Abstract
A recombinant inbred line (RIL) population, comprising 181 lines derived from ILC588 × ILC3279, was evaluated in 10 environments across three locations with different moisture gradients. A drought resistance score (DRS) and three phenology traits—plant height (PLHT), days to flowering (DFLR), and days to maturity (MAT)—were recorded along with seven yield-related traits—grain yield (GY), biological yield (BY), harvest index (HI), the number of pods/3 plants (Pod), percentage of empty pods (%Epod), 100 seed weight (100 sw), and seed number/3 plants (SN). Two RILs (152, 162) showed the best GYs and DRSs under stressed and non-stressed environments. The quantitative trait loci (QTLs) analyses detected 93 significant QTLs (LOD ≥ 2.0) across the genome × environment interactions. The highest phenotypic variation (>24 %) was explained by the QTLDFLR in Terbol-11. Four common possible pleiotropic QTLs on LG3 and LG4 were identified as associated with DFLR, DRS, GY, MAT, HI, SN, and Pod. No significant epistatic interactions were found between these QTLs and the other markers. However, the QTL for DRS was detected as a conserved QTL in three late planting environments. The markers H6C-07 (on LG3) and H5G01 (on LG4) were associated with QTLs for many traits in all environments studied except two. The allele ‘A’ of marker H6C07 (from the tolerant parent ILC588) explained 80 % of the yield increase under late planting and 29.8 % of that under dry environments. Concentrating on LG3 and LG4 in molecular breeding programs for drought could speed up improvement for these traits.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Drought is the most economically important abiotic constraint to crop production in the world (Boyer 1982). Drought can be defined as below normal precipitation that limits plant productivity (Kramer and Boyer 1995). Chickpea is a member of the Leguminosae family, which includes 18,000 species, grouped into 650 genera, grown in the semi-arid regions of the world (Polhill and Raven 1981). It is a major pulse crop in South Asia, the Middle East, East Africa, the western Mediterranean, Australia, and Mexico. Chickpea and other pulse crops, such as lentil (Lens culinaris Medik.), dry pea (Pisum sativum L.), and dry bean (Phaseolus vulgaris L.), are a major source of protein in human diets, particularly in low-income countries. In addition to being a major source of protein for human food in the semi-arid tropical regions, the chickpea crop plays an important role in the maintenance of soil fertility, particularly in the dry, rain-fed areas (Saxena 1990; Katerji et al. 2001). The time of sowing and the photoperiod vary among these regions, but generally most of the precipitation is received before or during the early crop season and the crops matures under progressively declining soil moisture and increasing temperature. The crop is a self-pollinated diploid (2x = 2n = 16 chromosomes) with a relatively small genome size of around 740 Mbp (Arumuganathan and Moss 1991). Chickpea faces diverse environments in these and other production areas in terms of the photoperiod, temperature, and precipitation, all of which have a profound effect on growth and development (Khanna-Chopra and Sinha 1987).
Chickpea frequently suffers from drought stress towards the end of the growing season under rain-fed conditions. Ninety percent of the world’s chickpea is produced in areas relying upon a decreasing, conserved soil moisture. Therefore, crop productivity is largely dependent on the efficient use of the available soil moisture (Kumar and Van Rheenen 2000). Although chickpea is known as being more drought tolerant than most of the other cool season legumes, drought still reduces yields and can even lead to total crop failure. In both Mediterranean and sub-tropical climates, chickpea often suffers from terminal drought, which delays flowering and seed filling, leading to a low seed yield (Turner et al. 2001). Plants adapt to drought stress either by escape, avoidance, or tolerance mechanisms. It is reported that chickpea adapts by developing a deep root system (Kashiwagi et al. 2005).
Conventional breeding methodologies for improving drought tolerance in crop plants are primarily based on screening germplasm and an empirical selection for GY under different drought stress conditions over several years. Because of the variability in drought patterns from year to year, and to overcome the low response to direct selection, substantial efforts have targeted the manipulation of morpho-physiological traits influencing drought resistance through escape, avoidance, and/or tolerance mechanisms (Ludlow and Muchow 1990; Blum 1996).
A deep root system capable of extracting additional soil moisture should positively affect yield under drought stress environments. A set of 257 RILs was developed from the cross of Annigeri × KC4958 at the International Crops Research Institute for the Semi-Arid Tropics (ICRISAT) and evaluated to identify molecular markers for root traits. A QTL, flanked by markers TAA170 and TR55 on LG4A, was identified for root length (Chandra et al. 2003), however, the yield advantage of a deep root system is still under investigation. A few researchers have studied the genetic basis of the flowering time of chickpea. Or et al. (1999) suggested a major photoperiod response gene (Ppd) affecting time to flowering. Cho et al. (2002) identified a single QTL for days to 50 % flowering on LG3. Lichtenzveig et al. (2006) identified two QTLs on LG3 and LG4 linked to time to first flower. Cho et al. (2002) also identified a QTL for seed weight on LG4 accounting for 52 % of the total phenotypic variation. These reports generated information on the QTLs of important traits measured under different conditions to establish their usefulness for stress breeding in chickpea. The basis of further development could lie in the integration of physiology, biotechnology, and plant breeding (Blum and Nguyen 2004).
At ICARDA, the principal criterion used to breed for drought tolerance is based on yield performance (empirical selection) under terminal drought and heat stress. The methodology involves late planting chickpea (around 20 March), as under Mediterranean environments the chances of rainfall occurring after late March are minimal. This exposes the crop to terminal drought and heat because as the season progresses, conserved soil moisture recedes and the temperature increases. This methodology led to the release of varieties which survived severe drought. For example, the variety Gökçe, released in Turkey, survived severe drought in 2006–07 when most of the crops, including wheat, were completely devastated. Therefore, in this study, we have attempted to understand the genetic basis of drought tolerance in elite genotypes developed for dry environments, by combining several phenological/morphological traits in varied environments and using molecular marker analysis to identify and discern QTLs related to drought tolerance in chickpea.
Materials and methods
Plant materials and DNA isolation
A mapping population of 181 (F8) RILs was used in this study. They were derived from an intra-specific cross between drought tolerant chickpea genotype ILC588 and a drought susceptible cultivar ILC3279. The single seed descent (SSD) method was used to advance the lines from F2 to F7. The DNA was extracted from the young leaves of four to 6 week old seedlings at the F8 generation. The DNA was extracted from five seedlings using the cetyltrimethylammonium bromide (CTAB) method, as described by Rogers and Bendich (1985). In brief, fresh leaf material from seedlings was frozen in liquid nitrogen and ground into a fine powder, which was subsequently added to a 2 mL Eppendorf tube with 1 mL pre-warmed 2 × CTAB buffer—2 % CTAB, 0.1 M Tris–HCl (pH 8.0), 1.4 M NaCl, 20 mM ethylenediaminetetraacetic acid (EDTA). The suspension was mixed and incubated at 65 °C for 30 min. The suspension was cooled at room temperature (RT) for 5 min, 1 mL chloroform-isoamyl alcohol (24:1) was added to the tube and the suspension gently mixed by shaking for 10 min. The suspension was centrifuged at 4,500 rpm (Beckmann YA-12) for 20 min at RT and the supernatant transferred to a new tube. The DNA was precipitated with 700 μL of cold isopropanol. The DNA was transferred into a micro-centrifuge tube and washed twice with a washing buffer (75 % ethanol and 200 mM sodium acetate) for 20 min. After air-drying for about 10–20 min, the DNA was dissolved in 100 μL of 1 × TE buffer—10 mM Tris–HCl, pH 8.0, 1 mM EDTA. The sequences of chickpea primers were obtained from the literature (Winter et al. 2000; Lichtenzveig et al. 2005; Gujaria et al. 2011). The PCR mixture (20 μL) contained 10 ng genomic DNA and 10 pmol of each forward and reverse primer, 0.2 mM dNTP, 1 × PCR buffer and 1 U Taq DNA polymerase (Invitrogen, Carlsbad, Calif.) Polymerase chain reaction (PCR) was performed in a thermocycler (ABI-GeneAmp® 2720) with the following profile: an initial denaturing cycle of 94 °C for 3 min followed by 35 cycles of 94 °C for 15 s (denaturation), a specific temperature depending upon the primer pair for 15 s (annealing) and 72 °C for 30 s (extension); followed by a final extension at 72 °C for 5 min. The PCR products were electrophoresed on 8 % polyacrylamide gel and stained by ethidium bromide as described by supplier (Electrophoresis User Guide-Invitrogen).
Environments/locations
Three different environments used in this study were Terbol (Lebanon); Tel Hadya (TH), Aleppo, Syria, and Breda, Syria. These locations were varied in their moisture gradients and the long-term annual average rainfalls are Terbol, 450 mm; TH, 350 mm; and Breda, 250 mm. At each location the lines were grown in multiple row plots and laid out in an alpha lattice design,—plot length 5 m, and spacing between rows 0.2 m—and two replications were used in all trials. Planting dates, years, and locations are summarized in Table 1.
Phenotyping in stressed and non-stressed environments
A RIL population, comprising 181 lines, was evaluated in 10 environments that were across 4 years and included early and late plantings (Table 1). The following traits were measured under both stressed and non-stressed environments as reported by Canci and Toker (2009): The DRS was estimated by using a rating scale ranging from 1 (plants with 95–100 % pod setting) to 9 (plants did not set any pods and died). The PLHT was measured as the average length of the plants from the ground to the top extremity of the plant at maturity. The DFLR was measured when 50 % of the plants had set the first flower. The MAT was recorded as the number of days after sowing when 95 % of the pods had reached physiological maturity. Five plants per plot were harvested and dried to a moisture content of about 14 % when their yield and yield components, including biomass, were recorded. The HI was measured as the GY divided by the BY. The whole plot yield was combined with the yield per five plants to obtain the total yield per genotype in high rainfall environments.
Statistical analysis
A correlation matrix between the DRS and yield components was calculated using the means of each trait across all environments including drought stress (rainfall <25 mm; TH-Dr08 and TH-Dr10), and non-stress environments (rainfall >400 mm; Terbol09 and Terbol-11). The correlation was performed by using GenStat 12th edition. An analysis of variance (ANOVA) was performed on the phenotype data collected from the different environments using GenStat 12th edition. GenStat was used to perform an additive main effects and multiplicative interactions (AMMI) analysis on the values of the DRSs obtained per plot across environments. The plot genotype and environment means were calculated against four interaction principal component axes (IPCAs). The AMMI model is described by Duarte and Vencovsky (1999) as follows:
where Y ij is the mean response of genotype i in environment j; μ is the overall mean of the test; g i is the fixed effect of genotype i (i = 1, 2,…g); a j is the fixed effect of environment j (j = 1, 2,…a); λk is the singular value of the kth IPCA, (k = 1, 2,…p, where p is the maximum number of estimable principal components); α ik is the singular value of the ith genotype in the kth IPCA; y jk is the singular value of the jth environment in the kth IPCA; r ij is the residue of the GEI or AMMI residue (data noise); k is the characteristic non-zero root, k = [1, 2,…min(g−1, e−1)].
After selecting the AMMI model the stability and adaptability were studied by biplot graphs obtained from combinations of the orthogonal IPCAs.
Mapping was performed using the JoinMap 4® software (Van Ooijen 2006). Loci were assigned to linkage groups based on a logarithm of odds score of greater than three (LOD > 3) and a recombination frequency less than 0.45. Map distances (centimorgan—cM) were calculated using the Kosambi mapping function (Kosambi 1944). QTL analysis (including the percentage explanation for variability and the additive effects) was performed using the MapQTL 6® software (Van Ooijen 2009). The significance of the QTL was estimated (LOD threshold ≥2) from a 2,000 permutation test by random sampling of phenotypic data. Epistasis and a two-locus model of epistatic gene interaction were carried out using the QTL IciMapping v3.1 software, which provided an improved statistical method for inclusive composite interval mapping-epistatic mapping (ICIM-EPI) showing the significant interactions between identified QTLs (Li et al. 2007).
Results
Phenotypic characterization
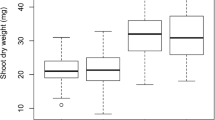
Four years of data (2008–2011) from three different locations, with different planting dates representing diverse environments receiving different rainfalls, were used in this study. The rainfall after the planting experiments ranged from 14.6 mm (late planting season 2008) in TH up to 594.7 mm in Terbol in 2011 (Table 2). In this study, 12 agronomic and morphological traits, including DRS, were measured for 181 RILs of a chickpea population derived from a cross between the drought tolerant parent, ILC588, and the drought sensitive one, ILC3279. The ANOVA showed significant differences (P < 0.001) for all the traits studied among the 181 RILs and the 10 environments used in this study (ANOVA tables are not shown). However, the mean values of these traits varied significantly among the ten environments (Fig. 1). These traits showed a good fit to the normal distribution in the population studied (data not shown). To study the interactions between the environments and drought tolerance, the estimated DRS, which showed significant negative correlation with GY (r = −0.65***), was selected as a major trait related to drought to be further analyzed by an ANOVA using the AMMI model.
Means with standard error of the data for drought agronomic and morphological traits of chickpea population ILC588 × ILC 3279 at 10 environments under drought in Syria during the 4 years 2008–2011. Horizontal axis (1–10) represents environments (1:TH-cont08, 2:TH-Dr08, 3:TH-cont09, 4:TH-Dr09, 5:Breda-09, 6:Terbol-09, 7:TH-con-10, 8:TH-Dr10, 9:Breda-10, 10:Terbol-11).). Vertical axis represented traits as following: DRS drought resistance score (1-9); PLHT plant height (cm), DFLR days to flowering (days), MAT days to maturity (days); GY grain yield (g); BY biological yield (g), HI harvest index, Pod pod number (pods), Epod empty pods (pods); %Epod percentage of empty pods (%); (100sw)100 seed weight (g); and SN seed number (seeds)
The AMMI ANOVA showed that 45.2 % of the total sum of squares was attributable to environmental effects, only 7.9 % to genotypic effects, and 17.9 % to genotype × environment interaction (GEI) effects (Table 3). A large sum of squares for environments indicated that the environments were diverse, with the large differences between the environmental means causing most of the variation in the DRS values. The magnitude of the GEI sum of squares was almost 2.3 times larger than that for genotypes, indicating that there were substantial differences in genotypic responses across environments. Results from the AMMI analysis also showed that the first principal component axis (PCA 1) of the interaction captured 39 % of the interaction sum of squares. Similarly, the second principal component axis (PCA 2) explained a further 18 % of the GEI sum of squares. Furthermore, PCA 1 and PCA 2 had sums of squares (1,752) greater than that for genotype (1,354). The mean squares for the PCA 1 and PCA 2 were significant at P < 0.001 and cumulatively contributed around 57 % of the total GEI. Therefore, the postdictive evaluation using an F-test at P < 0.001 suggested that two principal component axes of the interaction were significant for the model with 374 degrees of freedom. However, the interaction of the 181 lines with the 10 environments was best predicted by the first two principal components of genotypes and environments.
The AMMI model 2 biplot of the 10 environments is illustrated in Fig. 2. The late planted drought environments TH-Dr08 and TH-Dr10 were distributed evenly in quadrant IV and these received only 23.5 and 14.6 mm rainfall after planting respectively. The environments Terbol-11, which received 405.4 mm total rainfall, and Terbol-09, which received 594.2 mm, were sparsely distributed in quadrant I. Most of the other environments were close to the general mean of DRS with low PCA 1 values. One environment, TH-Dr09, located in quadrant III, was excluded from the data as the plants were severely infected by Fusarium wilt (caused by Fusarium oxysporum f.sp. ciceris). A comparison of the average of the GY with the DRS in stressed and non-stressed environments showed that at least two promising RILs (152, 162) had good performances under both contrasted environments, stressed (rainfall <25 mm) and non-stressed (rainfall >400 mm) (see Fig. 3).
a Comparison between RILs based on DRS and GY under drought-stressed environments (TH-Dr08 and TH-Dr10; rainfall <25 mm). b Comparison between GY averages under stressed (rainfall <25 mm) and non-stressed environments (rainfall >400 mm). The comparison shows that the performances of RILs (152 and 162) were high under drought stress and around average in non-stressed environments. Dashed lines indicate the average score of each trait
Using the mean of the data over all environments, the correlation coefficient between the traits ranged from r = 0.06** (between Epod and HI) to r = 0.99*** (between SN and GY) (see Table 4; set A). Except %Epod the DRS was negatively correlated with all traits, and these values ranged from r = −0.28*** for %Epod to r = −0.65*** for GY (Table 4; set A). A significant positive correlation (r = 0.78***) was observed between DFLR and MAT across different environments varying in their moisture gradients. The average estimates of broad-sense heritability of the 12 traits under 10 environments were low and ranged from 8 to 37 %. However, except for Epod, the values of heritability for the other traits increased when stressed (rainfall <25 mm) and non-stressed (rainfall >400 mm) environments were compared (Table 5). The highly stressed environments showed the highest heritability with the values for GY increasing from 0.13 to 0.78 (Table 5) in this study.
Mapping and QTL analyses
Of the 263 primers tested, only 110 (42 %) of the primer pairs revealed DNA polymorphism between the parents ILC588 and ILC3279. The RIL population was screened with these primer pairs. The linkage map comprised a total of 77 simple sequence repeat (SSR) markers spanning 520 cM on nine linkage groups. The average distance between the markers was 7.6 cM, however, 33 (30 %) of the markers remained unlinked. Interval mapping revealed significant QTLs for all the traits using the average of two replications at each location (Table 6).
Ninety-three significant QTLs (LOD ≥ 2.0) (Table 6) were detected for the 12 drought-related traits in the population studied. Most of the QTLs were environmentally specific, however eight QTLs expressed in more than one environment (Table 6). The highest phenotypic variation (greater than 24.2 %) was explained by the QTLs for DFLR in Terbol-11. Although QTLs were detected on linkage groups 1, 2, 3, 4, and 9, four regions on LG3 and LG4 contained QTL for several traits and may contain QTL with pleiotropic effects (Fig. 4). The first significant QTL region on LG3, was comprised four markers (H4G-07, H1B-04, H6C-07, and H1E-06) spanning 12.1 cM, and contained a significant QTL for DFLR, DRS, GY, MAT, HI, SN, BY and Pod (Fig. 4; Table 6). The second QTL comprised two markers (H3G-09 and TR-50 s) spanning 0.68 cM, and contained significant QTL for 100sw, Pod, PLHT, Epod, SN, and BY. Two common QTLs for different traits were observed on LG4. The first comprised two markers (H1G-20, and H5G-01) spanning 5.81 cM, indicating a common significant QTL for six traits DFLR, DRS, GY, MAT, HI, and Pod. The second QTL on LG4 comprised two markers (H1B-17 and SCY17) spanning 4.63 cM, indicating a common significant QTL for DFLR, MAT, and SN (Fig. 4). The pooled data of the stressed and non-stressed environments showed that the DRS QTL expressed strongly in the stressed environments, but was weak or not significant in the non-stressed ones (Fig. 5). The epistatic interactions were not significant between these markers at their location on the genome. The QTL for the DRS was detected as a stable QTL in three late planted environments—TH-Dr08, TH-Dr10, and TH-cont10 (Table 6). Interestingly, the marker H6C-07 was associated with the QTLs for many traits—DFLR, DRS, GY, MAT, HI, SN, and Pod—in all the environments except Breda-09 and Breda-10. The allele ‘A’ of marker H6C07 (from the tolerant parent ILC588) increased GY 80, 29.8 and 20.3 % under late planting environments (TH-Dr08, TH-Dr10, and TH-cont10, % respectively; Table 7). Although the PLHT QTL did not explain more than 6.1 % of the total variation, it was interesting to find that QTLPLHT was associated with the non-linked marker TA-76 in four environments—TH-cont09, Terbol-09, TH-cont10, and TH-Dr10.
Linkage map depicting QTLs for eight drought-related traits detected in a RIL (ILC588 × ILC3279) mapping population. The QTLs have been designated with the trait name abbreviations. Traits are DRS Drought resistance score, PLHT plant height at maturity, DFLR days to flowering, MAT days to maturity, Pod number of pods per 3 plants; GY grain yield; SN seed number per 3 plants, BY biological yield, 100sw 100 seed weight; HI harvest index
Discussion
In plant breeding programs, drought tolerance is an important objective. However, understanding the physiological mechanisms that contribute to variability in crop performance in drought environments remains limited (Ceccarelli and Grando 1996; Passioura 1996). One of the major mechanisms of drought resistance is drought escape, which involves plants with early flowering and pod initiation, as any change in the duration of various phenological phases (vegetative phase, days to flowering, grain development duration, etc.) helps plants to avoid exposure of the critical growth phases to stress conditions (Turner 1986). Therefore, drought and yield are important traits. Many different genes act throughout the life of the plant, plus there are the effect of the environments and interactions. In our study, we used 10 different environments, varying in rainfall (ranging from <25 mm to >400 mm; Table 2), temperature (10–25 °C) and planting dates (early December to end March), to study drought tolerance. (Drought tolerance was measured by the DRS score, phenology traits (PLHT, DFLR, and DMAT), yield, and yield components (GY, BY, HI, Pod, Epod, %Epod, 100sw, and SN) under moisture stress and optimum conditions.
In general, the broad-sense heritability estimates for the 12 traits in this study in 10 environments ranged from 3 % (%Epod) to 37 % (DFLR) with an average value of 16.8 %. These values are lower than those reported earlier for chickpea (Eser 1976; Muehlbauer and Singh 1987; Abbo et al. 2005; Cobos et al. 2007). However, the heritability estimate values increased when stressed and non-stressed environments were compared. For example, the environments TH-Dr08, and TH-Dr10, which received minimum rainfall (<25 mm after planting), showed higher heritability values, which averaged 63.4 %. Eser (1976) observed a 13 % narrow-sense heritability for single plant yield, 25 % for number of pods per plant, 30 % for plant height, and 78 % for seed weight in chickpea and concluded that seed weight was the least environmentally influenced trait. Our results showed that %Epod was the least environmentally influenced trait with 87 % heritability only under stressed environments. Abbo et al. (2005) reported high heritability values for seed weight (71 %). However, in the present study the heritability of seed weight did not exceed 37 % even in the environments receiving minimum rainfall values (<25 mm). The mean hereditability obtained for yield across the 10 environments was 13 %, which is very low (Table 5) and similar to the 14 % observed by Cobos et al. (2007). However, when the results for the non-stressed and stressed environments were compared, this heritability value for yield increased to 35 % (Terbol-09 and Terbol-10) and 78 % (TH-Dr08 and TH-Dr10). This indicated that for chickpea, the drought-related gene(s) are probably highly expressed under the moisture-stressed environment, and thus direct selection for yield under stressed conditions would give greater response than indirect selection under non-stressed conditions. No such comparisons for heritability have been made for chickpea under moisture stressed and non-stressed environments. Similar findings were reported by Mhike et al. (2012) for maize when they compared the heritability values of the anthesis-silking interval and ears per plant under optimum and drought environments. They found that the heritability values were increased under moisture stressed conditions when compared to the optimum environment.
Our results indicated a positive correlation (63 %) between yield and DFLR when we pooled the data for all environments. This correlation was negative and significant in the dry environments TH-Dr08 (−0.42***) and TH-Dr10 (−0.44***), but not significant in the non-stressed environments TH-cont08 (−0.07), TH-cont09 (−0.13), and TH-cont10 (−0.05). This result agreed with that found by Rubio et al. (2004), who performed a multi-location/year assay and found a significant and positive effect of earliness on yield in chickpea. Therefore, the detection of QTLs for seed yield associated with early flowering would help improve for chickpea yield in dry environments using MAS. In our study, a significant correlation (r = 0.52, P < 0.001) was detected between GY and MAT when we pooled the data across all 10 environments indicating that lae maturity could give greater yield. However, under drought-stressed environments, the correlation between these two traits (GY and MAT) is negative (Table 4, set B) indicating that plants with early maturity gave higher yield under moisture stress conditions and thus could facilitate the selection of high-yielding genotypes for drought tolerance. In contrast, no correlation between GY and MAT was reported by Kumar and Rao (1996) and Gowda et al. (2011). Toker and Cagirgan (1998) found significant correlations between the drought susceptibility index and seed yield (−0.65**), BY (−0.41**), and HI (−0.37**) for chickpea in drought-stressed environments. In the present study and by using the mean of the pooled data for all environments, the correlation between DRS and yield and its components indicated stronger significance between seed yield (−0.65***), BY (−0.58***), and HI (−0.48***). In this study, the RILs (152 and 162) had good performances in stressed and non-stressed environments in term of DRS and GY, which is the approach we are applying in our breeding program to develop drought tolerant and water responsive genotypes. Significant correlations between different drought stress-related traits indicated that different genes are controlling these traits and the interaction between these genes and the environment.
The AMMI, the genotype main effects, and GEI effects models are widely used for effective analysis and interpretation of a multi-environment data structure in breeding programs. The most accurate model for AMMI can be predicted by using the first two PCAs (Gauch and Zobel 1996; Yan and Rajcan 2002). Conversely, Sivapalan et al. (2000) recommended a predictive AMMI model with the first four PCAs. Ceccarelli (1989) exposed a number of genotypes to a set of contrasting environments to identify genotypes with a high average yield and low GEI. In our study, the GEI sum of squares was almost 2.3 times larger than that for the genotypes, indicating that there were substantial differences in genotypic response across environments. This is similar to Kaya et al. (2002) who studied the yield performances of 20 bread wheat genotypes across six environments and found that the GEI sum of squares was 3.4 times larger than that for genotypes. The AMMI ANOVA indicated highly significant variation for genotypes, environments, and GEI for DRS traits. The application of the AMMI model for partitioning GEI revealed that the first two IPCAs together with the sum of squares (1,752) was greater than that of the genotypes (1,354) and were highly significant (P < 0.001). This suggested the AMMI model, with first and second multiplicative IPCAs, was adequate for cross-validation of the drought tolerance variation explained by the GEI in the present data set since it excludes most of its actual noise.
In the present study, we initially performed composite interval mapping (CIM) to detect the QTLs using MapQTL 5. Ninety-three significant QTLs (LOD ≥ 2.0) were identified and mapped for the 12 traits in the population studied. Both temperature and photoperiod have been reported to affect DFLR in chickpea (Roberts et al. 1985; Kumar and Abbo 2001). Segregation of DFLR can be clearly observed in the data distribution of the RILs grown under different conditions and varied by planting date. In our study, we used different planting dates and found the DFLR ranged from 51 (late planting) to 136 (early planting). Using data from field and greenhouse experiments, Cobos et al. (2007) detected a highly significant QTL for DFLR (QTLDF1), LG4, closely linked to GAA47. They suggested that LG4 could be considered an interesting genomic region for agronomic traits in chickpea. We found four QTLs for DFLR; two, on LG3 closely linked markers, were H1F-14 and H6C-07, and two, on LG4 closely linked markers, H5G-01 and H1B-17. GAA47 was found to be monomorphic, thus making it difficult to confirm whether H1B-17 is genetically close to GAA47, because GAA47 was mapped on LG4 (Cobos et al. 2007), LG6 (Gaur et al. 2011), and LG8 (Rajesh and Muehlbauer 2008). Two QTLs for resistance to Ascochyta blight have also been reported on the LG4 flanking markers, TA72 and TA146 (Iruela et al. 2006; Cho et al. 2004), which are also flanking the QTLs detected in this study for seed size, yield, and DFLR. In our study, it was also observed that both LG3 and LG4 appeared to be pleiotropic, with QTLs for each of DRS, Pod, DFLR, GY, MAT, and HI traits.
Of the QTLDRS identified in our study, only one, identified on LG3 at marker H6C-07, gave high LOD scores; ranging from 5.19, 5.1, and 6.25 in the three environments TH-Dr08, TH-Dr10, and TH-cont10, respectively. These were late planted environments (late March), which means that the plants were exposed to high temperatures during a short growing period, in addition to being highly drought stressed in TH-Dr08 and TH-Dr10. Interestingly, QTLDSR on LG3 and LG4 was associated with the six traits, Pod, DFLR, GY, MAT, HI, and SN in the TH-Dr08, TH-Dr10, Terbol-09, Terbol-11, TH-cont08, and TH-cont10 environments (Table 6). This supports the significant positive correlations observed among these traits (Table 4). Based on the linked markers, it was interesting to note that LG3 and LG4 in our study correspond to the LG3 and LG4 of the chickpea reference map (Winter et al. 2000), containing QTLs for many traits, such as Pod, GY, 100sw, DRS, DFLR, HI, Epod, and MAT (this study); DFLR and Ascochyta blight resistance (Aryamanesh et al. 2010); seed number per plant, 100sw, and days to 50 % flowering (Cho et al. 2002); seed weight and lutein concentration (Abbo et al. 2005); and seed size, yield, and days to 50 % flowering (Cobos et al. 2007). Hence, concentrating on LG3 and LG4 in molecular breeding programs could speed up the improvements for these traits.
Four 100sw associated QTLs were identified on LGs 3, 4, 5 and 7. Among the 100sw QTLs on LG3, one common QTL was identified in the TH-Dr08 and TH-Dr10 environments, explaining 17.2 and 7.4 % of the total variation. However, this figure is much lower than that determined by Cho et al. (2002) who first reported a major QTL for seed weight which explained 52 % of the total phenotypic variation. In our study, this QTL was likely pleiotropic with a QTL for SN, BY, Pod, and Epod. Similarly, Gowda et al. (2011) found that this trait appeared to be pleiotropic with a QTL for yield per plant and plant height. Cobos et al. (2007) identified two QTLs for seed weight on LG4 and LG8 explaining 20.3 and 10.1 % of the variation. It is possible that one or more of the 100sw QTLs might be the same as the seed weight QTLs detected by Cho et al. (2002) and Cobos et al. (2007). However, fine mapping could reveal the true nature of the pleiotropism of these QTLs. In our study, the non-linked marker TA-76 was associated with plant height (QTLPLHT) in the four environments TH-cont09, Terbol-09, TH-cont10, and TH-Dr10. Another four QTLs for plant height were identified and the QTL on LG3 (TA-179) explained highest variation of 8.8 % in Breda-09.
The development of MAS for phenologic and yield traits related to drought will allow identification of tolerant lines at an early stage without the need for costly, environment-dependent, and repetitive phenotyping. The QTLs which were identified in the present study are likely to be useful for improving drought resistance in chickpea and provide confirmation for the genetic basis of correlations. They also suggest that selection for a small number of target genomic regions (especially QTLDRS and QTLGY on LG3 and LG4) may achieve benefits for multiple characteristics, especially yield. In late planting and dry environments, our results indicated an important allele, H6C-07 from the ILC588 parent, was linked to increased yield (80, 29.8 and 20.3 %, Table 7). This marker association allele must be validated in other mapping populations or in association studies to confirm that QTL will be useful for selecting for drought tolerance in chickpea. Likewise, inclusion of the QTLs expressed in a particular environment or physiological conditions, along with the stable QTLs, could be a breeding strategy for developing genotypes adapted to a wide range of environments. In the present study, the use of 10 environments and performing interaction analyses permitted detection of many QTLs, some of which were stable across environments while other were not, most likely pointing to the well know concept of specific versus wider adaptation of genotypes. Additionally, QTLs for many other important traits were located on these LGs by previous researchers. Hence, these LGs can be targeted in molecular breeding programs to achieve simultaneous and correlated improvement in these traits. Based on our study, it can also be concluded that drought escape triggered by moisture stress under late planting conditions has a major role in the development of drought tolerant chickpea varieties.
References
Abbo S, Molina C, Jungmann R, Grusak MA, Berkovitch Z, Reifen R, Kahl G, Winter P (2005) Quantitative trait loci governing carotenoid concentration and weight in seeds of chickpea (Cicer arietinum L.). Theor Appl Genet 111:185–195
Arumuganathan K, Moss JP (1991) Nuclear DNA content of some important plant species. Plant Mol Biol 9:208–218
Aryamanesh N, Nelson MN, Yan G, Clarke HJ, Siddique KHM (2010) Mapping a major gene for growth habit and QTLs for Ascochyta blight resistance and flowering time in a population between chickpea and Cicer reticulatum. Euphytica 173:307–319
Blum A (1996) Crop responses to drought and the interpretation of adaptation. Plant Growth Regul 20:135–148
Blum A, Nguyen HT (2004) Physiology and biotechnology integration for plant breeding: Epilogue. In: Nguyen HT, Blum A (eds) Physiology and biotechnology integration for plant breeding. Marcel Dekker, Inc New York, pp 611–618
Boyer JS (1982) Plant productivity and environment. Science 218:443–448
Canci H, Toker C (2009) Evaluation of yield criteria for drought and heat resistance in chickpea (Cicer arietinum L.). J Agron Crop Sci 195:47–54
Ceccarelli S (1989) Wide adaptation: how wide? Euphytica 40:197–205
Ceccarelli S, Grando S (1996) Drought as a challenge for the plant breeder. Plant Growth Regul 20:149–155
Chandra BR, Nguyen BD, Chamarerk V, Shanmugasundaram P, Chezhian P, Jeyaprakash P, Ganesh SK, Palchamy A, Sadasivam S, Sarkarung S, Wade LJ, Nguyen HT (2003) Genetic analysis of drought resistance in rice by molecular markers: association between secondary traits and field performance. Crop Sci 43:1457–1469
Cho S, Kumar J, Shultz JL, Anupama K, Tefera F, Muehlbauer FJ (2002) Mapping genes for double podding and other morphological traits in chickpea. Euphytica 128:285–292
Cho S, Chen W, Muehlbauer FJ (2004) Pathotype-specific genetic factors in chickpea (Cicer arietinum L.) for quantitative resistance to ascochyta blight. Theor Appl Genet 109:733–739
Cobos MJ, Rubio J, Fernández-Romero MD, Garza R, Moreno MT, Millán T, Gil J (2007) Genetic analysis of seed size, yield and days to flowering in a chickpea recombinant inbred line population derived from a Kabuli × Desi cross. Ann Appl Biol 151:33–42
Duarte JB, Vencovsky R (1999) Interação Genótipos × Ambientes: Uma introdução à análise. AMMI. Funpec Ribeirão Preto Sociedade Brasileira de Genética
Eser D (1976) Heritability of some important plant characters, their relationship with plant yield and inheritance of ascochyta blight resistance in chickpea (Cicer arietinum L.). Ankara Universitesi, Ziraat Fakultesi Yayinlari 620, Ankara, Turkey
Gauch GH, Zobel RW (1996) AMMI analysis of yield trials. In: Kang MS, Gauch HG (eds) Genotype by environment interaction. CRC Press, Boca Raton, pp 85–122
Gaur R, Sethy NK, Choudhary S, Shokeen B, Gupta V, Bhatia S (2011) Advancing the STMS genomic resources for defining new locations on the intraspecific genetic linkage map of chickpea (Cicer arietinum L.). BMC Genomics 12:117
Gowda SJM, Radhika P, Mhase LB, Jamadagni BM, Gupta VS, Kadoo NY (2011) Mapping of QTLs governing agronomic and yield traits in chickpea. J Appl Genet 52:9–21
Gujaria N, Kumar A, Dauthal P, Dubey A, Hiremath P, Prakash AB, Farmer A, Bhide M, Shah T, Gaur PM, Upadhyaya HD, Bhatia S, Cook DR, May GD, Varshney RK (2011) Development and use of genic molecular markers (GMMs) for construction of a transcript map of chickpea (Cicer arietinum L.). Theor Appl Genet 122:1577–1589
Iruela M, Rubio J, Barro F, Cubero JI, Millán T, Gil J (2006) Detection of two QTL for resistance to Ascochyta blight in an intra-specific cross of chickpea (Cicer arietinum L.): development of SCAR markers associated to resistance. Theor Appl Genet 112:278–287
Kashiwagi J, Krishnamurthy L, Crouch JH, Serraj R (2005) Variability of root length density and its contributions to seed yield in chickpea (Cicer arietinum L.) under terminal drought stress. Field Crop Res 95:171–181
Katerji N, Van Hoorn JW, Hamdy A, Mastrorilli M, Owies T, Malhotra RS (2001) Response to soil salinity of chickpea varieties differing in drought tolerance. Agr Water Manage 50:83–96
Kaya Y, Palta C, Taner S (2002) Additive main effects and multiplicative interactions analysis of yield performances in bread wheat genotypes across environments. Turk J Agric 26:275–279
Khanna-Chopra R, Sinha SK (1987) Chickpea: physiological aspects on growth and yield. In: Saxena MC, Singh KB (eds) The Chickpea. CAB International, Wallingford, pp 163–189
Kosambi DD (1944) The estimation of map distance from recombination values. Ann Eugen 12:172–175
Kramer PJ, Boyer JS (1995) Water relations of plants and soils. Academic Press, San Diego
Kumar J, Abbo S (2001) Genetics of flowering time in chickpea and its bearing on productivity in the semi-arid environments. Adv Agron 72:107–138
Kumar J, Rao BV (1996) Super early chickpea developed at ICRISAT Asia Center. Int Chickpea Pigeonpea Newslett 3:17–18
Kumar J, Van Rheenen HA (2000) A major gene for time of flowering in chickpea. J Hered 91:67–68
Li H, Ye GY, Wang JK (2007) A modified algorithm for the improvement of composite interval mapping. Genetics 175:361–374
Lichtenzveig J, Scheuring C, Dodge J, Abbo S, Zhang HB (2005) Construction of BAC and BIBAC libraries and their applications for generation of SSR markers for genome analysis of chickpea (Cicer arietinum L.). Theor Appl Genet 110:492–510
Lichtenzveig J, Bonfil DJ, Zhang H, Shtienberg D, Abbo S (2006) Mapping quantitative trait loci in chickpea associated with time to flowering and resistance to Didymella rabiei the causal agent of Ascochyta blight. Theor Appl Genet 113:1357–1369
Ludlow MM, Muchow RC (1990) A critical evaluation of traits for improving crop yields in water-limited environments. Adv Agron 43:107–153
Mhike X, Okori P, Magorokosho C, Ndlela T (2012) Validation of the use of secondary traits and selection indices for drought tolerance in tropical maize (Zea mays L.). Af J Plant Sci 6:96–102
Muehlbauer FJ, Singh KB (1987) Genetics of chickpea. In: Saxena MC, Singh KB (eds) The chickpea. CAB International, Wallingford, pp 99–125
Or E, Hovav R, Abbo S (1999) A major gene for flowering time in chickpea. Crop Sci 39:315–322
Passioura JB (1996) Drought and drought tolerance. Plant Growth Regul 20:79–83
Polhill RM, Raven PH (1981) Advances in legume systematics. Royal Botanic Gardens, Kew
Rajesh PN, Muehlbauer FJ (2008) Discovery and detection of single nucleotide polymorphism (SNP) in coding and genomic sequences in chickpea (Cicer arietinum L.). Euphytica 162:291–300
Roberts EH, Hadley P, Summerfield RJ (1985) Effects of temperature and photoperiod on flowering in chick peas (Cicer arietinum). Ann Bot-London 55:881–892
Rogers SO, Bendich AJ (1985) Extraction of DNA from milligram amounts of fresh, herbarium and mummified plant tissues. Plant Mol Biol 5:69–76
Rubio J, Flores F, Moreno MT, Cubero JI, Gil J (2004) Effects of the erect/bushy habit, single/double pod and late/early flowering genes on yield and seed size and their stability in chickpea. Field Crop Res 90:255–262
Saxena NP (1990) Status of chickpea in the Mediterranean basin. Present status and future prospects of chickpea crop production and improvement in the Mediterranean countries. Options Méditerranéennes Série Séminaire 9:17–24
Sivapalan S, Brien LO, Ferrara GO, Hollamby GL, Barclay I, Martin PJ (2000) An adaptation analysis of Australian and CIMMYT/ICARDA wheat germplasm in Australian production environments. Aust J Agric Res 51:903–915
Toker C, Çagirgan MI (1998) Assessment of response to drought stress of chickpea (Cicer arietinum L.) lines under rainfed conditions. Turk J Agric For 22:615–621
Turner NC (1986) Crop water deficits: a decade of progress. Adv Agron 39:1–51
Turner NC, Wright GC, Siddique KHM (2001) Adaptation of grain legumes (pulses) to water limited environments. Adv Agron 71:193–231
Van Ooijen JW (2006) JoinMap 4.0, software for the calculation of genetic linkage maps. Kyazma, B.V. Wageningen, Netherlands
Van Ooijen JW (2009) MapQTL 6, software for the mapping of quantitative trait loci in experimental populations of diploid species. Kyazma, B.V. Wageningen, Netherlands
Winter P, Benko-Iseppon AM, Hüttel B, Ratnaparkhe M, Tullu A, Sonnante G, Pfaff T, Tekeoglu M, Santra D, Sant VJ, Rajesh PN, Kahl G, Muehlbauer FJ (2000) A linkage map of the chickpea (Cicer arietinum L.) genome based on the recombinant inbred lines from a C. arietinum × C. reticulatum cross: localization of resistance genes for Fusarium races 4 and 5. Theor Appl Genet 101:1155–1163
Yan W, Rajcan I (2002) Biplots analysis of the test sites and trait relations of soybean in Ontario. Crop Sci 42:11–20
Acknowledgments
The authors thank Amani Khalifeh for genotyping the RIL population and the chickpea breeding program staff for field trials of the RIL population. This work was supported by Grains Research and Development Corporation (GRDC) Australia.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by B. Diers.
Rights and permissions
About this article
Cite this article
Hamwieh, A., Imtiaz, M. & Malhotra, R.S. Multi-environment QTL analyses for drought-related traits in a recombinant inbred population of chickpea (Cicer arientinum L.). Theor Appl Genet 126, 1025–1038 (2013). https://doi.org/10.1007/s00122-012-2034-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00122-012-2034-0