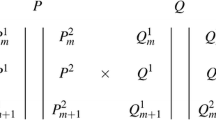Abstract
Key message
A mixed model framework was defined for QTL analysis of multiple traits across multiple environments for a RIL population in pepper. Detection power for QTLs increased considerably and detailed study of QTL by environment interactions and pleiotropy was facilitated.
Abstract
For many agronomic crops, yield is measured simultaneously with other traits across multiple environments. The study of yield can benefit from joint analysis with other traits and relations between yield and other traits can be exploited to develop indirect selection strategies. We compare the performance of three multi-response QTL approaches based on mixed models: a multi-trait approach (MT), a multi-environment approach (ME), and a multi-trait multi-environment approach (MTME). The data come from a multi-environment experiment in pepper, for which 15 traits were measured in four environments. The approaches were compared in terms of number of QTLs detected for each trait, the explained variance, and the accuracy of prediction for the final QTL model. For the four environments together, the superior MTME approach delivered a total of 47 regions containing putative QTLs. Many of these QTLs were pleiotropic and showed quantitative QTL by environment interaction. MTME was superior to ME and MT in the number of QTLs, the explained variance and accuracy of predictions. The large number of model parameters in the MTME approach was challenging and we propose several guidelines to help obtain a stable final QTL model. The results confirmed the feasibility and strengths of novel mixed model QTL methodology to study the architecture of complex traits.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Yield and other complex traits of agronomic importance are typically measured for collections of genotypes across multiple environments, and genotype by environment interactions is common (GEI)Footnote 1 (van Eeuwijk et al. 2010): superiority of genotypes can change in relation to the environment. The statistical genetic analyses of complex traits showing GEI can effectively be addressed by mixed model methodology with terms for QTL by Environment Interaction (QEI) (Boer et al. 2007). QTLs can then be categorized according to the stability of their effects across different environments. A ‘constitutive’ QTL is consistently detected across most environments, while an ‘adaptive’ QTL is detected only in specific environmental conditions, or increases in expression with the level of an environmental factor (Vargas et al. 2006).
For measurements obtained simultaneously for several traits, it is more appropriate to perform statistical analyses multivariately than univariately. This requirement is even stronger when biological processes are interdependent. Traits are genetically correlated and proper QTL mapping helps differentiating whether correlations are due to pleiotropic QTLs or closely linked QTLs. Analyzing correlated traits univariately, leads to higher sampling variances of estimated parameters and lower power for hypothesis tests. The joint analysis of multiple traits has been shown to improve the power and precision of QTL mapping. It has also helped in improving the selection of some primary traits with low heritabilities or that are difficult to measure by exploiting their genetic correlations with other traits (Jiang and Zeng 1995).
Recent advances in statistical genetics methodology have led to extensions of the traditional QTL mapping techniques and the mixed model is now the approach of choice (van Eeuwijk et al. 2010; Vilhjalmsson and Nordborg 2013). This is a result of the suitable framework offered by mixed models in handling many of the challenges present in QTL analysis, including simultaneous observations on many traits and across multiple environments, the possibility of unequal replication of genotypes either due to experimental design and/or missing observation and phenotypic measurements over time (Verbeke and Molenberghs 2000). Furthermore, mixed models do not rely on unrealistic assumptions, such as zero genetic correlations between environments and traits, and constant variance across environments. It can account for both intra- and inter-trial variability in the estimation of QTL effects and trait values prediction (van Eeuwijk et al. 2010). Mixed models have been extensively applied in many QTL mapping settings (Anhalt et al. 2009; Boer et al. 2007; Hackett et al. 2001; Klasen et al. 2012; Korte et al. 2012; MacMillan et al. 2006; Malosetti et al. 2004, 2006, 2008; Panozzo et al. 2007; Piepho 2000; Verbyla et al. 2003; Xu 2013), ranging from single trait single environment analysis up to the most complex setting of multi-trait multi-environment (MTME) with various interactions (traits, environments and/or environmental characterizations).
In pepper, GEI and QEI approaches have not been used previously to map multiple quantitative traits in multiple environments. Earlier studies focused mostly on univariate analyses of traits in single environments (Alimi et al. 2013; Barchi et al. 2009; Ben Chaim et al. 2006; Ben Chaim et al. 2001; Kargbo and Wang 2010; Lee et al. 2008; Lefebvre et al. 2003; Mimura et al. 2010; Rao et al. 2003; Zygier et al. 2005). In MTME analysis, the most challenging aspect often arises from the number of trait by environment combinations (TE’s) in relation to computational requirements. This paper contains a large implementation of MTME in QTL analysis with emphasis on how to circumvent some of the computational issues that may arise due to the increase in the number of parameters being estimated. In this paper, we implemented three different multivariate modelling strategies to analyse data on a recombinant inbred line (RIL) pepper population (Alimi et al. 2013; Voorrips et al. 2010; www.spicyweb.eu). These modelling strategies are multi environment (ME), multi trait (MT) and multi-trait multi-environment (MTME) analyses. We modelled genetic correlations within (between traits in a given environment) and between environments, and explicitly test the presence of QEI and pleiotropic QTLs. In the GEI stage, we performed multi-environment (ME) analysis for each trait to investigate GEI. In the multi-trait (MT) analysis, we combined the 15 traits for each trial in a joint analysis to investigate pleiotropic QTLs. We thereafter created factorial combinations of traits and environments for use in the MTME analysis. We employed unstructured covariance model which allowed each pair of TE combinations to have unique covariance. We then searched for main effect QTLs and QEI effects, by including genome-wide marker data. We investigated accuracy of predictions by the fitted QTL models from each of the three methods and discuss the relative improvements of the final QTL results. We further reduced the TE combinations through principal component analysis. QTL analysis was then performed on the selected components to investigate if QTLs similar to those from ME, MT and MTME analyses would be detected.
Materials and methods
Plant materials, marker data and phenotypic evaluation
We summarize the main features of the data here. A detailed description can be found in Alimi et al. (2013). The mapping population consists of sixth generation (F6) and still segregating recombinant inbred lines (RILs) of an intraspecific pepper cross between the large-fruited inbred cultivar ‘Yolo Wonder’ (YW) and the pungent small-fruited cultivar ‘Criollo de Morelos 334’ (CM 334). DNA was extracted from 149 RILs to produce information for 455 markers assembled into 12 pepper chromosomes, covering 1,705 cM (Fig. 1). The map used here is an improved version of the map used in Alimi et al. (2013) which had five chromosomes with two linkage groups each. All chromosomes now have only one linkage group each. The majority of markers used in the current map are SNP and SSR markers. Almost all the AFLP markers in the former map were discarded (Nicolaï et al. 2012). The percentage of missing genotype information across the full set of markers was 13.7 %. None of the markers showed segregation distortion.
Phenotypic evaluations of the RILs were carried out via designed greenhouse experiments across two locations; Spain (SP) and the Netherlands (NL). The trials were conducted under both spring (1) and autumn (2) weather conditions in 2009. This gave a total of four trials (i.e. environments); Netherlands trial in spring (NL1), Netherlands trial in autumn (NL2), Spain trial in spring (SP1) and Spain trial in autumn (SP2). A total of 15 traits (Table 1) were analysed, 13 of which were already detailed in Alimi et al. (2013). Two additional traits, increase rate of leaf area index (LAI) and light use efficiency (LUE), were added. LAI expresses mean increase in leaf area index per unit time, where time is expressed in degree-days. LUE is the dry matter production (g) per megajoule (MJ) of intercepted global radiation. LUE was estimated as the slope of a graph in which the increase in total plant biomass was plotted against the cumulative amount of intercepted light.
Multi-environment phenotypic and QTL analysis
Each trait was evaluated over the four trials with the aim of investigating genotype-by-environment interaction (GEI) and QTL-by-environment interaction (QEI). As data for this analysis, for each RIL, we used best linear unbiased estimates (BLUE) per environment from an earlier analysis reported in Alimi et al. (2013). To enhance numerical stability, for each trait scale effects were removed and the BLUE values were standardized such that they form a distribution with mean equal to zero and standard deviation equal to one.
Following Boer et al. (2007), the multi-environment phenotypic analysis and QTL estimation were combined. For QTL detection the so-called genetic predictors (functions of conditional QTL genotype probabilities) need to be calculated. The genetic predictors were calculated at all 455 marker positions and 184 intermediate positions for those marker intervals that were larger than 5 cM, genomic positions will be indexed by q, with q = 1, 2,…, 639. The genetic predictor for individual i at genomic evaluation point q is denoted by x iq . The genetic predictors for the additive QTL effect had the value x iq = −1 if both alleles at a fully informative marker arose from parent 1 (YW), or x iq = 1 if they arose from parent 2 (CM334). At intermediate positions and marker positions with missing marker genotypes, these integer values were replaced by linear combinations of conditional QTL genotype probabilities given marker information. Starting with fitting single QTL models using simple interval mapping (SIM) (Lander and Botstein 1989),
where \(y_{ij}\) denotes the standardized phenotype of the ith genotype (i = 1,…,149) in environment j (j = 1,…,4), E j is the environmental mean, \(g_{ij}\) represented the genetic effect of genotype i at environment j, and \(\varepsilon_{ij}\) represented the non-genetic component. We assumed that the vectors \(g_{i} = (g_{i1} , \ldots ,g_{ij} )\) follow a multivariate normal distribution with zero mean and an unstructured VCOV matrix G i.e. \(g_{i} \sim N(0,G)\,\alpha_{jq}\) was the environment-specific QTL main effect at evaluation point q. Testing for the significance of \(\alpha_{jq}\) was done through Wald tests (Verbeke and Molenberghs 2000) with \(H_{0} : \alpha_{1q} = \alpha_{2q} = \alpha_{3q} = \alpha_{4q} = 0,\) where α 1,…,α 4 refers to the QTL effect at each of the four environments. From the fit of model (1), the map positions showing significant deviations from H 0 were selected and the corresponding genetic predictors were set as cofactors in subsequent composite interval mapping (CIM) (Zeng 1994).
where C was the set of cofactors. The cofactor selection thresholds were determined using an approach described by Li and Ji (2005), with genome-wide significance level set at 0.05. CIM was run at least twice consecutively to confirm stability of the test statistic profiles. The full set of significant positions from CIM was subjected to a backward selection procedure to arrive at the final QTL model (Boer et al. 2007). The minimum distance between significant QTLs was assumed to be 20 cM for the final QTL model. In the final QTL model significant QEI effects were determined by testing significance of environment-specific deviations from the main environmental effect through a Wald test. In this case, an effect was called significant when its P value was below the significance level of 0.05, no correction for multiple testing was applied at this stage.
Multi-trait QTL estimation
The specification of multi-trait (MT) model is very similar to the ME model. In the case of MT model, instead of having environment (E) in QTL model (2), we have trait (T). Per environment, there were 15 traits, resulting in four MT analyses. With the inclusion of multiple QTLs as cofactors, the QTL model for CIM is:
where T p (p = 1, 2,…, 15) is the trait mean, α pq is the trait-specific QTL main effect at evaluation point q, \(g_{ip}\) represents the genetic effect of genotype i for trait p, and ε ip is the residual effect. This model allowed us to explicitly model genetic correlations between traits by specifying an unstructured VCOV matrix among each pair of traits giving a total of 120 parameters. It further allowed us to identify QTLs with pleiotropic effects. Synergistic pleiotropy refers to positive covariance between the effects of a gene or gene substitution on two or more traits, based upon correspondence in expression (sign of effects) with regards to the traits. This implies that the increasing alleles for all the traits being influenced by the pleiotropic QTL are from just one of the parents. In antagonistic pleiotropy, pleiotropic effects of a QTL are opposite in sign, positive in one context of expression and negative in another (West-Eberhard 2003).
Multi-traits multi-environments QTL estimation
Extension to multi-trait multi-environment (MTME) setting was achieved by combining traits across the four environments in a single mixed model analysis. ME and MT models are extended by allowing the response trait (y) to be a vector of the traits (T) and environments (E) combinations. The mean for the trait by environment combination, TE, is taken as fixed in the QTL analysis. We restricted ourselves to SIM method for the MTME as CIM could not be implemented successfully as a result of increase in the number of parameters after adding cofactors. The model for SIM is:
where TE z (z = 1, 2,…, 60) is the TE mean (z is the product of four environments and 15 traits = 60), α zq is the environment-specific and trait-specific QTL main effect at evaluation point q, \(g_{iz}\) represents the genetic effect of genotype i for TE z, and ε iz is the residual effect. We specified an unstructured VCOV matrix for all pairs of the TE combinations, giving a total of 1,830 parameters. With the MTME model, GEI and genetic correlations between traits were simultaneously modelled.
MTME final QTL selection and window size
We performed the SIM scan and carried out a backward selection on the significant positions. An initial step was taken to determine an optimal QTL peak window size for the final QTL model, that is, what should be the minimum distance between consecutive QTLs at a chromosome. We investigated QTL window sizes ranging from 5 to 40 cM. When QTL window sizes above 20 cM were used, some putative QTLs were missed. Using window sizes below 20 cM led to selecting some QTLs at very close distance that affected the same set of traits and thus looked as representing a single QTL. A window size of 20 cM was found to be optimum for our data and was used in the final QTL modelling step. The final QTLs were selected using a peak window size of 20 cM and taking into account changes in the signs of neighbouring QTLs. If for two QTLs next to each other, the signs for QTL effects remained unchanged over all TEs, the QTLs were interpreted to represent the same QTL and only the position showing the strongest effects was retained in the final QTL model.
The phenotypic and QTL analyses were performed using the QTL facilities in GenStat 15 (VSNi 2012).
Comparisons of ME, MT and MTME approaches
For the three QTL mapping methods, the number of significant QTLs and their explained variance for each of the TE combinations, e.g. Axl in NL1 (Axl.NL1) were compared. We also investigated whether the same QTL positions were detected for a given TE by the different methods. This enabled us to confirm if QTLs as detected by simpler methods were not lost in the more complex methods. Predictive accuracies of the models were also explored and compared. Predictive accuracy was defined conveniently, although slightly simplistically, as the correlation between BLUE and predicted phenotypic values from the final QTL models in the three approaches. (More in depth treatment of predictive accuracy of various QTL and genomic prediction methods will be submitted in a follow-up paper.)
Results
Genetic correlations between traits (within and between trials)
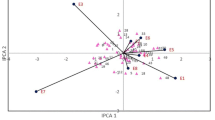
The genetic correlations of traits among environments are given in Table 6 in Appendix A, while the genetic correlations between traits within each trial are presented with the aid of biplots from the first two principal components of the traits (Appendix B, Fig. 9). The correlations between the four environments for individual traits were mostly comparable (uniform correlations) and were generally moderate to high, ranging from 0.30 for NI between NL2 and SP1 to 0.86 for NLE between NL1 and NL2. Overall mean of the genetic correlation was 0.62, with the majority of the correlations above 0.5. Trait variances differed over environments (Appendix A, Table 7,). Within trial correlations were consistent in sign within the trials (Appendix B, Fig. 9). Many of the correlations were according to physiological expectation, considering the relationships between traits, where one trait was computed from others (e.g. DWV from DWS and DWL), or traits related jointly to a part of the plant, e.g. fruit-related traits such as DWF, NF and pt_frt. There were some very high (e.g. between LAI and DWL) and very low (DWF and NLE) correlations, but most correlations between traits within environments were moderate. Some negative correlations were considered remarkable; they depicted resource allocation competitions between plant organs. For example pt_leaf was negatively correlated to fruit-related traits such as NF, DWF and pt_frt. These negative correlations were more pronounced in SP trials than in NL trials.
Multi-environment analyses
The plot of the CIM genome scan for DWF (yield) for the ME approach is given in Fig. 2. The plots of the CIM genome scans for the other traits are presented in Appendix C (Fig. 10). Table 8 in Appendix C presents the QTL positions and effects for all 15 traits. For DWF, three significant QTLs were detected on chromosomes 2, 4 and 7, respectively. Two of these QTLs (C4–35 cM and C7–79 cM) were constitutive i.e. these showed consistent significant effects across the four environments. The QTL on chromosome 2 showed QEI effects in magnitude, but not in direction (=non-crossovers). Such QEI are regarded as quantitative; i.e., the effects had the same sign in all environments. Generally for most traits, QEI effects were quantitative. However, one QTL on chromosome 11 (~70 cM) showed significant crossover interactions (i.e. qualitative QEI) for the traits LUE, Axl, SL and INL in SP1 and SP2 environments. This particular QTL may be categorized as location specific and adaptive as it was significant only in Spanish trials (Appendix C).
CIM profile plot of the multi-environment analyses for yield (DWF). The top section shows the P-values of tests for QTL main effects. The bottom section shows heat maps along the genome for each environment, where blue means that the YW allele had a significant positive effect and red means that the CM334 allele had a significant positive effect in that environment (the darker the colour, the higher the significance level of the QTL). Three QTLs were detected on chromosomes 2, 4 and 7. The QTLs showed no crossovers across environments (colour figure online)
Multi-trait analyses
The plots of CIM genome scans for the MT analysis in the four environments (Fig. 3) showed many significant QTLs across the genome, influencing different traits to different magnitude and direction. After applying backward selection on the CIM scan, a total of 13, 17, 16 and 15 QTL regions exceeded the significance threshold in NL1, NL2, SP1 and SP2, respectively. All QTLs showed pleiotropic effects, i.e., multiple traits were affected by the same QTL. A few of these pleiotropic QTLs displayed synergistic pleiotropic effects while many of them showed antagonistic pleiotropic effects. Clear examples of synergistic pleiotropic QTLs were found on chromosomes (4@70 cM in NL1, 4@11 cM in NL2, 7@35 cM in NL2 and 3@40 cM) in NL1. An example of an antagonistic QTL was present on chromosome 3 (~150 cM) in SP2. This QTL showed increasing effects from YW on fruit-related traits (DWF and pt_frt) and increasing effects from CM334 on other traits such as SL, NLE, NI, Axl and LUE. Many of these pleiotropic QTLs are consistent with genetic correlations among the traits. As an example, the QTLs on chromosomes 2 and 4, influencing pt_leaf and fruit traits such as DWF showed antagonistic pleiotropy especially in SP trials, which is consistent with the negative correlations that exist between pt_leaf and the fruit traits. For many traits, MT analyses revealed more QTLs than the ME analyses (Table 2). These QTLs also explained more genetic variations than those from ME analyses. In SP2, about 10 QTLs were detected for DWF including the three QTLs detected in ME analyses. These QTLs explained about 45 % of genetic variation against 29 % explained by the three QTLs from ME analyses. The MT QTL positions and effects for each of the environments are presented in Appendix D.
CIM Profile plots of the multi-trait analyses for the four environments. The top section shows the P-values of tests for QTL main effects. The bottom section shows heat maps along the genome for each trait, where blue means that the YW allele had a significant positive effect and red means that the CM334 allele had a significant positive effect on the given trait (the darker the colour, the higher the significance level of the QTL). Most of the QTLs showed pleiotropies which were most times antagonistic (colour figure online)
Multi-trait multi-environment analysis
The plot of the SIM genome scan for the MTME analysis using an unstructured VCOV is given in Fig. 4. A total of 47 regions were identified as harbouring putative QTLs. Chromosomes 4 and 10 had the smallest number of QTLs (=2) while chromosomes 1 and 3 had the highest number of QTLs (=6). Similar to the results from MT analyses, pleiotropic QTLs were observed for genetically correlated traits. The majority of the 47 QTLs showed antagonistic pleiotropic effects, i.e., the increasing alleles originated from both parents for different traits. Five QTL with synergistic pleiotropic effects for the YW parent (contributing the increasing allele) were found on chromosomes 2 (31 cM), 4 (53 cM), 7 (0 cM), 11 (20 cM), and 12 (75 cM). Also for parent CM334, five of these QTL were found on chromosomes 2 (128 cM), 3 (135 cM), 5 (38 cM), 6 (0 cM), and 8 (19 cM). The majority of the pleiotropic QTLs were not constitutive as they were not consistently affecting particular traits across all environments. This means that many of the QTLs displayed QEI. The QEI were mostly quantitative, but there were some qualitative QEI especially on chromosome 11 for LUE, Axl, SL and INL, similar to the results from ME analyses. Table 3 contains the list of QTL positions from chromosomes 1 and 2 as detected from MTME analysis after backward selection. Results for the remaining chromosomes are in Appendix E, Tables 10, 11, 12.
SIM profile plot for multi-trait multi-environment analysis. The top section shows the P-values of tests for QTL main effects across all trait-environment combinations with the bars on the x-axis indicating the 47 QTL positions after backward selection. The bars in red indicate QTL positions similar to significant positions from ME and MT analyses while those in black are unique to MTME. The bottom section shows heat maps along the genome for each trait, where blue means that the YW allele had a significant positive effect and red means that the CM334 allele had a significant positive effect on the given trait-environment (the darker the colour, the higher the significance level of the QTL) (colour figure online)
Comparison of MT, ME, and MTME results
In environment SP2, a total of 13 QTLs were detected for DWF in the MTME analysis, 3 and 10 more than those from MT and ME analyses, respectively. The percentages explained variances by these QTL jointly were 56, 45 and 29 % in the MTME, MT and ME analyses, respectively (Table 2). QTL effects for DWF on chromosomes 3 and 4 were significant in the four environments. DWF QTLs were in many cases pleiotropic to other yield-related traits such as pt_frt and NF. Such pleiotropic QTLs were observed on chromosomes 2, 3, 4, 6 and 12 (Fig. 5). Pleiotropy with other traits was also observed such as with Axl, NI and INL on chromosome 1; with DWL, DWS, DWV, LAI, LUE and INL on chromosome 2. Others were with LUE, SLA, SL and NI on chromosome 6 and NLE, NI and INL on chromosome 12.
Comparing QTL positions from SE, ME, MT and MTME analyses for yield-related traits (DWF, NF and pt_frt) across the four environments. Blue indicates QTLs with significant effect from YW allele while red indicates QTLs with significant effect from CM334 allele. QTLs detected in SE, ME and MT analyses were present in QTLs from MTME with additional QTLs only picked up in MTME (colour figure online)
Figure 6 shows the joint distribution of total percent of variation attributable to QTLs from the MTME model, which ranges from three QTLs explaining about 19 % to 13 QTLs explaining 60 %. This revealed varying contributions of different QTLs to the total amount of variation explained. In general, the proportions of variation explained were positively correlated to the number of detected QTLs. However, for some traits fewer QTLs explained similar percentages of variation as other traits with more QTLs. For example, eight QTLs for NLE.SP2 explained more variation (63.4 %) than 13 QTLs for INL.SP2 (61.4) and DWL.SP1 (60.4 %). This was consistent with the presence of a few QTLs with large effects for some traits and many QTLs of smaller effects for other traits. On average over the four environments, INL and NLE had the highest proportion of explained genetic variance (54 and 53 %, respectively), this proportion was 46 % for DWF while DWS and NF had the lowest proportions of 32 and 33 %, respectively (Table 2).
Joint distribution of total percent of variation attributable to significant QTLs from MTME. This revealed varying contributions of different QTLs to the total amount of variation explained. In general, the proportions of variation explained were positively correlated to the number of detected QTLs. However, some traits need a smaller number of QTLs to explain as much variation as other traits
Table 2 gives the number of QTLs together with their explained variance for each of the 15 traits in the four environments using ME, MT and MTME methods and also results from single trait single environment (SE) QTL analysis for comparison. As we used a different map in this study, the results for the SE analysis here was slightly different from those reported in Alimi et al. (2013). In principle, the QTL approach for SE is similar to other methods explained except that each trait in each environment was handled univariately. CIM was also used to account for multiple QTL. For each trait in each environment, there was a clear increase in the number of QTLs and explained variance going from ME to MT to MTME. There was also a clear gain in going from univariate analysis to multivariate analyses and in modelling correlations among environments and among traits within an environment. As an example, one, two, four and seven QTLs were identified for DWF in the NL1 trial using SE, ME, MT and MTME methods, respectively, explaining about 18, 22, 24 and 32 % of genetic variations, respectively. Ten QTLs explaining 44 % of the variance were detected for pt_frt in SP2 trials as against 5 (28 %), 3 (36 %) and 3 (26 %) QTLs for MT, ME and SE, respectively. The percentages explained variation by individual QTLs from ME, MT and MTME ranged from 3 to 35 % (Fig. 7). The MTME method yielded many QTLs of small effects (between 3 and 8 %) that were not detected in both ME and MT methods. Also, MT and ME had more QTLs that explained 10–20 % variation than MTME. This might be related to the “Beavis effect” (Beavis 1994, 1997) as simpler models failed to detect some QTLs with small effects and also resulted in overestimation of some effect sizes.
Almost all QTLs detected in simpler methods were also detected in more complex methods. Using fruit-related traits for illustration (Fig. 5), the three QTLs picked up for DWF in SP2 by SE method were also picked up by ME, MT and MTME methods. The positions of the three QTLs shifted slightly for MT and MTME as a result of their effects on other traits. The directions of their effects were also consistent. The QTL on chromosome 7 was significant in all environments under the ME method, but it disappeared for NL1, NL2 and SP1 trials using any of the other three methods. Many of the extra QTLs detected in MT were also detected in MTME. Similar patterns were observed for NF and pt_frt (Fig. 5).
The prediction accuracies of the final QTL models for each trait under ME model were largely similar across environments, though prediction accuracies from SP trials were slightly higher in most cases (Table 4). Highest prediction accuracy for DWF under the ME model (0.54) was obtained in SP environments. This agreed well with our earlier findings that the three QTLs found for DWF under the ME model explained far more variation in SP environments than in NL environments. This also indicated the presence of QEI for this trait. There was an improvement of trait predictions going from ME to MT and MTME models. The fitted QTL model from MTME predicted trait phenotypes better than MT and ME models. Prediction accuracies for DWF improved from about 0.54 under the ME model to about 0.7 under MT and 0.83 under MTME. Furthermore, the genetic correlations between predicted traits in each environment were similar to genetic correlations between BLUEs (Appendix B).
Discussion
Several studies have shown that multi-trait and/or multi-environment QTL analyses based on linear mixed models are more powerful and effective to map pleiotropic QTL and QTL by environment interactions than performing single trait and single environment analyses (Boer et al. 2007; Korte et al. 2012; Malosetti et al. 2008; Sukhwinder et al. 2012). We also showed that in situations such as the EU-SPICY project (Barócsi 2012; Nicolaï et al. 2012; van der Heijden et al. 2012; Voorrips et al. 2010; www.spicyweb.eu), where phenotypic data on a large number of traits have been collected in multiple environments, using QTL methods that properly model underlying VCOV structures among the traits and between environments led to improved power to detect more QTLs than performing individual trait/environment analyses. The joint analysis was especially suitable for complex traits (such as yield) whose genetic variations are usually due to a large number of QTLs of smaller effects which might go undetected with single trait/environment analysis.
We performed and compared three mixed modeling approaches that modeled correlations between environments and/or among traits within an environment. In multi-environment studies, independent analyses without explicit modeling of the correlation structure between environments would not allow to identify GEI and QEI. In multi-trait datasets, univariate analysis that do not account for possible correlations among the traits would not allow us to properly identify QTLs with pleiotropic effects. The probability of finding QEI and/or pleiotropic QTLs is influenced by the magnitude of genetic correlations between environments and between traits within each environment, respectively. It was expected that QTLs with identical effect directions will be detected for highly correlated traits while no common QTLs may be detected for non-correlated traits. Equally, high between-trial correlations would reduce the incidence of QEI. Pleiotropic QTLs that showed effects with trait increasing alleles from both parents are more likely to be detected for traits with negative correlations. The pepper traits considered showed positive and mostly uniform correlations between environments. This was also supported by the QEI results as most of the QEI observed were only due to differences in magnitude, and not different in direction. In our multi-trait analysis, synergistic pleiotropic QTLs were picked up for positively correlated traits. The pleiotropy was usually consistent across the four environments. Also, antagonistic pleiotropic QTLs were found for negatively correlated traits. These negative correlations depicted resource allocation competitions that exist between plant organs e.g. leaf- and fruit-related traits.
Factorial combinations of traits and environments and their joint analysis through the MTME method significantly increased the power of QTL detection with increased precision. This model fully utilizes covariance structures between environments and among traits within environments, and hence is better capable of mimicking biological process for complex traits than fitting ME and MT models separately. Considering yield, the results from SE and ME analyses showed that all the alleles increasing yield originated from the large fruited YW parental line. However, MT and MTME permitted to detect also favourable alleles from the small fruited parent CM334 on chromosomes 3, 5, 7, 11 and 12 (Fig. 5). All those QTLs displayed pleiotropic effects with number of fruits (NF) and/or proportion of partitioning to fruit (pt_frt). The detection of these QTLs with MTME will permit to take it into account when selecting recombinant individuals for high yield. This is more generally true since QTLs for vegetative traits were mainly restricted to chromosomes 1, 2 and 9, and to chromosomes 2, 3, 4 and 10 for fruit traits in the previous SE analyses (Alimi et al. 2013; Barchi et al. 2009; Ben Chaim et al. 2006; Rao et al. 2003). Since MTME model and also ME and MT models are based on mixed modelling technique, they are capable of handling unbalanced data in situation where not all traits are measured in all environments.
However, it is not in all situations that an MTME model can be successfully fitted. In situations where linear dependencies exist among some traits in the combination, some of these traits might need to be removed or transformed before an MTME fit can be successful. As an example, the total plant biomass (DWP) was partitioned to fruit (pt_frt), leaf (pt_leaf) and stem (pt_stem) components. We had to remove DWP and one of the partitioned components (pt_stem) before we could successfully fit the MTME model. However, we decided to leave some of the dependent traits such as DWV, DWS and DWL in our model as their presence did not affect the success of the MTME model. Also, this problem is more of combinatorial issue than correlation. As an example, DWF and pt_frt in our model are well correlated (about 0.9). When traits are well correlated, the method can still be successful unless the number of combinations to be handled are big with some linear dependencies among the traits.
MTME models might also prove difficult to fit due to the increase in the number of parameters to be estimated in the REML step as a result of large number of TE combinations. This becomes more laborious if markers (genetic predictors) are specified in the model as cofactors. If the problem occurs after adding cofactors, the result from the simple interval mapping could be subjected to backward selection before applying the final QTL model using appropriate QTL window size to separate the QTL positions. With a large QTL window size, some putative QTLs are lost while a small QTL window size could lead to declaration of duplicate QTLs. Duplicate QTLs could be detected via careful visual inspections of the QTL effect signs. If the signs of two neighbouring QTLs remain unchanged over all the traits, the QTLs can be regarded as one. For example, consider four traits T1, T2, T3 and T4 being influenced by three QTLs Q1, Q2 and Q3 that are very close to each other on a chromosome. If the effects of the three QTLs on the four traits follow these sequences: Q1 = {+, +, −, +}, Q2 = {+, +, −, +} and Q3 = {+, +, −, −}. Then Q1 and Q2 could be regarded as one QTL since the patterns are identical while Q3 is a different QTL from Q1 and Q2 because of the change in effect sign on T4. Furthermore, the appropriate QTL window size can be analytically checked using the Weller and Soller (2004) approach. In our case, the appropriateness of a 20-cM peak window size was confirmed by analytically calculating the required confidence intervals for QTL location for a RIL population of our size given the magnitude of QTL effects (Weller and Soller 2004). For the standardized traits, this was found to be around 15 cM assuming (standardized) effect size of 0.25 with sample size of 149 and heritability of 0.25. It should be noted that this calculation was for univariate analysis with no multivariate correction. The actual interval in the multivariate case would even be smaller. So taking the smallest interval across all traits and environments can be seen as the upper bound of the interval in the multivariate sense. In our case the effects from many of the detected QTLs were more than 0.25 with the highest being around 0.6. This means that 15 cM is like the upper bound for the interval.
In this study, we successfully applied the MTME approach to a dataset of 60 TE combinations. A simple approximating approach would have been to first apply data reduction techniques such as principal component analysis to reduce the number of variables and then perform a QTL analysis on the new set of variables, the principal component scores, just to identify the major genomic regions where DNA variation affects trait variation. We explored this approach—taking the scores of the first 10 principal components as trait values, and found that it produced most of the important QTLs underlying the original variables. That is, 16 QTLs were detected with high correspondence to significant QTLs from SE, ME, MT and MTME analyses (Fig. 8). A major drawback of the use of principal components is the biological interpretation of the results, but as method to identify the most interesting genomic regions, it performs well.
CIM Profile plot of the multi-trait analyses for scores from 10 PCs. Percentage of explained variance the top section shows the P-values of tests for QTL main effects. The bottom section shows heat maps along the genome for each PC, where blue means that the YW allele had a significant positive effect and red means that the CM334 allele had a significant positive effect on the given PC (colour figure online)
The QTL identified in this study will be aligned with eQTL results from a gene expression study in the same EU-SPICY project, (M. Vuylsteke, personal communication). The eQTL results will provide a set of candidate genes co-located with the QTL for yield and, hence, being likely involved in growth of pepper. Identifying these candidate genes would increase insight into the functioning of the pepper plant, and also increase efficiency of breeding, since this allows multiple alleles to be found within the gene, accounting for different phenotypes. Successful candidate genes, whose sequence position is related to QTL position, will be used to assess the marker-phenotype association in a core collection of pepper accessions (Nicolaï et al. 2012). Such an association genetics approach will be helpful in further selection of candidate genes, and will provide us with potential allelic values for phenotype prediction.
In conclusion, multivariate QTL mapping methods such as the MTME approach are instrumental to boost the power and accuracy of QTL detection for complex traits by successful identification of QTLs with relatively small effects. It would also lead to better detection of alleles in repulsion phase, differential allele expression according to environments and an increased explained variance for most complex traits. This would lead to improvement in the prediction of phenotype by the genotype and thus the genetic gain in genome-assisted breeding. This will ultimately increase our understanding of complex traits and our ability to use QTL in genome-assisted breeding.
Notes
The list of all abbreviations is given in Table 5 in Appendix A.
References
Alimi NA, Bink MCAM, Dieleman JA, Nicolaï M, Wubs M, Heuvelink E, Magan J, Voorrips RE, Jansen J, Rodrigues PC, Heijden GWAM, Vercauteren A, Vuylsteke M, Song Y, Glasbey C, Barocsi A, Lefebvre V, Palloix A, Eeuwijk FA (2013) Genetic and QTL analyses of yield and a set of physiological traits in pepper. Euphytica 190:181–201
Anhalt UCM, Heslop-Harrison JS, Piepho HP, Byrne S, Barth S (2009) Quantitative trait loci mapping for biomass yield traits in a Lolium inbred line derived F2 population. Euphytica 170:99–107
Barchi L, Lefebvre V, Sage-Palloix A-M, Lanteri S, Palloix A (2009) QTL analysis of plant development and fruit traits in pepper and performance of selective phenotyping. Theor Appl Genet 118:1157–1171
Barócsi A (2012) Intelligent, net or wireless enabled fluorosensors for high throughput monitoring of assorted crops. Meas Sci Technol 24:025701
Beavis WD (1994) The power and deceit of QTL experiments: Lessons from comparative QTL studies. In: Proceedings of the forty-ninth annual corn and sorghum research conference. American Seed Trade Association, Washington, pp 250–266
Beavis WD (1997) QTL analyses: power, precision, and accuracy. In: Paterson AH (ed) Molecular dissection of complex traits. CRC Press, Boca Raton, pp 145–162
Ben Chaim A, Paran I, Grube RC, Jahn M, van Wijk R, Peleman J (2001) QTL mapping of fruit-related traits in pepper (Capsicum annuum). Theor Appl Genet 102:1016–1028
Ben Chaim A, Borovsky Y, Rao G, Gur A, Zamir D, Paran I (2006) Comparative QTL mapping of fruit size and shape in tomato and pepper. Israel J Plant Sci 54:191–203
Boer MP, Wright D, Feng LZ, Podlich DW, Luo L, Cooper M, van Eeuwijk FA (2007) A mixed-model quantitative trait loci (QTL) analysis for multiple-environment trial data using environmental covariables for QTL-by-environment interactions, with an example in maize. Genetics 177:1801–1813
Hackett CA, Meyer RC, Thomas WTB (2001) Multi-trait QTL mapping in barley using multivariate regression. Genet Res 77:95–106
Jiang CJ, Zeng ZB (1995) Multiple-trait analysis of genetic-mapping for quantitative trait loci. Genetics 140:1111–1127
Kargbo A, Wang CY (2010) Complex traits mapping using introgression lines in pepper (Capsicum annuum). Afr J Agric Res 5:725–731
Klasen JR, Piepho HP, Stich B (2012) QTL detection power of multi-parental RIL populations in Arabidopsis thaliana. Heredity 108:626–632
Korte A, Vilhjalmsson BJ, Segura V, Platt A, Long Q, Nordborg M (2012) A mixed-model approach for genome-wide association studies of correlated traits in structured populations. Nat Genet 44:1066–1071
Lander ES, Botstein D (1989) Mapping mendelian factors underlying quantitative traits using RFLP linkage maps. Genetics 121:185–199
Lee HR, Cho MC, Kim HJ, Park SW, Kim BD (2008) Marker development for erect versus pendant-orientated fruit in Capsicum annuum L. Mol Cells 26:548–553
Lefebvre V, Daubeze AM, van der Voort JR, Peleman J, Bardin M, Palloix A (2003) QTLs for resistance to powdery mildew in pepper under natural and artificial infections. Theor Appl Genet 107:661–666
Li J, Ji L (2005) Adjusting multiple testing in multilocus analyses using the eigenvalues of a correlation matrix. Heredity 95:221–227
MacMillan K, Emrich K, Piepho HP, Mullins CE, Price AH (2006) Assessing the importance of genotype x environment interaction for root traits in rice using a mapping population II: conventional QTL analysis. TAG 113:953–964
Malosetti M, Voltas J, Romagosa I, Ullrich SE, van Eeuwijk FA (2004) Mixed models including environmental covariables for studying QTL by environment interaction. Euphytica 137:139–145
Malosetti M, Visser RGF, Celis-Gamboa C, Eeuwijk FA (2006) QTL methodology for response curves on the basis of non-linear mixed models, with an illustration to senescence in potato. Theor Appl 113:288–300
Malosetti M, Ribaut JM, Vargas M, Crossa J, van Eeuwijk FA (2008) A multi-trait multi-environment QTL mixed model with an application to drought and nitrogen stress trials in maize (Zea mays L.). Euphytica 161:241–257
Mimura Y, Minamiyama Y, Sano H, Hirai M (2010) Mapping for axillary shooting, flowering date, primary axis length, and number of leaves in pepper (Capsicum annuum). J Jpn Soc Hortic Sci 79:56–63
Nicolaï M, Pisani C, Bouchet J, Vuylsteke M, Palloix A (2012) Discovery of a large set of SNP and SSR genetic markers by high-throughput sequencing of pepper (Capsicum annuum). Genet Mol Res 11:2295–2300
Panozzo JF, Eckermann PJ, Mather DE, Moody DB, Black CK, Collins HM, Barr AR, Lim P, Cullis BR (2007) QTL analysis of malting quality traits in two barley populations. Aust J Agric Res 58:858–866
Piepho H-P (2000) A mixed-model approach to mapping quantitative trait loci in barley on the basis of multiple environment data. Genetics 156:2043–2050
Rao GU, Ben Chaim A, Borovsky Y, Paran I (2003) Mapping of yield-related QTLs in pepper in an interspecific cross of Capsicum annuum and C. frutescens. Theor Appl Genet 106:1457–1466
Sukhwinder S, Hernandez MV, Crossa J, Singh PK, Bains NS, Singh K, Sharma I (2012) Multi-trait and multi-environment QTL analyses for resistance to wheat diseases. PLoS One 7:e38008
van der Heijden G, Song Y, Horgan G, Polder G, Dieleman A, Bink M, Palloix A, van Eeuwijk F, Glasbey C (2012) SPICY: towards automated phenotyping of large pepper plants in the greenhouse. Funct Plant Biol 39:870–877
van Eeuwijk FA (ed) (2012) Smart tools for prediction and improvements of crop yield—KBBE 211347. http://www.spicyweb.eu
van Eeuwijk FA, Bink M, Chenu K, Chapman SC (2010) Detection and use of QTL for complex traits in multiple environments. Curr Opin Plant Biol 13:193–205
Vargas M, van Eeuwijk F, Crossa J, Ribaut J-M (2006) Mapping QTLs and QTL × environment interaction for CIMMYT maize drought stress program using factorial regression and partial least squares methods. Theor Appl Genet 112:1009–1023
Verbeke G, Molenberghs G (2000) Linear mixed models for longitudinal data. Springer, New York
Verbyla AP, Eckermann PJ, Thompson R, Cullis BR (2003) The analysis of quantitative trait loci in multi-environment trials using a multiplicative mixed model. Aust J Agric Res 54:1395–1408
Vilhjalmsson BJ, Nordborg M (2013) The nature of confounding in genome-wide association studies. Nat Rev Genet 14:1–2
Voorrips RE, Palloix A, Dieleman JA, Bink MCAM, Heuvelink E, Heijden GWAM, van der Vuylsteke M, Glasbey C, Barócsi A, Magán J, van Eeuwijk FA (2010) Crop growth models for the -omics era: the EU-SPICY project. In: Advances in genetics and breeding of capsicum and eggplant: Proceedings of the XIVth EUCARPIA meeting on genetics and breeding of capsicum and eggplant. Editorial Universidad Politécnica de Valencia, Valencia, pp 315–321
VSNi (2012) GenStat for Windows 15th Edition. VSN International, Hemel Hempstead
Weller J, Soller M (2004) An analytical formula to estimate confidence interval of QTL location with a saturated genetic map as a function of experimental design. Theor Appl Genet 109:1224–1229
West-Eberhard MJ (2003) Developmental plasticity and evolution. Oxford University Press, New York
Xu S (2013) Mapping QTL for multiple traits. In: Principles of statistical genomics. Springer, New York, pp 209–222
Zeng ZB (1994) Precision mapping of quantitative trait loci. Genetics 136:1457–1468
Zygier S, Chaim AB, Efrati A, Kaluzky G, Borovsky Y, Paran I (2005) QTLs mapping for fruit size and shape in chromosomes 2 and 4 in pepper and a comparison of the pepper QTL map with that of tomato. Theor Appl Genet 111:437–445
Acknowledgments
The research leading to these results has received funding from the European Community’s Seventh Framework Programme (FP7/2007-2013) under grant agreement n° 211347. We thank the EU-SPICY Industrial Advisory Board for support and discussions. Rik van Wijk and Syngenta are especially acknowledged for their highly valuable help in making available additional SNP markers that strongly improved the quality of the genetic map. Roeland Voorrips and other members of the EU-SPICY project are acknowledged for their contributions and helpful comments. We also thank Paul Keizer, Marcos Malosetti and Martin Boer of Biometris for their valuable insights.
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical standard
The authors declare that the experiments in this study comply with the current laws of the countries (Spain and Netherlands) in which the experiments were performed.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by I. Mackay.
Appendices
Appendix A
Appendix B
See Fig. 9.
Biplots for BLUEs and fitted trait values in each environment. NL1, NL2, SP1 and SP2 are the biplots of BLUE for the traits in each environment while NL1p, NL2p, SP1p and SP2p are the biplots for fitted values of each trait in each environment from the MTME QTL model. The cosine of the angle between the lines approximates the correlation between the traits they represent. The closer the angles are, the higher the correlations. Angles close to 90 or 270° reflect weaker correlations. In each environment, angles between traits are similar for biplots from BLUEs and fitted values. E.g. the biplot for NL1 and NL1p, show a strong relationship between DWF and NF, and a weak relationship between DWF and NLE. The lines enclosing the sample points in the biplots are known as convex hulls, representing the smallest convex set of the sample data
Appendix C: QTL by environment results from ME analyses
CIM Profile plot for all the traits in the multi-environment analyses. The top section shows the P-values of tests for QTL main effects. The bottom section shows heat maps along the genome for each environment, where blue means that the YW allele had a significant positive effect and red means that the CM334 allele had a significant positive effect in that environment. Many of the QTLs are constitutive i.e. consistent across environments with no crossover interaction except the QTL on chromosome 11 for LUE, Axl, SL and INL (colour figure online)
Appendix D: QTL effects from MT analyses
See Table 9.
Appendix E: QTL effects from MTME analyses: chromosomes 3–12
Rights and permissions
About this article
Cite this article
Alimi, N.A., Bink, M.C.A.M., Dieleman, J.A. et al. Multi-trait and multi-environment QTL analyses of yield and a set of physiological traits in pepper. Theor Appl Genet 126, 2597–2625 (2013). https://doi.org/10.1007/s00122-013-2160-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00122-013-2160-3