Abstract
Sweet melon cultivars contain a low level of organic acids and, therefore, the quality and flavor of sweet melon fruit is determined almost exclusively by fruit sugar content. However, genetic variability for fruit acid levels in the Cucumis melo species exists and sour fruit accessions are characterized by acidic fruit pH of <5, compared to the sweet cultivars that are generally characterized by mature fruit pH values of >6. In this paper, we report results from a mapping population based on recombinant inbred lines (RILs) derived from the cross between the non-sour ‘Dulce’ variety and the sour PI 414323 accession. Results show that a single major QTL for pH co-localizes with major QTLs for the two predominant organic acids in melon fruit, citric and malic, together with an additional metabolite which we identified as uridine. While the acidic recombinants were characterized by higher citric and malic acid levels, the non-acidic recombinants had a higher uridine content than did the acidic recombinants. Additional minor QTLs for pH, citric acid and malic acid were also identified and for these the increased acidity was unexpectedly contributed by the non-sour parent. To test for co-localization of these QTLs with genes encoding organic acid metabolism and transport, we mapped the genes encoding structural enzymes and proteins involved in organic acid metabolism, transport and vacuolar H+ pumps. None of these genes co-localized with the major pH QTL, indicating that the gene determining melon fruit pH is not one of the candidate genes encoding this primary metabolic pathway. Linked markers were tested in two additional inter-varietal populations and shown to be linked to the pH trait. The presence of the same QTL in such diverse segregating populations suggests that the trait is determined throughout the species by variability in the same gene and is indicative of a major role of the evolution of this gene in determining the important domestication trait of fruit acidity within the species.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The taste of most fruits is determined by a combination of sugar and organic acid levels, together with their characteristic volatile compounds. While most fruit have a strongly acidic pH, the sweet melons, Cucumis melo, are unique in that they have an unusual low acidity level in the mature fruit, and the pH values for cultivated sweet melons are in the near neutral range of >6. Therefore, melon fruit quality is determined primarily by sugar content alone (Yamaguchi et al. 1977). However, a tremendous range of genetic variability exists in this species and there are genotypes accumulating high levels of acid in the fruit, characterized by low pH of <5 (Burger et al. 2009; Kubicki 1962).
Genetic variability for both quantitative and qualitative levels of organic acids and fruit pH has been observed in other fruit species, as well. For example, genetic control for the level of fruit acid has been reported for peach (Etienne et al. 2002; Moing et al. 1998), citrus limes (Brune et al. 2002; Fang et al. 1997; Marsh et al. 2001) and tomato (Fulton et al. 2002; Stevens 1972). However, the genetic variability in melon is unique in terms of the extreme difference in acidity level between sour and non-sour genotypes (nearly two pH units, or [H+] differences of 102). The main organic acids that accumulate in most fruits, including melon (C. melo), are citric and malic acid (Ulrich 1970; Leach et al. 1989; Wang et al. 1996) and we have previously shown that the citric and malic acid levels of mature fruit from sour melon lines are indeed higher than the levels from near-isogenic non-sour counterparts (Burger et al. 2003).
The trait of fruit sourness in C. melo was initially described by Kubicki (1962), based on a cross between sour and sweet varieties and determined to be inherited as a single locus (So) with sour fruit dominant to non-sour. Danin-Poleg et al. (2002) mapped the trait of fruit pH as a single gene based on an F2 population derived from the inter-varietal cross of a sweet, non-acid cultivar ‘Dulce’ (reticulatus group) and a non-sweet, sour accession of the momordica group of melon, PI 414723.
We present here a detailed QTL analysis of fruit pH and organic acid contents based on a recently described F7–F8 RILs population (Harel-Beja et al. 2010) derived from the PI 414723 × ‘Dulce’ cross. Furthermore, we extend the significance of this major QTL to additional C. melo genotypes and show that the same locus determines fruit acidity in a broad spectrum of melon groups. In addition, we mapped nearly 60 genes encoding the complete metabolic pathway of organic acid metabolism, including the vacuolar proton transporters, and show that none co-localize with the major locus for melon fruit pH.
Materials and methods
Plant material
A population of recombinant inbred lines (RILs, designated 414xDul) was developed from a cross between PI 414723-S5 (C. melo var. momordica), an Indian cooking melon line with non-sweet acidic fruit, and ‘Dulce’ (C. melo var. reticulatus), an American muskmelon with non-sour, sweet fruit (Danin-Poleg et al. 2002). F2 individuals (n = 112) originating from two F1 plants were propagated by the single-seed descent method to create the population, consisting of 99 RI lines comprising a mix of F6, F7 and F8 generations, as described in Harel-Beja et al. (2010).
The two parents and their RI lines were grown in the open field in Newe Ya’ar, Israel, during the summer of 2006. Each line, in a completely randomized design, was represented by ten plants. The two parents were each grown in three replications of 10 to 12 plants. A single fruit per plant was harvested when the abscission layer developed.
Segregating F2 and F3 populations were also developed from the following two crosses: (1) ‘Noy Yizre’el’ (NY, C. melo var. cantaloupensis), a sweet, non-sour variety, crossed with ‘Faqqous’ (FAQ, C. melo var. chate), a non-sweet, sour variety; (2) ‘Sakata Sweet’ (SAS, C. melo var. conomon), a sweet, non-sour variety, crossed with ‘Doya’ (DOY, C. melo var. flexuosus), a non-sweet, sour variety.
Evaluation of fruit pH and organic acid analysis by HPLC
Harvested fruits were brought to the laboratory and sampled as follows. Approximately 1 g fresh weight of mesocarp tissue, taken from the center-equatorial portion of the fruit, was placed in 80% EtOH and stored at −20°C until extraction. The organic acids, together with the soluble sugars were extracted in 80% alcohol which was evaporated to dryness, and re-suspended in ddH2O. Following filtration through a 0.45 mm filter, analysis was performed by HPLC using an Aminex® Organic Acid Column (100 × 7.8 mm, BioRad Laboratories, Hercules, CA) with 0.008 N H2SO4 (pH 2.2) as running solvent at 1.0 ml min−1, according to manufacturer’s recommendations. The HPLC system consisted of an Agilent 1200 binary SL pump and PDA detector. Citric and malic acid were identified by retention time and quantified, compared to known standards.
An additional peak eluted on the Aminex® Organic Acid Column at the same retention time as a standard of succinic acid; however, an enzyme linked assay (Boehringer, succinic acid kit, UV method, cat # 176281) indicated that this compound was not succinic acid. To identify the compound it was sequentially purified from 40 g of cv. ‘Dulce’ mature fruit using the following three HPLC separations: (1) Bio-Rad Aminex® Organic Acid Column, described above and collecting the peak eluting at 14.6 min; (2) Restek Allure® Organic Acids Column (4.6 × 300 mm), 0.1 M phosphate buffer, pH 2.5, flow rate 0.5 ml min−1, and collecting the peak eluting at retention time of 28 min; (3) Dionex Acclaim® Mixed-Mode Wax-1 Column (4.6 × 150 mm), 50/50 v/v acetonitrile/50 mM phosphate buffer pH 6.0, flow rate 1 ml min−1 and collecting the peak eluting at 2.4 min. The UV/VIS spectrum was obtained by PDA of each of the collected peaks. The highly purified compound was identified as uridine, as follows.
Identification of uridine in ‘Dulce’ melon fruit
The identification of uridine in water extracts of melon flesh tissue (cv. Dulce), was performed by mass spectral analysis, carried out by the ultra-performance liquid chromatography coupled to a quadrupole time-of-flight (UPLC-qTOF) instrument [Waters, High Definition MS System (HDMS) Synapt], with the UPLC column connected on-line to a PDA detector (Waters, Acquity), and then to the MS detector, equipped with an electrospray probe. The settings of the apparatuses were as described by Itkin et al. (2011). Uridine was first putatively identified in the positive and negative modes, using the elemental composition, selected according to the accurate masses and the isotopic pattern using the MassLynx software, and MS/MS fragments were compared to those reported in the literature (Linden 2004; Kellner et al. 2011). Then the identification was verified by comparison of retention time, MS/MS fragments and UV spectral properties of the putative compound to those of the corresponding standard (Sigma, USA). The concentration of uridine in planta was quantified against a standard using the Bio-Rad Aminex® column, as above.
DNA preparation
DNA was extracted from young leaf tissue from ten plants of each of the parental lines and ten plants of each of the RI lines according to the preparation procedure described by Fulton et al. (1995), as recently described by Harel-Beja et al. (2010).
Organic acid metabolism genes: identification and mapping
Genes belonging to the organic acid metabolism pathway were selected based on the literature and the International Cucurbit Genomics Initiative database (Clepet et al. 2011; http://www.ICUGI.org). In addition, an exhaustive search for additional paralogues, based on BLAST analysis of a melon fruit EST database derived from a 454 transcriptome analysis (Portnoy et al. 2011) was also performed. Information related to these genes is summarized in Tables 1 and 2. Polymorphisms were identified based on comparative sequencing of the parental lines (Supplemental Table 1). In cases where no polymorphism was observed in the gene sequence, BAC clones harboring the gene were end-sequenced and polymorphisms identified in genomic regions along the BAC. To identify specific BACs that contained genes of interest, specific probes were designed for hybridization with the melon BAC library (MR1, CM_MBa, Clemson University, Clemson, SC). Standard hybridization procedures were used with a radioactively labeled probe on BAC library membranes (Luo et al. 2001). Positive BACs were re-checked by sequencing the BAC using the primers that had been used to prepare the probes. Representative PCR products were further sequenced to validate the results.
Single nucleotide polymorphisms (SNP) genotyping was carried out by the two methods described in Harel-Beja et al. (2010): dCAP (dCAPS Finder 2.0, Neff et al. 2002) or Sequenom (San Diego, CA) MassARRAY Compact Analyzer, at the Cancer Research Center and Pediatric Hematology-Oncology, Sheba Medical Center, Tel Hashomer, Israel. Assays and primers were designed using the MassARRAY assay Design 2 software.
Map construction and QTL analysis
Mapping was performed using JoinMap® 3.0 software (Van Ooijen and Voorrips 2001). Markers were grouped at a minimum logarithm of odds (LOD) score of 4.0, and a recombination frequency value of 0.4. Linkage group (LG) designation was according to the recently merged melon map (Diaz et al. 2011), which includes the 414xDul map. QTL analyses were performed by the MapQTL® 5 software (Van Ooijen 2004), including: interval mapping (IM), multiple QTL model (MQM) and permutation analysis. QTL was significant when LOD score was higher than the LOD threshold calculated by permutation tests (1,000 permutation at p = 0.05; ≥2.3 LOD for all traits).
SSR markers
Simple sequence repeat (SSR) markers that were found to be adjacent to the pH trait (Harel-Beja et al. 2010) were screened in 14 F2 plants of each of the additional populations: NY × FAQ and SAS × DOYA. Marker CMCTTN181 which was polymorphic in the NY × FAQ was screed in the plants of this population while marker CMAT141 was polymorphic and was screened in the plants of SAS × DOYA.
Seven of the plants were of high pH and seven low pH. SSR genotyping was performed using the fragment analysis protocol of the AB3130xl Genetic Analyzer. In short, microsatellite loci were amplified by PCR using fluorescently labeled forward and unlabeled reverse primers. The PCR amplicons were separated by size using electrophoresis. The dyes labeled products were identified by fluorescence detection. GeneMapper® Software version 4 (Applied Biosystems) was used to size and genotype the alleles.
Primers used to amplify the SSR markers:
-
CMCTTN181F: CTCTCTGCAATTCTCGCC;
-
CMCTTN181R: CAACCATCCGCTTCACTC;
-
CMAT141F: AAGCACACCACCACCCGTAA;
-
CMAT141R: GTGAATGGTATGTTATCCTTG
Results
QTLs for pH and organic acids of ripe melon fruit
The 414xDul RILs population was analyzed for fruit pH, as well as content of the major organic acids, determined by HPLC (Table 3). Significant variability was observed for all of the traits between families as indicated by Welch ANOVA test. The distribution mode of the accumulation of the three metabolites among the RI population was normal according to the Shapiro–Wilk goodness-of-fit test. The acidic parent PI414723 was characterized by pH values of the extracted juice of <5.0 while the sweet ‘Dulce’ variety has fruit pH values >6.0. The two parents did not significantly differ in either their citric or malic acid concentrations in the present experiment. Fruit of the same lines grown in different seasons have, at times, shown higher levels of malic acid in the sour PI 414723 parent, compared to the non-sour ‘Dulce’ parent (e.g., spring 2000 season: PI 414723: citric, 2.78 + 0.44 mg/gfw, malic, 0.86 + 0.22 mg/gfw; ‘Dulce’: citric, 2.84 + 0.30 mg/gfw, malic, 0.28 + 0.10 mg/gfw). The non-sour ‘Dulce’ had significantly higher levels of a third metabolite visible under our chromatographic conditions. The non-sour ‘Dulce’ had significantly higher levels of a third metabolite visible under our chromatographic conditions.
In light of this observation we undertook to identify the third metabolite. MS, as well as UV analysis identified the peak component as uridine and chromatographic co-elution of an uridine standard under three chromatographic separation systems, further confirmed the identification (Fig. 1). The uridine contents of the RILs were calculated according to the Aminex chromatographic separation of an uridine standard. Absolute amounts of uridine were ~three orders of magnitude lower than the major organic acids but there were nevertheless significant differences in uridine content between the acidic and non-acidic genotypes.
The characteristics of pH, citric, malic and uridine were analyzed in the RILs mapping population of nearly 100 F7 and F8 families and QTLs for the traits were mapped (Table 4). A major QTL, on LG8, and with a LOD score of >35 was observed for pH. Co-localizing with this QTL were major QTLs for each of the two organic acids (LOD of ~7–8 for both citric and malic) and a QTL with LOD of >10 for uridine. In addition, there were minor QTLs for citric acid on LG4, LG11 and LG12, and QTL for malic acid more proximal on LG 8. Interestingly, all the minor QTLs for organic acid levels were positively additive for the low acid ‘Dulce’ parent and only the major QTL on LG8 contributed acidity in correlation with the parental genotypes.
Mapping and co-localization of genes involved in organic acid metabolism and transport
An exhaustive list of 58 melon genes encoding approximately 20 enzymatic reactions of organic acid metabolism, together with genes for vacuolar proton transport and organic acid transport, was compiled. Polymorphisms between the two parents of the RILs mapping population were identified for 53 of the genes (Supplemental Table 1). Polymorphisms included both indels and SNPs and determination of the genotypes of the ~100 RILs were performed by a mix of gel separations, standard sequencing reactions and Sequenom pyrosequencing technology. In many cases, polymorphisms were not observed in the coding region of the gene and were identified in flanking genomic sequences, either following identification of BACs harboring the gene of interest or searching for flanking regions of the genes in the genome of the highly syntenous cucumber. For five genes (see Supplemental Table 1) no polymorphisms were identified in any of these regions.
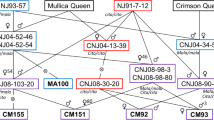
Each of the 53 genes clearly mapped to one of the 12 linkage groups of melon (Fig. 2). The genes were randomly distributed throughout the genome, showing no indication of clustering. With regard to co-localization with the pH and acid traits, none of the genes co-localized with the major QTL on LG8. A single instance of co-localization was observed, between a minor QTL for pH on LG11 and the gene for one of the ATP-citrate lyase paralogues, ACL2. None of the other minor QTLs showed evidence of co-localization with a structural gene for acid metabolism and transport. The identification of BAC clones harboring the genes allowed us also to map 15 BAC clones, contributing to the future physical map of melon (Supplemental Table 1).
Genetic map of the 414 × Dul RI population that includes QTLs for fruit acidity and candidate genes. Linkage group (LG) numbers are according to Diaz et al. (2011). Distances in centiMorgans from the top of each LG are marked on the left side and marker names are on the right side of each linkage group. Fruit acidity candidate genes are in red. Significant QTLs are presented by lines and for QTLs with LOD >5.0, LOD scores in the highest 10% are indicated by dark black lines within the broader QTL
Universality of the major QTL in C. melo
To determine whether the same locus determines fruit pH in other C. melo genetic backgrounds, we tested the closest linked SSR markers for the pH trait (CMCTTN181, CMAT141, located 2 and 3 cM from the trait, respectively; Harel-Beja et al. 2010) in two additional inter-varietal segregating populations. The F3 populations, derived from the low acid Noy Yizre’el and the acidic Faqqous, and from the low acid SAS and the acidic DOY, were analyzed for fruit pH and seven plants from each of the two phenotypes (high and low acidity) of each of the two populations were genotyped for the pH-linked markers. The CMAT141 marker was polymorphic and informative for the Noy Yizre’el and Faqqous population while the CMCTTN181 marker was polymorphic and informative for the SAS × DOY population. In both populations, there was strong linkage between the marker and mature fruit pH (Fig. 3) and the high pH segregants were all homozygous for the recessive allele derived from the high pH parent while the low pH segregants were either heterozygous or homozygous for the low pH allele.
Genotyping of SSR markers in 14 F2 plants and the parental lines using the fragment analysis option of 3130xl Genetic Analyzer. Each line is the amplicon of one F2 plant or parental line (indicated). On the left is an electrophoregram of the amplicons obtained using the primers of CMCTTN181 in the NY × FAQ population (red); on the right is an electrophoregram of the amplicons obtained using the primers of CMAT141 in the SAS × DOYA population (black). Peak location represents the size of the various alleles
Discussion
The results of this study clearly point to a single major gene controlling acid levels in melon fruit. The results of the analysis of three different segregating populations, whose parents derive from different subgroups of the C. melo species, further extends the significance of this locus to the species as a whole. The crosses between PI 414723 × Dul, SAS × DOY, NY × FAQ together comprise selections from both subspecies of C. melo, subspecies agrestis and subspecies melo. Together, the six parental varieties represent the melon groups reticulatus, momordica, conomon, flexuosus, cantalupensis, and chate, described by Pitrat et al. (2000). The presence of the same QTL in such diverse genetic backgrounds suggests that the trait is determined throughout the species by variability in the same gene and is indicative of a major role of the evolution of this gene in determining this important domestication trait of the sweet dessert melons.
Although there was a single major QTL on LG8 which showed very high LOD scores for all the traits involved in fruit pH, there were also QTLs for the traits at other map positions. However, these were minor QTLs with low LOD scores and their physiological and genetic significance is not clear. Interestingly, the minor QTLs on LG 4, 11 and 12 behave in the opposite direction as would be expected and in each case the ‘Dulce’ allele is correlated with an increase in acidity by increasing citric acid. This may indicate that there exists the possibility of transgressive modulation of fruit acidity by combining alleles from the two parents. This may be evidenced by the transgressive segregation in the RILs for pH values, as well as the three components assayed (Table 3). Nevertheless, the major QTL on LG8 is responsible for the overwhelming contribution to fruit acidity.
Previously a study of QTLs for pH and organic acids in melon based on near introgression lines (NILs) derived from the cross of a conomon variety, PI161375, and an inodorus cultivar, Piel de Sapo (Obando-Ulloa et al. 2009). Since these two parents are both low acid lines the QTL results did not indicate a major gene for this trait; nevertheless a minor QTL was reported (pHqf8.4, Diaz et al. 2011; Obando-Ulloa et al. 2009) in the region and may indicate genetic variation in the same locus even between lines of similar pH. However, since the report was based on NILs the introgression was relatively large and comparative co-localization cannot be performed.
The identification of well-defined QTLs for pH, accompanied by concomitant differences in organic acid content allowed for the performance of a co-localization test to determine whether any of the QTLs can be accounted for by enzymes of organic acid metabolism. In this study, we focused on a comprehensive list of enzymes involved in organic acid metabolism in melon, including all of the members of individual gene families of both enzymes and proton transporters (Tables 1, 2). The enzymes comprise 19 different reactions, many of which are composed of multiple protein subunits. Some of the enzyme families comprise multiple isozymes which are associated with distinct cellular compartmentalization (Schnarrenberger and Martin 2002). Finally, some of the enzyme families are also encoded for by gene paralogues with apparent tissue functionalization. The proton transporter genes comprise those encoding 13 subunits of the vacuolar ATP-dependent proton transporter, as well as two different PPi-dependent proton transporters.
We identified polymorphisms for practically all of these genes and mapped 53 of them, including the major genes which are significantly expressed in developing melon fruit, as indicated by the public melon EST database (http://www.ICuGI.org). To reduce the possibility that one of the five unmapped genes co-localizes with the pH trait we performed a synteny analysis between map positions of the corresponding cucumber homologs and the mapped melon genes (Supplemental Fig. 1, Supplemental Table 2). None of the five cucumber homologs map to a syntenous position of the melon pH locus on LG8 near the melon ICD1 and ICD2. Since there are numerous chromosomal rearrangements between melon and cucumber we cannot conclude that these genes do not co-localize with the pH locus; nevertheless, it would seem highly unlikely.
The results of this co-localization mapping study point to a striking absence of co-localization between the candidate genes coding for organic acid metabolism and the QTLs for the components of fruit acidity. Only a single instance of potential co-localization was observed, between the minor QTL for pH on LG11 and the gene for a ATP-citrate lyase paralogue. Further analysis of fine-mapping populations and sub-lines will be necessary to discern the contribution, if any, of this gene to fruit acidity. The closest instance of co-localization with the pH locus is with the citric acid cycle gene ICD1 (mitochondrial NAD-isocitrate dehydrogenase-α) but the QTL analysis based on the RILs population separates the two by nearly 10 cM.
The negative results with regard to the co-localization of candidate genes and phenotypic traits are not altogether surprising. In our previous study of melon fruit quality QTLs, which focused on QTLs for fruit carotenoid and sugar contents, a total of over 50 candidate genes encoding the enzymes of the sugar and carotenoid metabolic pathways were mapped and none co-localized with any of the significant QTLs for either carotenoid or sugar contents (Harel-Beja et al. 2010). This included 13 genes from the carotenoid biosynthetic pathway and over 30 genes for the sugar metabolism pathway (Cuevas et al. 2008, 2009; Harel-Beja et al. 2010). Similar results have been reported for tomato in which the quantitative component of carotenoid content variability was also not found to be associated with the structural carotenogenesis genes in tomato (Liu et al. 2003). However, genetic control of qualitative differences in carotenoid composition, such as in high β-carotene (Beta) or δ-carotene (Delta) lines, is determined by structural genes of the carotenoid biosynthetic pathway (Liu et al. 2003). Similarly, the strategy of co-localization of candidate genes was successful in identifying genes for the qualitative differences in anthocyanin metabolism and ascorbic acid accumulation in tomatoes (De Jong et al. 2004; Stevens et al. 2007). Thus, it appears more likely for genes encoding metabolic pathway enzymes to control qualitative, rather than quantitative, variation. Furthermore, many of the QTL-candidate gene co-localization studies, particularly in tomato (e.g., Baxter et al. 2005; Bermudez et al. 2008; Causse et al. 2004; Schauer et al. 2006) have been based on the analysis of NILs which allows for the co-localization at the level of bins, harboring many centimorgans of introgression. Our co-localization studies utilized a RILs population, allowing for a relatively fine-tuned co-localization analysis and the reduction of possible false-positives in candidate gene determination.
The identification of the nucleoside uridine as a significant component inversely associated with melon fruit acidity was unanticipated and, at present, we cannot explain its physiological significance, or the mechanism of the interaction with organic acid accumulation. Uridine has not previously been reported for melon, or for other fruits, to the best of our knowledge. There have been recent reports of uridine levels in ziziphus fruits (Guo et al. 2010), ginseng roots (Qian et al. 2008) and fritillaria tubers (Cao et al. 2010) and the absolute amounts are similar to those we observed, in the μg/gm range. In contrast, the levels of organic acids are in the mg/gm range, ~1,000-fold higher, and it is likely that the uridine content contributes little to the fruit pH but is rather an, as yet, unexplained effect of the pH gene. Alternatively, the possibility exists, based on the present data, that the uridine QTL is an independent QTL closely linked to the pH QTL. Further detailed comparative metabolic analyses, together with the functional identification of the pH gene should shed light on this surprising observation. Nevertheless, the results of this study suggest that the biological function of the pH gene is not at the level of organic acid metabolism. Together with the observation that the parental lines differ greatly in pH values but not necessarily in citric and malic acid levels, also supports this suggestion.
In conclusion, the mapping results indicate that an as yet unidentified gene, not one of the obvious candidates involved directly in acid metabolism or vacuolar proton transport, determines fruit acidity in the melon fruit in the broad range of C. melo germplasm. The identification of this gene will likely shed light on the elusive topic of fruit acidity and on the evolution under domestication of the sweet dessert melon.
References
Baxter CJ, Sabar M, Quick WP, Sweetlove LJ (2005) Comparison of changes in fruit gene expression in tomato introgression lines provides evidence of genome-wide transcriptional changes and reveals links to mapped QTLs and described traits. J Exp Bot 56:1591–1604
Bermudez L, Urias U, Milstein D, Kamenetzky L, Asis R, Fernie AR, Van Sluys MA, Carrari F, Rossi M (2008) A candidate gene survey of quantitative trait loci affecting chemical composition in tomato fruit. J Exp Bot 59:2875–2890
Brune A, Muller M, Taiz L, Gonzalez PC, Etxeberria E (2002) Vacuolar acidification in citrus fruit: comparison between acid lime (Citrus aurantifolia) and sweet lime (Citrus limettioides) juice cells. J Am Soc Hortic Sci 127:171–177
Burger Y, Sa’ar U, Distelfeld A, Katzir N, Yeselson Y, Shen S, Schaffer AA (2003) Development of sweet melon (Cucumis melo) genotypes combining high sucrose and organic acid content. J Am Soc Hortic Sci 128:537–540
Burger Y, Paris H, Cohen R, Katzir N, Tadmor Y, Lewinsohn E, Schaffer AA (2009) Genetic diversity of Cucumis melo. Hortic Rev 36:165–198
Cao XW, Li J, Chen SB, Li XB, Xiao PG, Chen SL, Yang DJ (2010) Simultaneous determination of nine nucleosides and nucleobases in different Fritillaria species by HPLC-diode array detector. J Sep Sci 33:1587–1594
Causse M, Duffe P, Gomez MC, Buret M, Damidaux R, Zamir D, Gur A, Chevalier C, Lemaire-Chamley M, Rothan C (2004) A genetic map of candidate genes and QTLs involved in tomato fruit size and composition. J Exp Bot 403:1671–1685
Clepet C, Joobeur T, Zheng Y, Jublot D, Huang M, Truniger V, Boualem A, Hernandez-Gonzalez ME, Dolcet -Sanjuan R, Portnoy V, Creus AM, Caño-Delgado A, Katzir N, Giovannoni JJ, Bendahmane A, Aranda MA, Garcia-Mas J, Fei Z (2011) Analysis of expressed sequence tags generated from full-length enriched cDNA libraries of melon. BMC Genomics 12:252. doi:10.1186/1471-2164-12-252
Cuevas HE, Staub JE, Simon PW, Zalapa JE, McCreight JD (2008) Mapping of genetic loci that regulate quantity of beta-carotene in fruit of US Western Shipping melon (Cucumis melo L.). Theor Appl Genet 117:1345–1359
Cuevas HE, Staub JE, Simon PW, Zalapa JE (2009) A consensus linkage map identifies genomic regions controlling fruit maturity and beta-carotene-associated flesh color in melon (Cucumis melo L.). Theor Appl Genet 119:741–756
Danin-Poleg Y, Tadmor Y, Tzuri G, Reis N, Hirschberg J, Katzir N (2002) Construction of a genetic map of melon with molecular markers and horticultural traits, and localization of genes associated with ZYMV resistance. Euphytica 125:373–384
De Jong WS, Eannetta NT, De Jong DM, Bodis M (2004) Candidate gene analysis of anthocyanin pigmentation loci in the Solanaceae. Theor Appl Genet 108:423–432
Diaz A, Fergany M, Formisano G, Ziarsolo P, Blanca J, Fei Z, Staub JE, Zalapa JE, Cuevas HE, Dace G, Oliver M, Boissot N, Dogimont C, Pitrat M, Hofstede R, van Koert P, Harel-Beja R, Tzuri G, Portnoy V, Cohen S, Schaffer A, Katzir N, Xu Y, Zhang H, Fukino N, Matsumoto S, Garcia-Mas J, Monforte AJ (2011) A consensus linkage map for molecular markers and quantitative trait loci associated with economically important traits in melon (Cucumis melo L.). BMC Plant Biol 11:111
Etienne C, Rothan C, Moing A, Plomion C, Bodenes C, Svanella-Dumas L, Cosson P, Pronier V, Monet R, Dirlewanger E (2002) Candidate genes and QTLs for sugar and organic acid content in peach [Prunus persica (L.) Batsch]. Theor Appl Genet 105:145–159
Fang DQ, Federici CT, Roose ML (1997) Development of molecular markers linked to a gene controlling fruit acidity in citrus. Genome 40:841–849
Fulton TM, Chunwongse J, Tanksley SD (1995) Microprep protocol for extraction of DNA from tomato and other herbaceous plants. Plant Mol Biol Rep 13:207–209
Fulton TM, Bucheli P, Viorol E, Lopez J, Petiard V, Tanksley SD (2002) Quantitative trait loci (QTL) affecting sugars, organic acids and other biochemical properties possibly contributing to flavor, identified in four advanced backcross populations of tomato. Euphytica 127:163–177
Guo S, Duan JA, Tang YP, Zhu ZH, Qian YF, Yang NY, Shang EX, Qian DW (2010) Characterization of nucleosides and nucleobases in fruits of Ziziphus jujuba by UPLC–DAD–MS. J Agric Food Chem 58:10774–10780
Harel-Beja R, Tzuri G, Portnoy V, Lotan-Pompan M, Lev S, Cohen S, Dai N, Yeselson L, Meir A, Libhaber SE, Avisar E, Melame T, van Koert P, Verbakel H, Hofstede R, Volpin H, Oliver M, Fougedoire A, Stalh C, Fauve J, Copes B, Fei Z, Giovannoni J, Ori N, Lewinsohn E, Sherman A, Burger Y, Tadmor Y, Schaffer AA, Katzir N (2010) A genetic map of melon highly enriched with fruit quality QTLs and EST markers, including sugar and carotenoid metabolism genes. Theor Appl Genet 121:511–533
Itkin M, Rogachev I, Alkan N, Rosenberg T, Malitsky S, Masini L, Meir S, Iijima Y, Aoki K, de Vos R, Prusky D, Burdman S, Beekwilder J, Aharoni A (2011) GLYCOALKALOID METABOLISM 1 is required for steroidal alkaloid glycosylation and prevention of phytotoxicity in tomato. Plant Cell. doi:10.1105/tpc.111.088732
Kellner S, Seidu-Larry S, Burhenne J, Motorin Y, Helm M (2011) A multifunctional bioconjugate module for versatile photoaffinity labeling and click chemistry of RNA. Nucl Acids Res. doi:10.1093/nar/gkr449
Kubicki B (1962) Inheritance of some characters in muskmelons (Cucumis melo). Genet Polonica 3:265–274
Leach DN, Sarafis V, Spooner-Hart R, Wyllie SG (1989) Chemical and biological parameters of some cultivars of C. melo. Acta Hortic 247:353–356
Linden K (2004) Disinfection efficiency and dose measurement of polychromatic UV light (Paperback). IWA Publishing, UK
Liu Y-S, Gur A, Ronen G, Causse M, Damidaux R, Buret M, Hirschberg J, Zamir D (2003) There is more to tomato fruit colour than candidate carotenoid genes. Plant Biotechnol J 1:195–207
Luo M, Wang YH, Frisch D, Joobeur T, Wing RA, Dean RA (2001) Melon bacterial artificial chromosome (BAC) library construction using improved methods and identification of clones linked to the locus conferring resistance to melon Fusarium wilt (Fom-2). Genome 44:154–162
Marsh K, González P, Echeverría E (2001) Partial characterization of H-translocating inorganic pyrophosphatase from 3 citrus varieties differing in vacuolar pH. Physiol Plant 111:519–526
Moing A, Svanella L, Rolin D, Gaudillere M, Gaudillere JP, Monet R (1998) Compositional changes during the fruit development of two peach cultivars differing in juice acidity. J Am Soc Hortic Sci 123:770–775
Neff MM, Turk E, Kalishman M (2002) Web-based primer design for single nucleotide polymorphism analysis. Trends Genet 2:613–615
Obando-Ulloa JM, Eduardo I, Monforte AJ, Fernandez-Trujillo JP (2009) Identification of QTLs related to sugar and organic acid composition in melon using near-isogenic lines. Sci Hortic 121:425–433
Pitrat M, Hanelt P, Hammer K (2000) Some comments on infraspecific classification of cultivars of melon. Acta Hortic 510:29–36
Portnoy V, Diber A, Pollock S, Forer R, Lev S, Tzuri G, Harel-Beja R, Portnoy VH, Lewinsohn E, Tadmor Y, Burger J, Schaffer AA, Katzir N (2011) Use of non-normalized, non-amplified cDNA for 454-based RNA sequencing of fleshy melon fruit. Plant Genome 4:36–46
Qian ZM, Wan JB, Zhang QW, Li SP (2008) Simultaneous determination of nucleobases, nucleosides and saponins in Panax notoginseng using multiple columns high performance liquid chromatography. J Pharm Biomed Anal 48:1361–1367
Schauer N, Semel Y, Roessner U, Gur A, Balbo I, Carrari F, Pleban T, Perez-Melis A, Bruedigam C, Kopka J, Willmitzer L, Zamir D, Fernie AR (2006) Comprehensive metabolic profiling and phenotyping of interspecific introgression lines for tomato improvement. Nat Biotechnol 24:447–454
Schnarrenberger C, Martin W (2002) Evolution of the enzymes of the citric acid cycle and the glyoxylate cycle of higher plants: a case study of endosymbiotic gene transfer. Eur J Biochem 269:868–883
Stevens MA (1972) Citrate and malate concentrations in tomato fruits: genetic control and maturational effects. J Am Soc Hortic Sci 97:655–658
Stevens R, Buret M, Duffé P, Garchery C, Baldet P, Rothan C, Causse M (2007) Candidate genes and quantitative trait loci affecting fruit ascorbic acid content in three tomato populations. Plant Physiol 143:1943–1953
Ulrich J (1970) Organic acids. In: Hulme AC (ed) The biochemistry of fruits and their products. Academic Press, London, pp 89–117
Van Ooijen JW (2004) MapQTL® 5, software for the mapping of quantitative trait loci in experimental populations. Kyazma B.V, Wageningen
Van Ooijen JW, Voorrips RE (2001) JoinMap® 3.0, software for the calculation of genetic linkage maps. Plant Research International, Wageningen
Wang YM, Wyllie SG, Leach DN (1996) Chemical changes during the development and ripening of the fruit of Cucumis melo (Cv. Makdimon). J Agric Food Chem 44:210–216
Yamaguchi M, Hughes DL, Yabumoto K, Jennings WG (1977) Quality of cantaloupe muskmelons: variability and attributes. Sci Hortic 6:59–64
Acknowledgments
The authors gratefully acknowledge financial support from of the Chief Scientist, Ministry of Agriculture; The Israel Bio-Tov Consortium & MAGNET program, Israeli Ministry of Industry, Trade and Labor; Binational Agriculture Research and Development (BARD) Grant IS-2270-94 and IS-3877-06; Israel Science Foundation Grant No. 386/06. This work was supported by the EU Framework Programme 6 project Meta-Phor (grant no. FOOD-CT-2006-036220). The work in AA lab was supported by the European Research Council (ERC) SAMIT project. This paper is journal series #003-12 of the Agricultural Research Organization.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by H. Nybom.
S. Cohen, G. Tzuri, and R. Harel-Beja contributed equally.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Cohen, S., Tzuri, G., Harel-Beja, R. et al. Co-mapping studies of QTLs for fruit acidity and candidate genes of organic acid metabolism and proton transport in sweet melon (Cucumis melo L.). Theor Appl Genet 125, 343–353 (2012). https://doi.org/10.1007/s00122-012-1837-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00122-012-1837-3