisting 241 recombinant-inbred lines (RILs) was constructed. These RILs were assessed for sharp eyespot resistance by conducting five field and greenhouse trials during the period from 2008 to 2012, and they were genotyped with 549 simple-sequence repeat DNA markers. Seven quantitative trait loci (QTL) were detected consistently across the five trial environments to be associated with the sharp eyespot resistance. They were mapped on chromosomes 1A, 2B, 3B, 4A, 5D, 6B, and 7B. Four of these QTL are unequivocally novel, while it is possible that the other three might also be novel. Plant height and heading date of the 241 RILs were recorded in the four field trials. All of the seven disease resistance QTL were independent of plant height and heading time except one that was significantly associated with plant heading time. This association might be attributed genetically to a single QTL, or to different but closely linked QTL. In the case of single QTL, pleiotropism might be involved or the sharp eyespot resistance might be conferred in a physical instead of physiological nature.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Bread wheat (Triticum aestivum L.) is a staple food crop for the human population (http://faostat.fao.org). Sharp eyespot disease has escalated into a major threat to wheat production in some regions of the world (Lemańczyk and Kwaśna 2013; McBeath and McBeath 2010). The main agent of wheat sharp eyespot is Rhizoctonia cerealis van der Hoeven (teleomorph Ceratobasidium cereale D Murray and LL Burpee, anastomosis group 1, CAG 1), a soil-borne fungus with bi-nucleate hyphal cells (Burpee et al. 1980; Lipps and Herr 1982). R. solani Kuhn, another soil-borne fungus with multi-nucleate hyphal cells and several anastomosis groups (AGs), is the other agent that can be isolated from wheat leaf sheaths and stems showing sharp eyespot lesions (Boerema and Verhoeven 1977; Lemańczyk 2010). Pathogen surveys, conducted in China over the last years (e.g., Chen et al. 2009; Guo et al. 2012; Wang et al. 2011), showed that the isolates from wheat leaf sheaths and stems showing sharp eyespot lesions were predominantly R. cerealis CAG 1 (>90 %). R. solani isolates, involving AG-1-IB, AG-2, AG-4 and AG-5, were occasionally observed (<10 %). In the latter case, R. solani was found to exist concurrently with or separately from R. cerealis in sharp eyespot lesions. These results were consistent across several major wheat-growing provinces. The isolates of R. cerealis CAG 1 were much more virulent than those of R. solani (e.g., Chen et al. 2009).
Wheat sharp eyespot is particularly associated with temperate wheat-growing regions such as in China (McBeath and McBeath 2010; Wang et al. 1994), Egypt (Hammouda 2003), England and Wales (Clarkson and Cook1983; Polley and Thomas 1991), New Zealand (Cromey et al. 2006), Poland (Lemańczyk 2010; Lemańczyk and Kwaśna 2013), and the USA. (Lipps and Herr 1982; Mazzola et al. 1996). Severe sharp eyespot can considerably decrease wheat grain yield (Clarkson and Cook 1983; Lemańczyk and Kwaśna 2013). In terms of wheat acreage affected by sharp eyespot, China is the largest epidemic region in the world, as exemplified by the 8.1 million hectares of winter wheat affected in 2005 (McBeath and McBeath 2010). Although winter wheat can be attacked by the fungi as early as when seeds begin to germinate in autumn, the attack occurs more frequently in the coming spring and early summer as manifested by lesions on plant basal leaf sheaths and stems. Typical lesions are of elliptical or ‘eye’ shape with sharply defined dark brown borders and a pale yellow center area, and may coalesce into large patches girdling the stem up to a height of about 30 cm. Sclerotia can be observed between the leaf sheath and stem or within the stem lumen. Severe infection of shoots may cause premature ripening (whiteheads) or plant lodging, or shoots may be killed before the ear can emerge from the sheath. In commercial winter wheat fields, sharp eyespot lesions can be readily observed beginning from the early tillering growth stages of wheat plants when the temperature is about 10 °C in early spring. The disease then rapidly increases from mid or late stem elongation to ear emergence when the temperature becomes higher than 15 °C, and lesions steadily extend from ear emergence onwards (Wang et al. 1994).
Fungicides, intensive or deep tillage, and crop rotation are currently used to manage the disease (Wang et al. 1994; Hamada et al. 2011). Wheat resistance to sharp eyespot can be a potential means to reduce the need for application of fungicides and the need for fuel of farm machines for intensive tillage. Reports of resistance mainly came from China (e.g., Cai et al. 2006; Huo 2002; Ren et al. 2010; Zhang et al. 2005). Extensive screening of bread wheat germplasm resources has identified no confirmed immunity or complete resistance to sharp eyespot. However, certain wheat cultivars or breeding lines or Chinese landraces such as ARz, Baimian 3, Chuan 35050, Shanhong Mai, and Shannong 0431 were proven to possess quantitative resistance.
Five RIL populations involving those resistance resources have been constructed and used to study the inheritance of resistance (Cai et al. 2006; Huo 2002; Ren et al. 2004, 2007; Tang et al. 2004; Zhang et al. 2005). A QTL for the resistance was detected consistently across different trial environments and mapped to chromosome 7D (Cai et al. 2006). Several suggestive QTL for the resistances were mapped to chromosomes 1A, 2B, 2D, 3A, 3B, 3D, 5A, 6A, and 6B. The numbers of DNA markers for mapping used in those populations were small (<160 for individual populations), giving a limited power for QTL detection. The QTL number thus might be underestimated in the five populations.
The wheat cross population of Opata85 × W-7984, which possesses a large number of DNA markers (>1,700), was used to detect QTL for sharp eyespot resistance by Huo (2002), although both parents of this cross were below the level of moderate sharp eyespot resistance. A minor QTL for resistance at the adult plant growth stage was suggested on chromosome 7B with the resistance allele originating from Opata 85 (Huo 2002). In comparison with some other plant diseases such as wheat stripe rust (Mallard et al. 2005; Paillard et al. 2012) and rice Rhizoctonia sheath blight (Sharma et al. 2009; Srinivasachary et al. 2011), wheat sharp eyespot has been much less genetically studied. In responding to the escalation of wheat sharp eyespot, it becomes increasingly important to detect novel resistance genes/QTL and to precisely map them to facilitate transfer of the resistances to wheat varieties.
We identified, by considerable screening of bread wheat germplasm resources over the past years (Zhang and Chen, unpublished), several winter wheat lines including Luke and AQ24788-83 (hereafter referred to as AQ) that possess quantitative resistance to sharp eyespot, and constructed a RIL population of Luke × AQ. The objectives of this study were to map QTL associated with the resistance to sharp eyespot in the Luke × AQ population at the adult plant growth stage, and to examine the relationships of these QTL with plant height and heading time.
Materials and methods
Plant materials and population development
Two hundred and forty-one RILs of the winter wheat cross Luke × AQ were used as the mapping population in this study. The crossing was done in May 2002, and F2 plants derived from an individual F1 plant were advanced to the F10 generation during the period from 2004 to 2012, via single-seed descent. Luke (pedigree: PI 178383/2* Burt//CItr 13438) has the accession number CItr 14586 at the National Small Grain Collection, Aberdeen, ID, 83210, USA. AQ was selected in our program for accumulating quantitative resistance to multiple diseases including sharp eyespot from the progeny of a double cross involving four Chinese winter wheat land races: Ma Zhamai/Bai Qimai//Hong Qimai/Qing Shoumai. AQ was in its F14 generation when it was crossed with Luke in 2002. The wheat line Yumai 49 (hereafter referred to as Y49) was used as susceptible control, and has been reported to be highly susceptible to sharp eyespot (e.g., Ren et al. 2010; Wang et al. 2011).
Pathogen material
R0301, an isolate of R. cerealis CAG 1, was used as the pathogen material, being kindly provided by professor Huaigu Chen at the Institute of Plant Protection, Jiangsu Academy of Agricultural Sciences, Nanjing 210014, P. R. China. R0301 has been reported to be highly virulent on wheat (Ren et al. 2010). An aliquot of R0301 stock, stored in mineral oil, was activated on freshly prepared potato-dextrose agar (PDA) just before use. Inoculum was produced on sterilized wheat kernels using a method similar to Lipps and Herr (1982). Briefly, wheat kernels soaked in distilled water for 24 h were added to Erlenmeyer flasks and autoclaved (121 °C for 60 min) twice on two consecutive days, with each 500-ml flask containing 300 ml of the wheat kernels. Agar discs (5–7 mm in diameter) cut from the margins of actively growing R0301 colonies on PDA were transferred into the flasks, which were then incubated at 25 °C for 3 weeks and shaken once every 2 days. The colonized wheat kernels were prepared just before use for inoculation in the field and greenhouse trials.
Resistance assessment in field trials with artificial inoculum
The trials were conducted during the 2008–09 and 2009–10 winter wheat seasons at the China Agricultural University Shang Zhuang Farm (39°54′20′′N, 116°25′29′′E; approximately 120 km northwest of the center of Beijing city; silt loam soil). The weather factor of low humidity in spring in this area is unfavorable to stem-base diseases, and such diseases have been rarely observed to occur naturally. The 241 F7 and F8 RILs of Luke × AQ were sown during the early Octobers of 2008 and 2009, in a randomized complete block design with three replicates in the 2008–09 season and a single replicate in the 2009–10 season. Three plots each of Luke, AQ, and Y49 were included in each replicate. An individual plot consisted of a single 80-cm row sown with 20 seeds. Rows were spaced 30 cm apart. The trial field was equipped with water sprinklers. In mid-Aprils 2009 and 2010, when wheat was at the tillering growth stage, the plants were inoculated by placing R0301-colonized wheat kernels (80 ml in each row) on the soil surface in contact with the plant bases. Such an inoculation dosage was determined on the basis of our previous pilot trials. To enhance humidity and thus to facilitate R. cerealis infection and development, the plants were sprinkled with water twice a day for the first 20 days and then with varied frequency depending on rainfall and soil moisture until final disease recording. Eight shoots randomly sampled from each plot were labeled with grease pen (oil-based ink marker), and were then individually assessed for disease intensity three times at 10-day interval beginning at the heading stage (about 4 or 5 weeks after inoculation) on a 0–9 scale as follows.
0: No symptom of sharp eyespot.
1: One or more sharp eyespot lesions on lower sheaths but no symptoms on the stem.
2: One or more sharp eyespot lesions on upper (as well as lower) sheaths but no symptoms on the stem.
3, 4, 5, 6, or 7: One or more sharp eyespot lesions girdling in total less than or equal to 1/5, 2/5, 3/5, 4/5, or 5/5 of the stem circumference, respectively, with the stem remaining un-softened.
8: Stem softened with sharp eyespot lesions on the stem.
9: Head prematurely ripened (whitehead) or plant collapsed with sharp eyespot lesions on the stem, or shoot killed as the ear emerges from the sheath with sharp eyespot lesions on the shoot.
For each of the three recording times, the mean score for an individual plot entry was obtained by averaging over the eight shoots in the plot, and the area under the disease progress curve (AUDPC) was calculated for each plot entry using the three mean scores with the formula: \( {\text{AUDPC}} = \sum {\left[ {\left( {x_{i} + x_{i + 1} } \right)/2} \right]} \times \left( {t_{i + 1} - t_{i} } \right) \), where x i and x (i+1) are scores for a plot entry on date t i and date t (i+1), respectively, and t (i+1)−t i = the number of days between date t (i+1) and date t i .
Resistance assessment in field trial with natural inoculum
The trials, conducted during the 2010–11 and 2011–2012 wheat seasons, were located in a commercial wheat field in Sheng Zhuang (36°18′09′′N, 117°13′05′′E; silt loam soil), a village approximately 10 km east of the town of Taishan in Shandong province of China. This site has been observed to be a ‘hotspot’ of wheat sharp eyespot where the environmental conditions, especially humidity and temperature, are quite conducive to the disease. Particularly, evidence was observed of sharp eyespot lesions on wheat plant residues prior to seeding in Septembers 2010 and 2011, suggesting that inoculum was present. To keep the residues on the soil surface and thus to enhance inoculation, no tillage was carried out, and the fields were directly seeded during the early Octobers of 2010 and 2011. The 241 F9 and F10 RILs of Luke × AQ were arranged in a randomized complete block design in four replicates, and three plots each of Luke, AQ, and Y49 were included in each replicate. An individual plot consisted of three 1-m rows (25 cm apart) sown with 50 seeds in each row. The wheat plants were infected naturally with the inoculum harbored in the soil and the infected wheat plant residues. To facilitate R. cerealis infection and development, the plants were sprinkled with water with varied frequency depending on rainfall during the springs and early summers of 2011 and 2012. The disease score and AUDPC calculations were calculated as detailed above.
Resistance assessment in a greenhouse trial with artificial inoculum
A greenhouse trial was conducted during the 2011–12 season, sown in mid-October 2011. The 241 F10 RILs of Luke × AQ were arranged in a randomized complete block design with three replicates, and three plots each of Luke, AQ, and Y49 were included in each replicate. An individual plot consisted of a single 23-cm-diameter pot filled with steam-sterilized (82–85 °C for 30 min) silt loam soil collected from the top 10 cm of the wheat fields at the China Agricultural University Shang Zhuang Farm, sown with eight seeds in each pot. During winter, windows of the greenhouse were kept open and heaters were closed to give low-temperature conditions for vernalizing the wheat seedlings. A rack (12 m long, 4 m wide, and 1.5 m high) was constructed with steel tubes over the plants and water sprinklers were fixed under the rack. In early March 2012, when wheat plants were at the tillering growth stage, thinning was done and four plants were retained in each pot. The plants were then inoculated by placing eight R0301-colonized wheat kernels on the soil surface in contact with the plant bases. To facilitate R. cerealis infection and development, the plants were sprinkled with water and covered by placing plastic film around the steel rack to maintain a high relative humidity. Temperature of the greenhouse was controlled between 15 and 24 °C during a 1-week incubation. After the incubation, the films were removed and then the plants were conditioned with a temperature between 15 and 30 °C with adequately frequent sprinkling until final disease assessment. The disease score and AUDPC calculation were calculated as detailed above.
Recording of plant height and heading date, and identification of fungal isolates
To determine if the resistance to sharp eyespot was related to plant height and heading date, both traits were recorded for the labeled shoots in the four field trials during the period from 2008 to 2012. Heading date was recorded as the number of days between sowing date and the date at which the first spikelet of an ear emerged from the flag leaf sheath. Plant height was measured in centimeters from the soil surface to the tip of the ear excluding the awns at the milk-ripening stage.
To determine if the recorded diseases were caused by R. cerealis or by R. solani, fungus samples were examined for hyphal nucleus number. The samples were isolated from the wheat stems showing sharp eyespot lesions collected along a diagonal transect across the trial areas on the date when the final disease recording was done. Fifteen stems were collected from each of the two naturally infected trials and five stems were collected from each of the three artificially inoculated trials. Rhizoctonia hyphae were isolated from the stems, and the hyphal cell nuclei were stained using the conventional procedures similar to the previous report (Lipps and Herr 1982). Briefly, stem segments (5–8 mm long) were surface sterilized in 5 % sodium hypochlorite for 5 min, rinsed in sterile distilled water, placed on 2 % water agar, and incubated at 23–25 °C. Hyphal tips were then transferred to PDA. Bi- and multi-nucleate Rhizoctonia hyphae were differentiated by HCl-Giemsa staining.
Genetic linkage map construction and QTL analyses
The 241 F9 RILs of Luke × AQ were used for constructing the genetic linkage map, and F10 RILs were used for confirming samples and replacing missing data. DNA was extracted from fresh leaves of Luke, AQ, and each RIL with the method using cetyl trimethyl ammonium bromide (CTAB) (Rogers and Bendich 1985). The DNAs of Luke and AQ were used to screen 1673 SSR and EST-SSR primer pairs for polymorphism. Sequences of these primers were acquired from the public domain including BARC (Song et al. 2002; http://www.scabusa.org), CFA, CFB, CFD (Guyomarc’h et al. 2002; Sourdille et al. 2001, 2003), EST-SSR (http://wheat.pw.usda.gov), GDM (Pestsova et al. 2000), GPW (Génoplante, http://wheat.pw.usda.gov), GWM (Röder et al. 1998), and WMC (Wheat Microsatellite Consortium, P. Isaac, IDnagenetics, Norwich, UK; Somers et al. 2004). The products of PCR, amplified in GeneAmp PCR System 9700 and Biometra cyclers, were separated in 6 % denaturing polyacrylamide gels and visualized using the silver staining method (Bassam et al. 1991). The primer pairs that yielded clearly distinct and readily repeatable polymorphic bands were selected to genotype the RILs. Nearly all loci in the F9 RILs were homozygous. Some RIL × locus combinations that remained heterozygous were considered as missing values.
Linkage analysis was conducted using the software MAPMAKER/EXP 3.0 (Lander et al. 1987) set to Kosambi mapping function in centiMorgan (cM) with the other working parameters/procedures similar to previous reports (e.g., Mallard et al. 2005). An individual linkage group was claimed if any distance between two adjacent markers was less than 50 cM. A framework was first constructed with a logarithm of the odds (LOD) threshold of 5.0, and the remaining markers were added to the framework using the ‘try’ command with the arbitrary LOD threshold of 3.0. Linkage groups were assigned to chromosomes by referring to the ITMI (International Triticae Mapping Initiative) SSR maps (Röder et al. 1998; Somers et al. 2004), and marker order within a linkage group was established on the basis of linkage within the Luke × AQ population. QTL analysis was conducted on the AUDPC as described above using composite interval mapping (CIM) in the Windows QTL Cartographer 2.0 program (Wang et al. 2010). The threshold LOD score to declare significant QTL was arbitrarily set at 2.5 with the other working parameters/procedures similar to those in the previous report (e.g., Mallard et al. 2005). Multiple-interval mapping (MIM) was conducted with the QTL from CIM analyses as the initial model to detect possible additional QTL. The percentage of phenotypic variance explained by the whole model (total R 2) was estimated using the ‘summary’ option of MIM, and the R 2 for individual QTL was estimated using CIM.
Statistical analysis
Data were analyzed using the SAS statistics package (SAS Institute Inc., v. 8.2., Cary, NC, USA). ANOVA was conducted on AUDPC data for each replicated trial using the PROC GLM procedure with genotype and replicate as variance factors. The normality test of residual distribution, Chi square test, and correlation calculation were performed using the PROC UNIVARIATE, PROC FREQ, and PROC CORR procedures, respectively.
Results
Phenotypic assessments of sharp eyespot infection
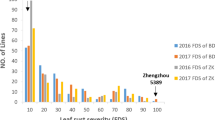
Typical and coalesced sharp eyespot lesions were readily observed beginning from the stem-elongation stage of the wheat plants in the trials (2 or 3 weeks after inoculation in the artificially inoculated trials). Severe diseases were induced in the trials of 2008–09, 2009–10, and 2011–12, as shown in Fig. 1 by the high AUDPC values (i.e., 158–171) of the susceptible control wheat line Y49. These values were considerably near to 180 that was the maximum possible AUDPC calculated by multiplying 9 (the maximum disease score) with 20 (the time in days between the first and the last recordings). The disease was less severe in the naturally infected trial of 2010–11 (Fig. 1). All of the 15 fungus samples isolated from the artificially inoculated plants and 28 of the 30 isolates from the naturally infected trials were identified as bi-nucleate Rhizoctonia, implying that the recorded diseases were mainly caused by R. cerealis. The other two isolates from the naturally infected plants were multi-nucleate Rhizoctonia. The AUDPC values (139–171) of Y49 were significantly higher than those (e.g., ≤60) of some RILs (Fig. 1), suggesting that the disease pressure was high enough to reveal the difference in resistance/susceptibility between genotypes. An ANOVA (Table 1) showed that the AUDPC variance of RIL genotypes was highly significant (F > 40, P < 0.0001; R 2 > 0.78), while the variance of replicates within each trial was comparatively low (F < 4.3, P > 0.015). The spatial uniformity of disease pressure within trials was better in the artificially inoculated trials (P > 0.069 and <0.094 for replicates) than in the naturally infected trial (P > 0.015 and <0.032 for replicates). The mean AUDPC values, calculated for each of the 241 RILs by averaging over the replicates within each trial, showed that no RIL was immune to the disease (i.e., all means >0). The RILs were distributed continuously over the range of AUDPC values from 1 to 180 in an approximately normal shape in each of the five trials (Fig. 1), suggesting a quantitative nature of the resistance in the Luke × AQ population. Correlations for AUDPC among the five trial environments were all positively significant at the P = 0.0001 test level (Table 2), suggesting that the expression of the disease resistance was consistent across the environments and the data were qualified well for further analyses.
The distributions of the 241 recombinant-inbred lines (RILs), derived from the winter wheat cross Luke × AQ24788-83, over the range of sharp eyespot disease intensity measured by the area under the disease progress curve (AUDPC). The RIL plants were artificially inoculated with Rhizoctonia cerealis CAG 1 isolate R0301 in the field trials in 2008–09 and 2009–10 (a) and in the greenhouse trial in 2011–12 (b), and naturally infected with the sharp eyespot inoculum harbored in the soil and wheat plant residues in the field trials in 2010–11 and 2011–12 (c). Approximate position of AUDPC mean (M) and standard deviation (SD) of the susceptible control wheat line Yumai49 were indicated with arrow and bar for each trial. The M and SD were calculated using 12, 9, and 3 plot data, respectively, for the trials of 2010–11 and 2011–12 in fields, 2008–09 in field and 2011–12 in greenhouse, and 2009–10 in field (see “Materials and methods”)
Linkage map construction and QTL mapping
From the SSR marker loci amplified with the 1,673 primer pairs, 613 were selected for genotyping the 241 RILs. Of the 613 markers, 596 were used to construct linkage groups with the exclusion of the other 17 that were proved distorted from 1:1 segregating at P = 0.01 of the Chi square test. Finally, 549 markers were mapped to 21 linkage groups that were then assigned to the 21 wheat chromosomes by referring to the previously published wheat SSR maps (Röder et al. 1998; Somers et al. 2004), with each chromosome having between 16 (chromosome 5D) and 47 (chromosome 3B) markers. On all 21 chromosomes, the distance between adjacent markers was less than 50 cM with the exception of 7D on which a gap of 59.4 cM existed. The other 47 markers were mapped on some small linkage groups or could not be linked to any group, and they were ignored in the subsequent analyses. The 21 assigned linkage groups spanned a total length of 4458 cM with average marker spacing of 8.1 cM and with six large distances of 39.8–45.9 cM as well as the 59.4-cM gap on 7D. The 21 chromosome maps were scanned via CIM for mapping QTL (for the sake of simplicity, maps were shown here only for the chromosomes involving the identified QTL). The AUDPC data of the 241 RILs as shown in Fig. 1 were used for the mapping that was done separately for the trials of 2010–11 and 2011–12. The AUDPC data of 2009–10 were, however, combined with those of 2008–09, i.e., mean AUDPC of each RIL was calculated by averaging across the four replicates: three in 2008–09 and one in 2009–10. The means were used for mapping with the consideration that the 2009–10 trial had only a single replicate and thus might be inappropriate for a separate mapping. Seven QTL were mapped as shown in Fig. 2 and Table 3. They were all significant at the LOD threshold of 2.5 for every artificially inoculated trial and for the naturally infected trial of 2011–12, though the significance was marginal for some QTL × trial environment combinations (Table 3). However, for the naturally infected trial of 2010–11, four of the seven QTL were below the threshold. Of the seven QTL, three were mapped to the short arms of chromosomes 1A, 2B, and 3B, and the other four to the long arms of 4A, 5D, 6B, and 7B. They were designated as QSe.cau-1AS, QSe.cau-2BS, QSe.cau-3BS, QSe.cau-4AL, QSe.cau-5DL, QSe.cau-6BL, and QSe.cau-7BL, respectively, explaining 5–29 % of AUDPC phenotypic variance individually and 38–68 % collectively. The resistance alleles were contributed by Luke at QSe.cau-1AS, QSe.cau-2BS, and QSe.cau-7BL, and by AQ at QSe.cau-3BS, QSe.cau-4AL, QSe.cau-5DL, and QSe.cau-6BL. Four additional QTL were detected with their peak LOD scores ranging from 2.57 to 5.66, explaining 7–15 % (i.e., R 2 %) of the AUDPC phenotypic variance. These four QTL were, however, consistent across only some of the five trial environments, and might be involved in some interactions with environments. They were omitted here from subsequent analyses, and are being subjected to further study.
Linkage groups assigned to chromosomes 1A (a), 2B (b), 3B (c), 4A (d), 5D (e), 6B (f), and 7B (g) showing the logarithm of the odds (LOD) curves for the respective quantitative trait loci (QTL) QSe.cau-1AS, QSe.cau-2BS, QSe.cau-3BS, QSe.cau-4AL, QSe.cau-5DL, QSe.cau-6BL, and QSe.cau-7BL conferring resistance to sharp eyespot in the wheat cross population of Luke × AQ24788-83. These QTL were detected consistently across the artificially inoculated field and greenhouse trials, and the naturally infected field trials. Distances are in Kosambi centiMorgans (cM). Each of the chromosomes is oriented with the telomere of the short arm to the left positioned at 0 cM. Upward arrows indicate the approximate positions of centromeres. Downward arrows indicate the approximate positions of previously reported QTL. The underlined SSR markers were common for and order consistent between the Luke × AQ map and Röder’s map or Somers’ map (Röder et al. 1998; Somers et al. 2004). The positions of the microsatellite (simple sequence repeat, SSR) markers are shown along the chromosomes. The horizontal lines indicate the threshold LOD of 2.5
Relationship of sharp eyespot resistance with plant height and heading time
The phenotypic correlation coefficients of sharp eyespot AUDPC with plant height were not significant at the P = 0.05 test level (r = 0.05–0.07, P = 0.43–0.26, degrees of freedom = 239). Moderate correlations were detected between AUDPC and heading time (r = −0.14 to −0.15 and P = 0.03–0.01 depending on trial; degrees of freedom = 239). To examine the relationships of individual QTL with plant height and heading time, ANOVA was carried out for the effect of each QTL on both traits. For simplicity, the two artificially inoculated trials (2008–09 and 2009–10) were combined by calculating mean AUDPC averaged over four replicates (i.e., three in 2008–09 and one in 2009–10) for each RIL, and in the same way, the two naturally infected trials (2010–11 and 2011–12) were combined over eight replicates (i.e., four in 2010–11 and four in 2011–12). As shown in Table 4, all of the QTL as represented by their nearest SSR markers were not significantly associated with plant height and heading time at the P = 0.05 test level, with the exception of Qse.cau-2BS that was associated with heading time (F > 12, P < 0.001). Spearman rank correlation analyses were carried out between heading time and Qse.cau-2BS with the resistance allele as rank ‘1’ and the susceptible allele as ‘0’, resulting in coefficients of 0.32–0.36 (P < 0.0001). These results revealed that heading dates were delayed in the RILs that had the resistance allele at Qse.cau-2BS. In the same interval of wmc154-barc200 as for QSe.cau-2BS, a QTL was detected which was associated with heading time with peak LOD scores of 3.7–7.8 (LOD score curves not shown).
Discussion
The wheat resistance to sharp eyespot was quantitative in the Luke × AQ population. It has been indicated that environments generally impose a significant influence on quantitative traits (Paterson et al. 1991). In agreement with this generality, the expression of the sharp eyespot resistance in this study could be affected and complicated by inoculum load, micro-climate, and the uniformity of disease pressure. These factors could increase the residual variation within a trial and reduce the power of QTL mapping. We, therefore, enhanced the trial environments for disease development and thus for QTL expression using a pathogen isolate of strong virulence, by accurately quantifying the inoculums, and by frequently sprinkling the inoculated plants with water. We believe that the application of these methods/techniques resulted in the heavy disease pressure (Fig. 1) and the adequate spatial uniformity of the disease as evidenced by the relatively limited variance (P > 0.069) among replicates within every artificially inoculated trial and the naturally infected trial of 2011–12 (Table 1). Consequently, the seven QTL were consistently detected across these trial environments (Fig. 2). However, in the naturally infected trial of 2010–11, four of the seven QTL did not express very well as shown by the sub-threshold LOD scores (Fig. 2b, e, f, g; Table 3). This situation could be, we supposed, attributable to the shortage of rainfall in Shandong province of China in the spring and early summer of 2011. Low humidity restricted the disease infection and development, and the disease pressure was not high enough to reveal the resistance/susceptibility well.
The present study had better coverage of the 21 chromosomes of bread wheat by the 549 SSR markers than the previously reported QTL studies of resistance to sharp eyespot (Cai et al. 2006; Huo 2002; Ren et al. 2004, 2007; Tang et al. 2004; Zhang et al. 2005). In those reports, no more than 159 markers were used in any of the five wheat cross populations. The enhanced genome coverage in the present study might increase the power of QTL detection. Consequently, as many as 11 QTL were found in the Luke × AQ population, of which seven were consistently detected across all of the five trial environments. Four of the seven QTL were delimited to an interval of less than 5 cM (Fig. 2a, c, d, e; Table 3).
Of the seven consistent QTL (Fig. 2; Table 3), the four of QSe.cau-1AS, QSe.cau-3BS, QSe.cau-4AL, and QSe.cau-5DL (on chromosomes 1A, 3B, 4A, and 5D, respectively) are unquestionably novel. No sharp eyespot resistance has yet been reported to be associated with chromosomes 4A and 5D. The QSe.cau-1AS and QSe.cau-3BS (on the short arms of 1A and 3B, respectively) can be readily distinguished, by referring to the linked SSR markers, from the three QTL that have been suggested, respectively, by Zhang et al. (2005) on the long arm of 1A in the Chuan35050 × Shannong483 population (hereafter referred to as C/S 1AL QTL), by Tang et al. (2004) on the long arm of 3B in the ARz × Yangmai 158 population (A/Y 3BL QTL), and by Ren et al. (2007) on the long arm of 3B in the Sumai 3 × Baimian 3 population (S/B 3BL QTL). On chromosome 1A, the SSR marker gwm135 was located between QSe.cau-1AS and the C/S 1AL QTL. The LOD score peak of QSe.cau-1AS was away from gwm135 by more than 27 cM (Fig. 2a), and the C/S 1AL QTL was distant from gwm135 by more than 130 cM (Zhang et al. 2005). Therefore, a distance of at least 157 cM existed between the two QTL. On chromosome 3B, the marker gwm77 was closely linked (<2 cM) to the LOD score peak of QSe.cau-3BS (Fig. 2c), and the marker gwm181 was quite near (<4 cM) to the A/Y 3BL QTL (Tang et al. 2004). A distance of more than 163 cM existed between the two markers (Fig. 2c). The S/B 3BL QTL was near (<8 cM) to the marker gwm533 (Ren et al. 2007). The QSe.cau-3BS resistance as represented by locus gwm77 was distant from gwm533 by at least 77 cM (Fig. 2c).
The three QTL of QSe.cau-2BS, QSe.cau-6BL, and QSe.cau-7BL (on the short arm of 2B, long arm of 6B, and long arm of 7B, respectively) could not be unequivocally differentiated from the four QTL reported, respectively, by Huo (2002) on the short arm of 2B in the Wenmai 6 × Shanhongmai population (W/S 2BS QTL), by Huo (2002) on the short arm of 6B in the W/S population (W/S 6BS QTL), by Ren et al. (2007) on the long arm of 6B in the S/B population (S/B 6BL QTL), and by Huo (2002) on the long arm of 7B in the Opata 85 × W-7984 population (O/W 7BL QTL). Nevertheless, the spatial relationships of QSe.cau-2BS, QSe.cau-6BL, and QSe.cau-7BL with the four QTL of W/S 2BS, W/S 6BS, S/B 6BL, and O/W 7BL can be examined by referring to the ITMI SSR map and the wheat SSR consensus map (Röder et al. 1998; Somers et al. 2004) that could be a bridge between the map of Luke × AC and the maps of W/S, S/B, and O/W. To begin with chromosome 2B, 15 SSR markers that were underlined as shown in Fig. 2b were common for and order consistent between the Luke × AQ map and Somers’ map, including wmc382, wmc154, barc200, centromere, and gwm526 listed in order from the distal part of the short arm to the centromere and further to the distal part of the long arm. The W/S 2BS QTL was proximal to wmc382 by less than 26 cM, whereas QSe.cau-2BS was between wmc154 and barc200 which was proximal to wmc382 by more than 47 cM (Fig. 2b). A distance of about 21 cM existed between the two QTL. For chromosome 6B, 10 markers that were underlined as shown in Fig. 2f were shared by the Luke × AQ map and Somers’ map, including wmc104, wmc132, wmc398, centromere, and wmc417 listed in order from the distal part of the short arm via the centromere to the distal part of the long arm. The W/S 6BS QTL was proximal to wmc104 (gwm132) by less than 30 cM, whereas the LOD score peak of QSe.cau-6BL was proximal to wmc104 (gwm132) by more than 48 cM. There was a space of about 18 cM between the two QTL. The S/B 6BL QTL was proximal to wmc398 by less than 11 cM, while the LOD score peak of QSe.cau-6BL was proximal to wmc398 by more than 19 cM (Fig. 2f). A distance of about 8 cM was observed between the two QTL. For chromosome 7B, six markers that were underlined as shown in Fig. 2g were common for the Luke × AQ map and Röder’s map, including gwm537, centromere, and gwm611 oriented from the short arm to the distal part of the long arm. The O/W 7BL QTL was proximal to DNA marker fbb189 that was proximal to gwm611 (Huo 2002; Röder et al. 1998), while the LOD score peak of QSe.cau-7BL was distal to gwm611 by about 16 cM (Fig. 2g). These comparisons suggest the possibility that QSe.cau-2BS, QSe.cau-6BL, and QSe.cau-7BL might be different from the four previously reported QTL, while we realized that these data provide no solid proof for difference since distances in different populations even with common markers can vary considerably.
Rice resistance to Rhizoctonia sheath blight has been reported to occur concurrently with tall plant height and/or late-heading time in several cases, and the QTL of those traits were localized in the same chromosome regions (Li et al. 1995; Sharma et al. 2009; Srinivasachary et al. 2011). It has been a concern that those resistance QTL, when being used practically in breeding programs, may impose some extra complications on selection since neither tallness nor late maturity is desired in most breeding programs (Sharma et al. 2009). No study has been previously reported on the relationship of wheat sharp eyespot resistance with plant morphological and developmental characters. In the present study, six of the seven QTL showed no correlation with plant height and heading time (Table 4). The independence of these resistance QTL from the undesired tallness and late maturity should be conducive to breeding.
The resistance allele at QSe.cau-2BS, however, was correlated with the late-heading character, which may constitute an inconvenience with respect to practical use in breeding. The mechanism underlying this correlation is unknown. QSe.cau-2BS and a QTL for heading time were co-localized in the same chromosome interval that was flanked by SSR loci wmc154 and barc200 (Fig. 2b). These two QTL might be at the same locus that had pleiotropic effects on sharp eyespot disease development and plant heading time. Alternatively, an allele at the single locus might primarily lengthen heading time. The late-heading character in turn influenced sharp eyespot infection. R. cerealis hyphae invade a wheat shoot at the base through the space between its sheath and stem. In spring, temperatures above 15 °C could rapidly speed up the invasion beginning from wheat stem elongation when the tightness of the closing of stems by sheaths began to vary among RILs. In comparison with early-heading RILs, late-heading ones may have rendered their stems more tightly closed by sheaths, leading to a physical situation less conducive to invasion and consequent appearance of lower disease. Another explanation may be that QSe.cau-2BS actually physiologically conferred sharp eyespot resistance but it was tightly linked to the QTL in the same wmc154-barc200 interval which could delay heading date. The current data were inadequate for spatially differentiating between the two possible QTL, and further study is needed using a larger RIL population and increased DNA markers.
All of the rice QTL (except one) for resistance to Rhizoctonia sheath blight could individually explain less than 30 % of the disease phenotypic variance (i.e., R 2 % < 30) (Srinivasachary et al. 2011). Similarly, the effects of the wheat sharp eyespot resistance QTL also were not high in the Luke × AQ population, although they were statistically significant at the arbitrary LOD threshold of 2.5. None of the 11 QTL had a LOD score higher than 7 or a R 2 % larger than 30. This situation also held for those wheat sharp eyespot resistance QTL reported previously (Cai et al. 2006; Huo 2002; Ren et al. 2004, 2007; Tang et al. 2004; Zhang et al. 2005). This is in contrast to some other diseases such as wheat stripe rust resistance QTL that can have a LOD score higher than 20 and a R 2 % larger than 30 (e.g., Mallard et al. 2005; Paillard et al. 2012). ‘Major’ QTL, therefore, have not yet been found in bread wheat for resistance to sharp eyespot. This raises quite a need for enhancement of extensive screening of wheat germplasm resources/wheat wild relatives for major resistance effects, efforts to elucidate the mechanisms underlying the resistance, efforts to attain useful genotypes by genetic engineering, and effective accumulation or pyramiding of minor resistance QTL with respect to agricultural value.
References
Bassam BJ, Caetano-Anolles G, Gresshoff PM (1991) Fast and sensitive silver staining of DNA in polyacrylamide gels. Anal Biochem 196:80–83
Boerema GH, Verhoeven AA (1977) Check-list for scientific names of common parasitic fungi. Series 26: fungi on field crops: Cereal and grasses. Neth J Plant Pathol 83:165–204
Burpee LL, Sanders PL, Cole H Jr, Sherwood RT (1980) Aastomosis groups among isolates of Ceratobasidium cornigerum and related fungi. Mycologia 72:689–701
Cai SB, Ren LJ, Yan W, Wu JZ, Chen HG, Wu XY, Zhang XY (2006) Germplasm development and mapping of resistance to sharp eyespot (Rhizoctonia cerealis) in wheat. (In Chinese with English abstract). Sci Agric Sin 39:928–934
Chen Y, Li W, Zhang XX, Zhang BQ, Yu HS, Chen HG (2009) Composition and virulence of pathogen of wheat sharp eyespot in north latitude 33° of China. (In Chinese with English abstract). J Triticeae Crops 29:1110–1114
Clarkson JDS, Cook RJ (1983) Effect of sharp eyespot (Rhizoctonia cereatis) on yield loss in winter wheat. Plant Pathol 32:421–428
Cromey MG, Parkes RA, Fraser PM (2006) Factors associated with stem base and root diseases of New Zealand wheat and barley crops. Australas Plant Pathol 35:391–400
Guo YP, Li W, Sun HY, Wang N, Yu SH, Chen HG (2012) Detection and quantification of Rhizoctonia cerealis in soil using real-time PCR. J Gen Plant Pathol 78:247–254
Guyomarc’h H, Sourdille P, Charmet G, Edwards KJ, Bernard M (2002) Characterization of polymorphic microsatellite markers from Aegilops tauschii and transferability to the d-genome of bread wheat. Theor Appl Genet 104:1164–1172
Hamada MS, Yin YN, Ma ZH (2011) Sensitivity to iprodione, difenoconazole and fludioxonil of Rhizoctonia cerealis isolates collected from wheat in China. Crop Prot 30:1028–1033
Hammouda AM (2003) First report of sharp eyespot of wheat in Egypt. Plant Dis 87:598
Huo NX (2002) QTL analysis of resistance to diseases caused by Rhizoctonia cerealis and Blumeris graminis. (In Chinese with English abstract) Ph. D. Dissertation, Graduate School of Chinese Academy of Agricultural Sciences
Lander ES, Green P, Abrahamson J, Barlow A, Daly MJ, Lincoln SE, Newburg L (1987) MAPMAKER: an interactive computer package for constructing primary genetic linkage maps of experimental and natural populations. Genomics 1:174–181
Lemańczyk G (2010) Occurrence of sharp eyespot in spring cereals grown in some regions of Poland. J Plant Prot Res 50:505–512
Lemańczyk G, Kwaśna H (2013) Effects of sharp eyespot (Rhizoctonia cerealis) on yield and grain quality of winter wheat. Eur J Plant Pathol 135:187–200
Li ZK, Pinson MSR, Marchetti MA, Stansel JW, Park WD (1995) Characterization of quantitative trait loci (QTLs) in cultivated rice contributing to field resistance to sheath blight (Rhizoctonia solam). Theor Appl Genet 91:382–388
Lipps PE, Herr LJ (1982) Etiology of Rhizoctonia cerealis in sharp eyespot of wheat. Phytopathology 72:1574–1577
Mallard S, Gaudet D, Aldeia A, Abelard C, Besnard AL, Sourdille P, Dedryver F (2005) Genetic analysis of durable resistance to yellow rust in bread wheat. Theor Appl Genet 110:401–1409
Mazzola M, Smiley RW, Rovira AD, Cook RJ (1996) Characterization of Rhizoctonia isolates, disease occurrence and management in cereals. In: Sneh B, Jabaji-Hare S, Neate S, Dijst G (eds) Rhizoctonia species: taxonomy, molecular biology, ecology, pathology and disease control. Kluwer Academic Publishers, Dordrecht, pp 259–267
McBeath JH, McBeath J (2010) Plant diseases, pests and food security. In: Martin B (ed) Environmental change and food security in china. Springer Technology and Engineering. Springer, Dordrecht, p 136
Paillard S, Trotoux-Verplancke G, Perretant MR, Mohamadi F, Leconte M, Coëdel S, de Vallavieille-Pope C, Dedryver F (2012) Durable resistance to stripe rust is due to three specific resistance genes in French bread wheat cultivar Apache. Theor Appl Genet 125:955–965
Paterson AH, Damon S, Hewitt JD, Zamir D, Rabinowitch HD, Lincoln SE, Lander EC, Tanksley SD (1991) Resolution of Mendelian factors underlying quantitative traits in tomato: comparison across species, generations and environments. Genetics 127:181–197
Pestsova E, Ganal MW, Röder MS (2000) Isolation and mapping of microsatellite markers specific for the D genome of bread wheat. Genome 43:689–697
Polley RW, Thomas MR (1991) Surveys of diseases of winter wheat in England and Wales, 1976–1988. Ann Appl Biol 119:1–20
Ren LJ, Cai SB, Tang T, Wu JZ, Zhou MP, Yan W, Ma HX, Lu WZ (2004) SSR markers linked resistance QTLs to sharp eyespot (Rhizoctonia cerealis) in wheat. (In Chinese with English abstract). J Yangzhou Univ 25:16–19
Ren LJ, Zhang X, Zhou MP, Lu WZ, Ma HX (2007) QTL analysis of sharp eyespot (Rhizoctonia cerealis) and Fusarium head blight in wheat. (In Chinese with English abstract). J Triticeae Crops 27:416–420
Ren LJ, Chen PD, Chen HG, Ma HX (2010) Screening of resistance to sharp eyespot in wheat. (In Chinese with English abstract). J Plant Genet Resour 11:108–111
Röder MS, Korzun V, Wendehake K, Plaschke J, Tixier MH, Leroy P, Ganal M (1998) A microsatellite map of wheat. Genetics 149:2007–2023
Rogers SO, Bendich AJ (1985) Extraction of DNA from milligram amounts of fresh, herbarium and mummified plant tissues. Plant Mol Biol 5:69–76
Sharma A, McClung AM, Pinson SRM, Kepiro JL, Shank AR, Tabien RE, Fjellstrom R (2009) Genetic mapping of sheath blight resistance QTLs within tropical Japonica rice cultivars. Crop Sci 49:256–264
Somers DJ, Isaac P, Edwards K (2004) A high-density microsatellite consensus map for bread wheat (Triticum aestivum L.). Theor Appl Genet 109:1105–1114
Song QJ, Fickus EW, Cregan PB (2002) Characterization of trinucleotide SSR motifs in wheat. Theor Appl Genet 104:286–293
Sourdille P, Tavaud M, Charmet G, Bernard M (2001) Transferability of wheat microsatellites to diploid Triticeae species carrying the A, B and D genomes. Theor Appl Genet 103:346–352
Sourdille P, Cadalen T, Guyomarc’h H, Snape JW, Perretant MR, Charmet G, Boeuf C, Bernard S, Bernard M (2003) An update of the Courtot × Chinese Spring intervarietal molecular marker linkage map for the QTL detection of agronomic traits in wheat. Theor Appl Genet 106:530–538
Srinivasachary, Willocquet L, Savary S (2011) Resistance to rice sheath blight (Rhizoctonia solani Kühn) [teleomorph: Thanatephorus cucumeris (A.B. Frank) Donk.] disease: current status and perspectives. Euphytica 178:1–22
Tang T, Ren LJ, Cai SB, Wu JZ, Lu WZ, Chen JM, Ma HX (2004) Study on QTL mapping of sharp eyespot resistance in wheat. (In Chinese with English abstract). J Triticeae Crops 24:11–16
Wang YZ, Wu ZF, Shi JR, Chen HG (1994) Study on occurrence of wheat sharp eyespot in Jiangsu province and the factors influencing its development in fields. (In Chinese with English abstract). Acta Phytophy Sin 21:109–114
Wang S, Basten JC, Zeng ZB (2010) Windows QTL Cartographer 2.5. Department of Statistics, North Carolina State University, Raleigh, NC. http://statgen.ncsu.edu/qtlcart/WQTLCart.htm. Accessed 10 Mar 2013
Wang M, Lu BL, Xing XP, Li HL (2011) Composition and virulence variation of the pathogen of wheat sharp eyespot from Henan Province. (In Chinese with English abstract). Acta Phytopathol Sin 41:556–560
Zhang XC, Li SS, Zhao XH, Fan YD, Li RJ (2005) QTL and molecular markers for resistance of wheat to sharp eyespot (In Chinese with English abstract). J Plant Genet Resour 6:276–279
Acknowledgments
This study was supported by the National Natural Science Foundation of China (30871612), the Commonweal Specialized Research Fund of China Agriculture (200903035, nyhyzx3-16), the National Basic Research Program of China (2013CB127700), and the Program for Changjiang Scholars and Innovative Research Team (IRT1042).
Conflict of interest
The authors declare no conflict of interest.
Ethical standards
The experiments comply with the current laws of P. R. China.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by B. Keller.
G. Li and Z. Du contributed equally to this paper.
Rights and permissions
About this article
Cite this article
Chen, J., Li, G.H., Du, Z.Y. et al. Mapping of QTL conferring resistance to sharp eyespot (Rhizoctonia cerealis) in bread wheat at the adult plant growth stage. Theor Appl Genet 126, 2865–2878 (2013). https://doi.org/10.1007/s00122-013-2178-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00122-013-2178-6