Abstract
Blackberry primocane fruiting, fruiting on first-year canes, has the potential to expand blackberry production both seasonally and geographically. The incorporation of the primocane-fruiting trait into cultivars with desirable horticultural attributes is challenging due to its recessive nature and tetrasomic inheritance. Molecular marker-assisted selection has high potential to facilitate incorporation, because breeders already use morphological marker-assisted selection of seedlings without marginal cotyledonary hairs to identify progeny that will be thornless when mature. The development of a genetic linkage map with these two traits is the first step to utilizing molecular markers in breeding for thornless primocane-fruiting blackberry cultivars. A full-sib family segregating for thornlessness and primocane fruiting, from a cross between ‘APF-12’ and ‘Arapaho’, was used to construct the first genetic map of tetraploid blackberry. Segregation patterns of several dominant markers and the two phenotypic traits fit those expected uniquely with tetrasomic inheritance (e.g., 5:1, 11:1 and 35:1). Some loci showed significant double reduction frequencies, but genotypes that could have originated only from double reduction were not found. The map consists of seven linkage groups (LG) in each parent, consistent with the basic number of chromosomes (2n = 4x = 28). Naming of LG1-LG6 followed that of the recently revised system for raspberry using SSR markers in common between blackberry and raspberry, and LG7 was tentatively defined by default. The loci controlling primocane fruiting and thornlessness were not linked to each other; thornless/thorny, the S Locus, was mapped on LG4, and the primocane-/floricane-fruiting locus, named in this work the F Locus, on LG7.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Blackberry (Rubus subgenus Rubus Watson.) is an economically important member of the Rosaceae plant family, cultivated mainly in Europe and North America. Its production has been expanding geographically and seasonally, due in part to demand, but also to the release of three primocane-fruiting blackberry cultivars (Clark et al. 2004; Clark and Perkins-Veazie 2011). Primocane-fruiting cultivars produce fruit on floricanes and then a second crop in late summer and early fall on canes that emerge in spring and are just a few months old, potentially extending fruit production. In warmer climates, this yields two crops. In colder climates where winter injury to canes had previously excluded blackberry production, canes can be mowed to the ground in late fall and the year’s entire crop could potentially be produced on canes that emerge the following spring. This practice not only eliminates concerns of winter injury to canes, but also helps manage weeds (Clark et al. 2007). New primocane-fruiting cultivars are being developed to meet the growing demand.
Knowledge of the inheritance of key horticultural traits is a basic requirement for cultivar development. Clonally propagated, blackberry is highly heterozygous. More than one species of blackberry has been cultivated, but the bulk of blackberry cultivars in the USA are tetraploid (2n = 4x = 28) (Clark et al. 2007). Tetrasomic segregation in blackberry and recessive inheritance were observed with primocane fruiting (Lopez-Medina et al. 2000) and thornlessness (Scott et al. 1957).
Selection of thornless breeding progeny can be done at the seedling stage based on the absence of marginal cotyledonary hairs. Thus, a sort of morphological marker-assisted selection has been used by blackberry breeders for many years, suggesting that molecular marker-assisted selection for a trait like primocane fruiting has the potential for adoption (Lewers et al. 2008). Clark et al. (2007) reported that, to obtain thornless primocane-fruiting progeny with acceptable fruit quality and yield, populations as large as 1,000 seedlings were required; this estimate may be low, considering the inheritance of the two traits. Expression of the trait is affected by environment and plant vigor, so it is not always expressed in the first or even the second year (Lopez-Medina et al. 2000). Maintaining and evaluating populations of this size for a minimum of 2 years to make selections may be less efficient than marker-assisted selection. The development of a genetic linkage map with these two traits is the first step to utilizing molecular markers in breeding to accelerate the development of new thornless primocane-fruiting blackberry cultivars, and so that is the objective of this work.
Blackberry-derived EST-SSRs were developed to increase the number of SSRs available for blackberry map development (Lewers et al. 2008). EST-SSRs have been described for a number of cultivated Rosaceae species (Lewers et al. 2005; Graham et al. 2006). Although some EST-SSRs may be transferable across Rosaceae species and have been used to associate linkage groups in one species with those in a related species (Bushakra et al. 2012; Spigler et al. 2010), a low percentage of SSR primer pairs developed from other species have amplified products from blackberry (Graham et al. 2004; Lewers et al. 2005; Stafne et al. 2005).
Construction of a linkage map is more difficult for the autopolyploid blackberry due to the complexity of polysomic inheritance. Until recently, mapping procedures in autopolyploid plants were based on SDRF (single-dose restriction fragment) segregation in mapping populations analyzed as pseudo-test crosses (Wu et al. 1992). Genetic maps were obtained in several autotetraploid species (Bradshaw et al. 1998; Hackett et al. 2000; Sledge et al. 2005, among others). The disadvantage of using only SDRFs for map construction is that only a small fraction of any available markers, only those derived from single simplex genotypes by nulliplex genotypes (Aaaa × aaaa) segregate in a way suitable for SDRF mapping.
More recently, progress has been made in developing the theory of linkage analysis and mapping for full-sib families of autotetraploid species (Luo et al. 2001; Hackett et al. 2001). This theory is based on the simplest situation that can give tetrasomic inheritance, namely random pairing of four homologous chromosomes to give two pairs of bivalents at meiosis. Though many departures from this simple situation could occur, such as multivalents and double reduction, lack of complete homology between chromosomes and hence departures from random pairing, and distorted segregation due to differential fertility and viability, which also occurs in diploids (Bradshaw et al. 2008), the software TetraploidMap (Hackett and Luo 2003; Hackett et al. 2007) was developed from the basic theory and is the only software that has been developed for autotetraploid mapping. Using TetraploidMap, it is now possible to develop a map in autotetraploid species, based on all co-dominant or dominant markers, and to calculate linkage between pairs of markers, either in coupling phase or in repulsion phase. The suitability of this software has been demonstrated in the tetraploid species of alfalfa (Julier et al. 2003), potato (Bradshaw et al. 2008), and rose (Gar et al. 2011) so far.
Materials and methods
Plant material
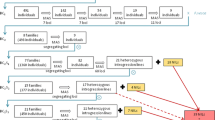
A full-sib family of 188 individuals segregating for thornlessness and primocane fruiting was developed from the cross between the parents ‘APF-12’ (marketed as Prime-Jim®) and ‘Arapaho’. ‘APF-12’ has thorns and is primocane fruiting. ‘Arapaho’ is thornless and not primocane fruiting, producing fruit only on floricanes. ‘APF-12’ is a selection from the cross of ‘Arapaho’ × Ark.830 (Clark et al. 2005), so the full-sib family also is a BC1 family. ‘Arapaho’ is nulliplex for thorny (ssss) and ‘APF-12’ is duplex (SSss) (Clark et al. 2007). ‘Arapaho’ is duplex (AAaa) for floricane fruiting and ‘APF-12’ is nulliplex (aaaa); the floricane-/primocane-fruiting locus has not yet been named. Progeny from ‘APF-12’ × ‘Arapaho’ were expected to segregate 5 thorny:1 thornless and 5 floricane fruiting:1 primocane fruiting.
Phenotypic evaluation
The parents and 188 full-sib progeny were individually evaluated at the University of Arkansas qualitatively as thornlessness or thorny, and primocane fruiting or floricane fruiting. Briefly, culture of the plants consisted of standard cultural practices for erect blackberry production including annual preemergence and postemergence herbicide applications, and annual spring nitrogen fertilization (56 kg ≅ ha−1 N) using ammonium nitrate. All seedlings had primocanes tipped at 1.1 m height two times each season, usually in mid-June and late July or early August. Dormant pruning of seedlings consisted of removing dead floricanes and also primocane tissue to a point below the fruiting/flowering area on the primocanes. All primocane-fruiting genotypes were allowed to produce both floricane and primocane crops. All seedlings received a single application of liquid lime sulfur (94 l ≅ ha−1) at bud break for control of anthracnose [Elsinoë veneta (Burkh.) Jenkins]. All plots were irrigated as needed using overhead sprinkler irrigation.
The phenotypic data were translated into present (1)/absent (0) data. Because thornlessness and primocane fruiting are conferred by recessive alleles (Scott et al. 1957; Lopez-Medina et al. 2000), progeny with those traits were given the “absent” (0) score, while thorny and floricane-fruiting individuals received the “present” (1) score. A Chi-square test was used to determine goodness of fit to the expected segregation ratio of 5:1 (5 thorny:1 thornless, and 5 floricane fruiting:1 primocane fruiting).
DNA extraction
DNA was extracted from young leaves harvested from each of the two parents and each of the progeny plants. DNA was extracted from the parents using a DNeasy Plant Maxi Kit (Qiagen, Valencia, CA) and from the progeny with a DNeasy 96 Plant Kit (Qiagen, Valencia, CA). DNA was quantified using the Spectramax 190 spectrophotometer (Molecular Devices, Sunnyvale, CA), and diluted with 1× TE buffer to 30 ng μl−1 for use in PCR.
Molecular markers
The 310 primer pairs tested for utility in the mapping population included 51 published SSR markers (Amsellem et al. 2001; James et al. 2003; Graham et al. 2004; Lewers et al. 2005), previously tested in blackberry by Stafne et al. (2005) and 259 SSR markers developed from a blackberry EST library (Lewers et al. 2008). For cost efficiency, the M13 sequence (5′TGTAAAACGACGGCCAGT3′) was added to the 5′ ends of the forward primers as described by Schuelke (2000), and separate M13 primers were labeled with the fluorescent proteins FAM, VIC, NED, and PET (Applied Biosystems, Foster City, CA, USA). PCR amplification was performed as described by Schuelke (2000). Samples for electrophoresis were prepared combining 1.5 μl of PCR amplification product 1:10 dilution 8.8 μl of Hi-Di formamide and 0.2 μl GS500 LIZ standard (Applied Biosystems) and denaturing for 5 min at 94 °C. PCR products were separated by capillary electrophoresis on an ABI 3730 DNA Analyzer (Applied Biosystems). The size standard used during electrophoresis was GENESCAN 500HD (LIZ). Sizing data were analyzed with GeneMapper 3.5 software (Applied Biosystems).
Prior to screening the progeny used for linkage analysis, the primer pairs were tested for amplification of a polymorphic product from the mapping parents. Because blackberry segregates tetrasomically, some SSR primers that did not detect polymorphisms between the parents would still segregate among the progeny in a way that would be useful for mapping with TetraploidMap software, so that parental screening was done first to ensure the primer pairs would amplify a product from at least one parent. For example, the primer pair EMFxaCAD1B generates a 285 base pair (bp) product that is present in both parents and segregates in a 3:1 ratio in the progeny. Hence, three types of polymorphism were potentially useful: products present in ‘APF-12’ and absent in ‘Arapaho’, products absent in ‘APF-12’ and present in ‘Arapaho’, and products present in both parents and segregating in the progeny.
To increase throughput, PCR reactions for markers detecting polymorphisms on the entire mapping population were generated using FAM, VIC, NED, and PET labels and pooled together after PCR so that amplification product sizes would not overlap. Preparation of PCR amplification products, capillary electrophoresis, and sizing data were done as described above. All PCR products (=peaks) were scored binarily as 1 (present) and 0 (absent). Initially, all PCR products from a primer pair were treated as co-dominant alleles.
Map construction
TetraploidMap software (Hackett and Luo 2003) was used to construct a genetic linkage map for each of the parents. Four main steps were carried out to construct the linkage map. (1) Analysis of single marker segregation. The “Findegno” subroutine from the TetraploidMap software gives the segregation type for each marker. It identifies the parental genotypes from the observed parent and offspring phenotypes at each locus, without or with double reduction. When the parental genotype identification could not be determined by the software, the PCR products amplified by a primer pair were re-scored as dominant markers. (2) Clustering into linkage groups. The “Cluster” subroutine was used to create, for each parent, groups of markers that do not segregate independently. (3) Estimation of recombination frequency between all pairs of markers within a linkage group (“Twopoint” subroutine). For each linkage group (LG), recombination frequencies and LOD scores were calculated between every pair of markers using the Expectation Maximization (EM) algorithm. (4) Ordering of the markers within each linkage group (“Simanneal” subroutine). A simulated annealing algorithm was used to identify the order with the minimum value of the weighted least squares criterion and to calculate map distances between the markers (Hackett and Luo 2003; Bradshaw et al. 2008; Hackett et al. 2007). Comparisons between the parental maps were performed manually using markers in common between new and named linkage groups (Bushakra et al. 2012; Graham et al. 2004, 2009; Sargent et al. 2007; Woodhead et al. 2008).
TetraploidMap software predicts the most probable genotypes of the parental lines at each of the markers based on marker phenotypes of the parents and their offspring. It also provides information about the most likely linkage phase (coupling or repulsion), but only between two markers. Each pair of markers has to be inspected directly, the phase has to be determined based on the information given for all the markers and manually inferred (see TetraploidMap for Windows manual), which can sometimes be tedious. Hence, to determine the linkage phase between each trait (thorny/thornless or floricane/primocane fruiting) and the markers linked to the trait, we manually tested different linkage phase possibilities, selecting the phase for each trait/marker pair or marker/marker pair which was supported by the observed segregation data. Alleles from the trait and the markers can be on the same or in different homologous chromosomes. If they are in the same homolog, they are in coupling, and if they are in different homologs, they are in repulsion. For the alleles in repulsion, there are different options; one allele is in one of the four homologous chromosomes and the other allele can be in any of the other three homologous chromosomes. To establish the phases, we located the alleles from the trait in the homologous chromosomes according to the genotype of each parent. Next, we randomly chose the location in the chromosomes of each marker considering their genotype. For example, marker M1 and M2 are both present as a single allele in one parent and absent in the other and they can be in the same or in different homologous chromosomes. Based on the location and on the genotype for the trait and the markers at each of the parents, we determined the gametes of each parent and the numbers of progeny expected for the possible genotypes among their progeny. These progeny genotypes were sorted into phenotypic groups by summing up the individuals with the same phenotype and were compared to the observed phenotypes. The Chi-square test was used to determine goodness of fit.
Results
Phenotypic segregation
Among the 188 progeny evaluated for the morphological traits thornlessness and primocane fruiting, 161 had thorns and 27 were thornless, and 154 were evaluated as floricane- and 34 as primocane fruiting. These data fit the expected segregation ratio 5:1 (presence vs. absence) for both traits (χ2 = 0.72, P = 0.39; χ2 = 0.27, P = 0.60 for thorny and floricane fruiting, respectively). This ratio corresponds to the segregation expected in duplex (AAaa) by nulliplex (aaaa) crosses, supports what is known from the pedigrees of the parents, and confirms that primocane fruiting and thornlessness in this population are each conferred by recessive alleles present in the respective parent expressing the trait as nulliplex.
Molecular marker analyses
A total of 310 primer pairs (including 51 SSRs selected from previous works and 259 from the blackberry EST-SSR library) were screened against the two parents of the mapping population (Table 1). The number of primer pairs detecting polymorphisms was around 30 %. One hundred and forty-three (46 %) did not amplify a PCR product or gave a non-specific amplification. Of the 167 (54 %) primer pairs that gave amplification, 74 (24 %) were monomorphic and 93 (30 %) were polymorphic between ‘APF-12’ and ‘Arapaho’. The 74 monomorphic primers were tested in the two parents plus six individuals from the population, and 25 of them showed one or more product sizes present in both parents but segregating in the population, consistent with tetrasomic segregation. In total, 107 primer pairs were used for genotyping the mapping population. These primers amplified up to 4 products per parent (except RH_MEa0007cG07 that amplified 5 products in ‘Arapaho’), with an average of 2.1 and 2.3 products from‘APF-12’ and ‘Arapaho’, respectively. Supplementary Table 1 supplies the details of these primer pairs (sequences and product sizes produced in each parent).
From the 107 primer pairs, 293 PCR products were scored, 261 of them detecting polymorphisms in the mapping population. For only 57 of these primer pairs, the parental genotypes could be determined from the segregation of their PCR products when read as co-dominant. For the remaining primer pairs, it was impossible to determine the parental genotype when the PCR products were treated as co-dominant, so each PCR product from these remaining primer pairs was employed separately as a dominant marker. Out of these 123 potential markers, the parental genotypes could be determined for 104, giving a total number of 161 markers to be used for map construction.
From the 57 polymorphic loci with co-dominant segregation, the average number of alleles per locus in both parents combined was 2.9. Most of the dominant markers segregated as double-simplex (3:1 ratio from two Aaaa parents), but a large number of markers had other segregation ratios, mainly single-simplex (1:1 from Aaaa × aaaa) and double-duplex (35:1 from two AAaa parents) genetic constituencies (Table 2). Among these markers, the percentage of markers with segregation ratios deviating significantly from the expected ratios (P < 0.05) reached 49 %, with 41 % of them showing a highly significant deviation (P < 0.001).
Marker segregation was analyzed to detect evidence of double reduction using TetraploidMap software, which includes a test for double reduction. It estimates the coefficient of double reduction (α) and calculates the significance of the test for double reduction (LR). An LR test lower than a χ2 with 1 degree of freedom (d.f.) is not significant at P = 0.05. For 41 loci, α was significantly different from zero, but the genotypes of the parents were also found under the absence of double reduction. The configuration of the parental genotypes that could have originated only from double reduction was not found. These would have been evidenced by combinations of alleles in some of the progeny that can only occur as a result of double reduction. The double reduction observed here is not different from segregation distortion.
Map construction
A total of 119 molecular markers and the two morphological traits, thorny and floricane fruiting, were used to construct a linkage map. All markers except those segregating in an 11:1 or 35:1 ratio and the highly distorted markers were used to build the map. Markers segregating in an 11:1 (Aaaa × AAaa) or 35:1 (AAaa × AAaa) ratio cannot be used by TetraploidMap. Highly distorted markers (P < 0.001) can be used, but we chose not to, because this is the first genetic map of blackberry. Cluster analysis of all markers where parental genotypes were identifiable generated nine linkage groups for ‘APF-12’ and eight for ‘Arapaho’.
Markers in common between the ‘APF-12’ and ‘Arapaho’ maps, the bi-parental markers, facilitated the identification of homologous linkage groups between both blackberry parental maps. Out of the 85 bi-parental markers, 83 appeared in the same linkage group in both parents. In addition, the linear order was maintained for over 69 % of these markers. LG3 consisted of the lowest number of markers following the same linear order in both parents. Three of the nine linkage groups can be paired by markers segregating in both parents, enabling us to connect these linkage groups so that there were seven linkage groups in each parent, a number consistent with the basic chromosome number of blackberry (Fig. 1). LGs 4 and 5 in ‘APF-12’ and LG5 in ‘Arapaho’ are divided in two parts.
Genetic linkage map of ‘APF-12’ × ‘Arapaho’ tetraploid blackberry. The number after the name of the markers indicates the molecular weight (in base pair) of the product sizes produced by the primer pairs treated as dominant. Boxed markers were present in the diploid raspberry map recently published by Bushakra et al. (2012) and used to name the linkage groups of the two blackberry parental maps
Markers in common between the ‘APF-12’ and ‘Arapaho’ maps, as well as published diploid raspberry maps, were used to identify and name the blackberry linkage groups. Linkage groups were compared to several previously published raspberry maps using the positions of SSR markers in common. Naming of LG1–LG6 followed that of the recently revised system for raspberry (Bushakra et al. 2012) using SSR markers in common between raspberry and blackberry, and LG7 was tentatively defined by default, as it was the only group without any markers in common with published raspberry maps. Table 3 presents the linkage groups in which SSR markers in common between blackberry and raspberry are located.
For ‘APF-12’, the map length was 768 cM, with 100 markers, 13 of which were unique to ‘APF-12’, and an average distance between markers of 7.7 cM. The map of ‘Arapaho’ covered 788 cM, with 99 markers positioned, nine of which were unique to ‘Arapaho’, and an average distance between markers of 7.9 cM. In both maps, the markers with segregation ratios that deviated significantly from expected were spread over the genome.
Markers linked to the traits
The two traits in this study were not linked to each other. The thornless/thorny trait, Locus S in blackberry, is on LG4, flanked by the markers RH_MEa0005aH07 and RH_MEa0011dG03b-265 at a distance of 4 and 21 cM. The primocane-/floricane-fruiting trait is on LG7, flanked by the markers RH_MEa0006aC04-175 and RH_MEa0007aG06-152 at a distance of 7 and 1 cM, respectively. This primocane-/floricane-fruiting locus is named in this work as the F Locus of blackberry.
The marker RH_MEa0005aH07 is the closest to the ‘APF-12’ thorny allele’s locus at 4 cM and is an EST-SSR without significant sequence similarity to any accession currently in GenBank. This co-dominant marker has two different alleles (RH_MEa0005aH07-144 and RH_MEa0005aH07-154), one (RH_MEa0005aH07-154) present in both parents and the other only present in ‘APF-12’. RH_MEa0005aH07-144 is linked in coupling to the thorny allele, meaning the amplification site for the PCR primer pair was on the same chromosome as the gene causing the dominant trait of thorny, while RH_MEa0005aH07-154 was linked in coupling to thornlessness (Fig. 2). When the marker data were compared to the phenotypic data, 23 out of the 24 individuals (96 %) carrying only the allele RH_MEa0005aH07-154 were thornless.
Possible location in the chromosomes of the marker linked to the S Locus controlling the trait thorny/thornless. ‘APF-12’ has a double dose for thorny (SSss), and thornless ‘Arapaho’ is nulliplex (ssss). B corresponds to the co-dominant marker RH_MEa0005aH07. The number after the name of the marker corresponds to the molecular weight of the allele in base pairs
The primocane-fruiting allele is linked in coupling with three markers, but for only one of the homologs containing the primocane-fruiting allele in each of the two parents (Fig. 3). The primocane-fruiting allele is flanked on one side by RH_MEa0006aC04-175, which is 7 cM away and has sequence similarity to a putative multi copper oxidase gene, and, on the other side, by RH_MEa0007aG06-149 and RH_MEa0014cG07-159, 14, and 71 cM away. Two of the homologs in ‘APF-12’ and one of the homologs in ‘Arapaho’ that contained the primocane-fruiting allele had no SSR markers associated with them, making them difficult to follow in breeding with the current level of marker saturation. The ‘Arapaho’ floricane-fruiting allele is in coupling with three SSR alleles, but for only one of the homologs, as no markers were linked in coupling with the other homolog containing a floricane-fruiting allele. The floricane-fruiting allele was 1 cM away from RH_MEa0007aG06-152, which has sequence similarity to a predicted E3 ubiquitin-protein ligase AIP2, an additional 6 cM away from marker RH_MEa0003bF12-148 and yet another 27 cM away from marker RH_MEa00a5cE06-264. These same three markers were linked in coupling with the primocane-fruiting allele in one of the four homologs of this linkage group in ‘APF-12’, but not the other three homologs, each containing the primocane-fruiting allele. This indicates a crossover event between these three markers and the locus determining floricane/primocane fruiting during meiosis in the generation of ‘APF-12’. Had these markers been used in marker-assisted selection in the process of breeding ‘APF-12’, this selection would have been discarded as highly unlikely to be primocane fruiting.
Possible location in the chromosomes of the markers linked to the trait floricane fruiting. ‘Arapaho’ has a double dose for floricane fruiting (FFff), and primocane-fruiting ‘APF-12’ is nulliplex (ffff). C, D, E, G, H, and I correspond to the markers RH_ME0006aC04-175, RH_ME0007aG06-152, RH_ME0003bF12-148, RH_ME0007aG06-149, and RH_ME0015cE06-264 RH_ME0014cG07-159, respectively. The number after the name of the marker corresponds to the molecular weight of the allele in base pairs
Discussion
The objective of this study was to develop a genetic map of blackberry as a first step toward marker-assisted breeding of thornless primocane-fruiting blackberry cultivars. The map presented here is the first tetraploid genetic linkage map based on a cross between tetraploid blackberry cultivars (‘APF-12 and ‘Arapaho’) and was developed assuming tetrasomic inheritance. Genetic segregation in autopolyploids is a reflection of meiosis with a combination of bivalent and multivalent pairing with multiple alleles per locus (de Winton and Haldane 1931). In this case, all homologous chromosomes can pair during meiosis resulting in tetrasomic inheritance. Tetraploids can also be the result of the union of the genomes of two different diploid species and subsequent doubling of chromosomes resulting in so-called allotetraploids (Ronfort et al. 1998). If the two parental genomes are sufficiently dissimilar, then, in meiosis, often only pairing of homologous chromosomes occurs and not of homoeologous chromosomes, and there is no multivalent formation. In such types of tetraploids, the mode of inheritance will be disomic. If there is some degree of pairing between homoeologous chromosomes (Sybenga 1996), the mode of inheritance will be intermediate between disomic and tetrasomic. With dominant markers, segregation patterns fitting the 1:1 and 3:1 ratios are expected whether the parental genome constitution is of the autotetraploid or allotetraploid type. In this study, a number of genomic loci showing segregation patterns detectable uniquely under tetrasomic inheritance (e.g., 5:1, 11:1 and 35:1) were also found, supporting tetrasomic inheritance in blackberry.
Evidence of double reduction was also analyzed. Double reduction is a phenomenon associated with multivalent formation in meiosis and refers to the fact that parts of sister chromatids come together in the same gamete during the second meiotic division. For it to occur, multivalent pairing must take place with a crossover between a locus and its centromere followed by the two pairs of chromatids passing to the same pole in anaphase I. Double reduction creates new types of gametes, modifies the probabilities of each type of gamete in a segregating population (Bever and Felber 1992), and is typical for autopolyploids (Bradshaw and Mackay 1994; Ronfort et al. 1998; Stift et al. 2010). The double reduction observed in this work is not different from segregation distortion. Similar results also were reported in alfalfa (Julier et al. 2003) and do not exclude the possibility of double reduction in blackberry.
In this study, the percentage of primer pairs detecting polymorphisms (30 %) agreed with what was expected (Lewers et al. 2008). This percentage is lower than reviews reporting ranges of 80–90 % (Ellis and Burke 2007), but most studies test new primer pairs on several different accessions of the same species instead of parents intended for linkage mapping. Some of these markers showed segregation ratios deviating significantly from the expected ratios. Such deviation is of interest, because, if linked, such markers may indicate preferential transmission due to an allele in the linkage group conferring seedling lethality or reduced vigor, or, in a tetrasomic species, a certain degree of pairing preference (Ronfort et al. 1998). However, these markers were spread over the genome, failing to support the supposition that the deviation was from lethal alleles or pairing preference.
As mentioned in “Results”, the blackberry linkage groups were compared to previously published raspberry maps using SSR markers in common. The blackberry map and the Bushakra et al. (2012) raspberry map have nine markers in common: EMFxaCAD1B, Rubus285a, Rubus126b, mRaCIRRIV2A8, Rubus117b, RH_MEa0007aB01, Rubus2a, EMFXaACO1B, and RH_MEa0001dD07. Several markers employed in mapping blackberry (Rubus237b, Rubus262a, Rubus24a, Rubus251a, Rubus107a, Rubus270a, Rubus145a, Rubus110a, Rubus24a, Rubus45c, Rubus98d, Rubus275a, Rubus277a) were not present in the maps developed by Bushakra et al. (2012), but were located in previous raspberry maps (Graham et al. 2004, 2009; Sargent et al. 2007; Woodhead et al. 2008). Most of them linked together in these raspberry maps and further supported the naming of the blackberry linkage groups. An exception was Rubus285a, which was on LG1 in the Bushakra et al. (2012) raspberry map, but is in LG3 in this blackberry map. This is because Rubus285a mapped with the raspberry-LG3 markers Rubus24a, Rubus251a, Rubus107a, Rubus270a, and Rubus194h. These markers have mapped together in previous works (Graham et al. 2004; Woodhead et al. 2008), supporting the naming of this group as LG3 for blackberry.
The two traits, thorny and primocane fruiting, mapped in this study were not linked to each other. This was suspected, based on experience with breeding progeny, but had not previously been supported with genetic segregation data. Independent segregation of these traits will make the task of combining them in breeding progeny simpler than if they had been linked in repulsion. In raspberry, Graham et al. (2004) mapped the spines trait on LG2 (which corresponds with LG3 in this work). Later research on the same population (Graham et al. 2006) also mapped this trait on LG2, close to gene H, which determines cane pubescence, and LG3 (which corresponds with LG6 in this work). More recently, Molina-Bravo (2009) reported two QTLs, located on LG7 and LG3, for spine density in a different population of raspberry. The QTL on LG7 explained the largest percentage of the phenotypic variation (around 75 %). LG7 corresponds with the LG4 in our linkage map, providing an interesting comparison for future research between a genomic region in blackberry conferring presence/absence of thorns versus a similar genomic region in controlling spine density in raspberry. There are several major genes conferring the spineless phenotype in raspberry: s (Lewis 1939), Sf (Rosati et al. 1986), STE (Hall et al. 1986), and SfL (McPheeters and Skirvin 1983; Rosati et al. 1988). A number of other genes are known to promote or suppress spines in raspberry (Jennings 1988). These different genes could provide potential genetic explanations for the different QTLs found in the multiple raspberry mapping studies. Further research will give insight into the genetic control of this trait in Rubus.
Because selection of thornless breeding progeny can be done at the seedling stage based on the absence of cotyledon marginal hairs, markers linked to the blackberry S Locus may seem to have limited value in marker-assisted breeding, but it should be remembered that these markers may still have value in selecting against other undesirable traits. Strong linkage has been reported between the s allele for thornlessness in ‘Merton Thornless’ and trailing growth habit, late harvest season, and acid fruit (Moore 1984). The University of Arkansas breeding program reported difficulty combining thornlessness and erect plant habit (Clark et al. 2007), so these molecular markers associated with thornlessness and new ones associated with upright plant habit may help in future efforts.
Primocane fruiting in raspberry was recently mapped to raspberry LG7 (Spencer 2012). That work reported the primocane-fruiting trait in the same linkage group where Molina-Bravo (2009) located a QTL for spine density in a different population of raspberry. According to Bushakra et al. (2012), this linkage group corresponds with our LG4. However, as previously mentioned, the two traits in this study were not linked to each other. Rubus116a and Rubus26a were the only SSR mapped on LG7 by Spencer (2012). These SSRs were not used in our map because they did not amplify products from either ‘APF-12’ or ‘Arapaho’. So, it was not possible to determine if these SSRs might be in a different linkage group, which would indicate a possible chromosomal rearrangement in blackberry. On the other hand, Spencer (2012) suggested that there were three genes for the primocane-fruiting trait in raspberry: one which controls the primocane fruiting ability and two which control whether fruits appear on the tips and/or lateral branches of the canes. In this blackberry study, only the ability to produce fruit on primocanes was studied and not the modifying traits. It is possible, then, that qualitative mapping of both the main trait and its modifiers may be possible with one or more populations designed toward this objective.
How the information from this study should be used may vary depending on the breeding population, the user, and the resources available, but the following is a suggestion. Because the two traits were found to be unlinked, thornless progeny can be identified most efficiently in the way it currently is, by identification of seedlings lacking marginal hairs on the cotyledons. Using the markers found in this study to be linked to primocane fruiting will require preliminary work with parental genotyping. To prepare for marker-assisted breeding, leaf tissue should be collected from all parents for each generation of parents and dried for later DNA extraction. It is presumed that data will have been collected for multiple traits, including thorn production and floricane/primocane fruiting, before selecting the genotype as a parent. To maintain genetic diversity, parents should be selected for complementary desired traits and not as candidates for marker-assisted selection. Once the parents for each cross are chosen, they would be evaluated to determine if their primocane-fruiting progeny could be identified through the use of the markers linked to the locus in this map.
For example, one of the homologs in ‘Arapaho’ associated with the primocane-fruiting allele is traceable by molecular markers (Fig. 3) and so is one of the homologs associated with floricane fruiting. If ‘Arapaho’ is scheduled to be cross-pollinated with a primocane-fruiting selection that does not have any of the markers associated with the ‘Arapaho’ floricane homolog, then at least half the floricane-fruiting progeny could be eliminated from the resulting family. When it is known that both floricane-associated homologs of a parent are labeled with these markers, then all of the floricane-fruiting progeny could be eliminated.
It is more likely that the primocane-fruiting ‘APF-12’ and other primocane-fruiting genotypes would be used as parents with floricane-fruiting genotypes with no primocane fruiting alleles, and then backcrosses would be made to a primocane-fruiting parent to obtain primocane-fruiting progeny. Progeny from the first cross in the process (with a floricane-fruiting parent) could be selected with either set of markers known to be associated with a primocane-fruiting allele. These progeny (FFff) could in turn be paired with parents that either contain none of the markers associated with the primocane-fruiting allele in the FFff genotypes or with parents known to contain the same markers linked in coupling with the primocane-fruiting allele. At first, the markers would only be useful in increasing the number of primocane-fruiting progeny from which to select other desirable traits, but in a few generations, the primocane-fruiting alleles of all the primocane-fruiting breeding parents would have known associations with molecular markers and could be followed in future families. This would be true, though in some families primocane fruiting would be associated with one set of markers and in others with a different set of markers.
Use of marker-assisted selection without knowledge of markers associated with all four homologs has the potential to reduce genetic diversity in this linkage group to the homologs currently associated with one or the other set of three markers. To help prevent a genetic bottleneck caused by selection for primocane fruiting, additional marker saturation in the area is needed until all four homologs of LG7 can be traced with molecular markers around the F Locus before marker-assisted selection is employed. Few of the SNP markers used to saturate the genetic map of diploid red raspberry (Ward et al. 2013) will be of direct use in tetraploid tetrasomic blackberry, but they will be of tremendous value in developing the R. idaeus genome sequence from which additional markers for blackberry can be designed.
Low marker saturation in this first molecular marker map of blackberry may also be the reason that some of the linkage groups identified in this study (LG4 and LG5 in ‘APF-12’ and LG5 in ‘Arapaho’) are divided into two groups. Another less likely but still plausible explanation is that they represent different arms of their respective chromosomes. As additional markers are added to the blackberry map, and maps additional to this first map are developed, especially maps using different parents, it is expected that, instead of seven linkage groups, twice that number eventually will become evident, representing each arm of the seven chromosomes, due to double reduction.
References
Amsellem L, Noyer JL, Le Bourgeois T, Hossaert-McKey M (2001) Isolation and characterization of polymorphic microsatellite loci in Rubus alceifolius Poir. (Rosaceae), an invasive weed in La Reunion Island. Mol Ecol Notes 1:33–35
Bever JD, Felber F (1992) The theoretical population genetics of autopolyploidy. Oxf Surv Evol Biol 8:185–217
Bradshaw JE, Mackay GR (1994) Breeding strategies for clonally propagated potatoes. In: Bradshaw JE, Mackay GR (eds) Potato genetics. CAB International Wallingford, UK, pp 467–497
Bradshaw JE, Hackett CA, Meyer RC, Milbourne D, McNicol JW, Phillips MS, Waugh R (1998) Identification of AFLP and SSR markers associated with quantitative resistance to Globodera pallid (stone) in tetraploid potato (Solanum tuberosum subsp. tuberosum) with a view to marker-assisted selection. Theor Appl Genet 97:202–210
Bradshaw JE, Hackett CA, Pande B, Waugh R, Bryan GJ (2008) QTL mapping of yield, agronomic and quality traits in tetraploid potato (Solanum tuberosum subsp. tuberosum). Theor Appl Genet 116:193–211
Bushakra JM, Stephens MJ, Atmadjaja AN, Lewers KS, Symonds VV, Udall JA, Chagne D, Buck EJ, Gardiner SE (2012) Construction of black (Rubus occidentalis) and red (R. idaeus) raspberry linkage maps and their comparison to the genomes of strawberry, apple and peach. Theor Appl Genet. doi:10.1007/s00122-012-1835-5
Clark JR, Perkins-Veazie P (2011) Prime-Ark® 45 primocane-fruiting blackberry. HortScience 46:670–673
Clark JR, Moore JN, Lopez-Medina J (2004) Primocane-fruiting blackberry cultivar releases from the University of Arkansas. HortScience 39:662
Clark JR, Moore JN, Lopez-Medina JL (2005) ‘Prime-Jan’ (‘APF-8’) and ‘Prime-Jim’ (‘APF-12’) Primocane-fruiting blackberries. HortScience 40:852–855
Clark JR, Stafne ET, Hall HK, Finn CE (2007) Blackberry breeding and genetics. Plant Breed Rev 29:19–144
de Winton D, Haldane JBS (1931) Linkage in the tetraploid Primula sinensis. J Genet 24:121–124
Ellis JR, Burke JM (2007) EST-SSRs as a resource for population genetic analyses. Heredity 99:125–132
Gar O, Sargent DJ, Tsai C-J, Pleban T, Shalev G et al (2011) An autotetraploid linkage map of rose (Rosa hybrida) validated using the strawberry (Fragaria vesca) genome sequence. PLoS ONE 6(5):e20463. doi:10.1371/journal.pone.0020463
Graham J, Smith K, MacKenzie K, Jorgenson L, Hackett C, Powell W (2004) The construction of a genetic linkage map of red raspberry (Rubus idaeus subsp. idaeus) based on AFLPs, genomic-SSR and EST-SSR markers. Theor Appl Genet 109:740–749
Graham J, Smith K, Tierney I, MacKenzie K, Hackett C (2006) Mapping gene H controlling cane pubescence in raspberry and its association with resistance to cane botrytis and spur blight rust and cane spot. Theor Appl Genet 112:818–831
Graham J, Hackett C, Smith K, Woodhead M, Hein I, McCallum S (2009) Mapping QTLs for developmental traits in raspberry from bud break to ripe fruit. Theor Appl Genet 118:1143–1155
Hackett CA, Luo ZW (2003) TetraploidMap: construction of a linkage map in autotetraploid species. J Hered 94:358–359
Hackett CA, Wachira FN, Paul S, Powell W, Waugh R (2000) Construction of a genetic linkage map for Camellia sinensis (tea). Heredity 85:346–355
Hackett CA, Bradshaw JE, McNicol JW (2001) Interval mapping of quantitative trait loci in autotetraploid species. Genetics 159:1819–1832
Hackett CA, Milne I, Bradshaw JE, Luo Z (2007) TetraploidMap for windows: linkage map construction and QTL mapping in autotetraploid species. J Hered 98:727–729
Hall HK, Cohen D, Skirvin RM (1986) The inheritance of thornlessness from tissue culture derived ‘Thornless Evergreen’ blackberry. Euphytica 54:891–898
James CM, Wilson F, Hadonou AM, Tobutt KR (2003) Isolation and characterization of polyploid microsatellites in diploid strawberry (Fragaria vesca L.) for mapping, diversity studies and clone identification. Mol Ecol Notes 3:171–173
Jennings DL (1988) Raspberries and blackberries: their breeding, diseases and growth. Academic Press, London
Julier B, Flajoulot S, Barre P, Cardinet G, Santoni S et al (2003) Construction of two genetic linkage maps in cultivated tetraploid alfalfa (Medicago sativa) using microsatellite and AFLP markers. BMC Plant Biol 3:9. http://www.biomecentral.com/1471-2229/3/9
Lewers KS, Styan SMN, Hokanson SC, Bassil NV (2005) Strawberry GenBank-derived and genomic simple sequence repeat (SSR) markers and their utility with strawberry, blackberry, and red and black raspberry. J Am Soc Hortic Sci 130:102–115
Lewers KS, Saski CA, Cuthbertson BJ, Henry DC, Staton ME, Main DS, Dhanaraj AL, Rowland LJ, Tomkins JP (2008) A blackberry (Rubus L.) expressed sequence tag library for the development of simple sequence repeat markers. BMC Plant Biol 8:69–76
Lewis D (1939) Genetical studies in cultivated raspberries. I. Inheritance and linkage. J Genet 38:367–379
Lopez-Medina J, Moore JN, McNew RW (2000) A proposed model for inheritance of primocane fruiting in tetraploid erect blackberry. J Am Soc Hortic Sci 125:217–221
Luo ZW, Hackett CA, Bradshaw JE, McNicol JW, Milbourne D (2001) Construction of a genetic linkage map in tetraploid species using molecular markers. Genetics 157:1369–1385
McPheeters K, Skirvin RM (1983) Histogenic layer manipulation in chimeral ‘Thornless Evergreen’ trailing blackberry. Euphytica 32:351–360
Molina-Bravo R (2009) Genetic and quantitative analysis of red raspberry (Rubus idaeus) for heat tolerance and longer chilling requirement. Dissertation, North Carolina State University
Moore JN (1984) Blackberry breeding. HortScience 19:183–185
Ronfort J, Jenczewski E, Bataillon T, Rousset F (1998) Analysis of population structure in autotetraploid species. Genetics 150:921–930
Rosati P, Gaggioli D, Giunchi L (1986) Genetic stability of micropropagated Loganberry plants. J HortSci 61:33–41
Rosati P, Hall HK, Jennings DL, Gaggiolo D (1988) A dominant gene for thornlessness obtained from the chimera thornless Loganberry. HortScience 23:899–902
Sargent DJ, Fernandez-Fernandez F, Rys A, Knight VH, Simpson DW, Tobutt KR (2007) Mapping of A1 conferring resistance to the aphid Amphorophora idaei and dw (dwarfing habit) in red raspberry (Rubus idaeus L.) using AFLP and microsatellite markers. BMC Plant Biol 7:15
Schuelke M (2000) An economic method for the fluorescent labeling of PCR fragments: a poor man’s approach to genotyping for research and high-throughput diagnostics. Nat Biotech 18:233–234
Scott DH, Darrow GM, Ink DP (1957) Merton Thornless as a parent in breeding thornless blackberries. Proc Am Soc Hortic Sci 69:268–277
Sledge MK, Ray IM, Jiang G (2005) An expressed sequence tag SSR map of tetraploid alfalfa (Medicago sativa L.). Theor Appl Genet 111:980–992
Spencer JA (2012) Molecular marker analysis of primocane-fruiting traits in raspberry. MS Thesis, North Carolina State University, Raleigh
Spigler R, Lewers K, Johnson A, Ashman T-L (2010) Comparative mapping reveals autosomal origin of sex chromosome in octoploid Fragaria virginiana. J Hered 101:S107–S117
Stafne ET, Clark JR, Weber CA, Graham J, Lewers KS (2005) Simple sequence repeat (SSR) markers for genetic mapping of raspberry and blackberry. J Am Soc Hortic Sci 130:722–728
Stift M, Reeve R, van Tienderen PH (2010) Inheritance in tetraploid yeast revisited: segregation patterns and statistical power under different inheritance models. J Evol Biol 23:1570–1578
Sybenga J (1996) Chromosome pairing affinity and quadrivalent formation in polyploids: do segmental allopolyploids exist? Genome 39:1176–1184
Ward JA, Bhangoo J, Fernández-Fernández F, Moore P, Swanson JD, Viola R, Velasco R, Bassil N, Weber CA, Sargent DJ (2013) Saturated linkage map construction in Rubus idaeus using genotyping by sequencing and genome-independent imputation. BMC Genomics 14:2
Woodhead M, McCallum S, Smith K, Cardle L, Mazzitelli L, Graham J (2008) Identification, characterisation and mapping of simple sequence repeat (SSR) markers from raspberry root and bud ESTs. Mol Breed 22:555–563
Wu KK, Burnquist W, Sorrells ME, Tew TL, Moore PH, Tanksley SD (1992) The detection and estimation of linkage in polyploids using single-dose restriction fragments. Theor Appl Genet 83:294–300
Acknowledgments
P. Castro is grateful to Andalusian Institute of Agricultural Research and Training (IFAPA) and European Social Fund (ESF) for a postdoctoral fellowship and the hosting institution of the fellowship, USDA-ARS-GIFVL. This project was partially funded by USDA-ARS Project 1245-21220-185-00 and by the University of Arkansas fruit breeding program. Mention of trade names or commercial products in this publication is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the U.S. Department of Agriculture or any of the other agencies involved in this research.
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical standards
The experiments of this study comply with the current laws of USA.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by D. A. Lightfoot.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Castro, P., Stafne, E.T., Clark, J.R. et al. Genetic map of the primocane-fruiting and thornless traits of tetraploid blackberry. Theor Appl Genet 126, 2521–2532 (2013). https://doi.org/10.1007/s00122-013-2152-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00122-013-2152-3