Abstract
Yellow-seeded Brassica napus was for the first time developed from interspecific crosses using yellow-seeded B. juncea (AABB), yellow-seeded B. oleracea (CC), and black-seeded artificial B. napus (AACC). Three different mating approaches were undertaken to eliminate B-genome chromosomes after trigenomic hexaploids (AABBCC) were generated. Hybrids (AABCC, ABCC) from crosses AABBCC × AACC, AABBCC × CC and ABCC × AACC were advanced by continuous selfing in approach 1, 2 and 3, respectively. To provide more insight into Brassica genome evolution and the cytological basis for B. napus resynthesis in each approach, B-genome chromosome pairing and segregation were intensively analyzed in AABCC and ABCC plants using genomic in situ hybridization methods. The frequencies at which B-genome chromosomes underwent autosyndesis and allosyndesis were generally higher in ABCC than in AABCC plants. The difference was statistically significant for allosyndesis but not autosyndesis. Abnormal distributions of B-genome chromosomes were encountered at anaphase I, including chromosome lagging and precocious sister centromere separation of univalents. These abnormalities were observed at a significantly higher frequency in AABCC than in ABCC plants, which resulted in more rapid B-genome chromosome elimination in the AABCC derivatives. Yellow or yellow-brown seeds were obtained in all approaches, although true-breeding yellow-seeded B. napus was developed only in approaches 2 and 3. The efficiency of the B. napus construction approaches was in the order 1 > 3 > 2 whereas this order was 3 > 2 > 1 with respect to the construction of yellow-seeded B. napus. The results are discussed in relation to Brassica genome evolution and the development and utilization of the yellow-seeded B. napus obtained here.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The most economically important Brassica species are three basic diploid species, specifically, B. rapa (AA, 2n = 20), B. nigra (BB, 2n = 16) and B. oleracea (CC, 2n = 18), and three amphidiploids, each of which evolved as a natural allopolyploid following hybridization between pairs of the three diploids, viz., B. napus (AACC, 2n = 38), B. juncea (AABB, 2n = 36) and B. carinata (BBCC, 2n = 34) (U 1935). It is believed that these three diploids descended from a now-extinct ancestor (Röbbelen 1960). There is strong evidence that the ancestor was hexaploid (Parkin et al. 2005; Lysak et al. 2007). However, the origin and evolution of the ancestral karyotype remains somewhat controversial.
Brassica interspecific hybrids are ideal organisms for elucidating genomic homoeology, and they are essential as bridges and breeding materials (Chen et al. 2011). Homologies between the A, B and C genomes were initially revealed by conventional cytogenetic analyses in digenomic diploids, e.g., AC, BC and AB, digenomic triploids such as AAB, BBC and BCC, and trigenomic haploid ABC, all obtained from interspecific crosses in Brassica (U 1935; Attia and Röbbelen 1986; Attia et al. 1987; Busso et al. 1987; Choudhary et al. 2002). In recent years, researchers have shown increasing interest in Brassica trigenomic hybrids, especially trigenomic tetraploids, for cytogenetic analysis and assessing gene introgression utilizing modern genomic in situ hybridization (GISH) and molecular marker techniques (Ge and Li 2007; Nelson et al. 2009; Mason et al. 2010, 2011; Navabi et al. 2011). A small number of studies has been published on Brassica trigenomic hybrids, where GISH has been used to study autosyndesis within B-genome and allosyndetic pairing between B and A/C genome chromosomes, e.g., trigenomic haploids (ABC) derived from B. carinata × B. rapa (ArBcCc), natural B. napus × B. nigra (AnBnCn), synthetic B. napus × B. nigra (ArBnCo), trigenomic tetraploids derived from B. juncea × B. napus (AjAnBjCn), B. napus × B. carinata (AnBcCcCn), B. juncea × B. carinata (AjBjBcCc), and pentaploid hybrids (AnArBcCcCn) derived from hybridizations between B. carinata, B. rapa and B. napus (Li et al. 2004, 2005; Ge and Li 2007; Mason et al. 2010). There is evidence that remarkable genomic diversity existed in the subgenomes of different original Brassica species (Jiang et al. 2011), and this diversity may be responsible for the significant differences in the chromosome pairing configurations reported in several ABC hybrids derived from different interspecific hybridizations (Busso et al. 1987; Ge and Li 2007). Therefore, a complete study of chromosome pairing in all the hybrid forms with different subgenomes but the same genomic constitution could provide additional insight into the evolutionary relationships and homologies among the A, B and C genomes. Moreover, from a breeding perspective, it would be instructive to conduct a detailed meiotic analysis, including an evaluation of anaphase stages, of the trigenomic hybrids, which may allow the monitoring of target chromatin during introgression and prediction of the stability of the hybrid offspring.
Brassica juncea (AABB, 2n = 36) is currently grown as an oilseed in India, China, Eastern Europe and Pakistan. This species offers many desirable and complementary traits, including tolerance to heat and drought and resistance to blackleg and pod shattering (Woods et al. 1991; Roy 1984; Getinet et al. 1996). The transfer of blackleg and pod-shattering resistance from B. juncea to B. napus has been achieved (Roy 1984; Prakash and Chopra 1988). B. oleracea L. contains a wide array of wild taxa and many cultivated races that exhibit extensive morphological and physiological diversity (Gladis and Hammer 2001). Several favorable traits have been identified in B. oleracea, such as cold tolerance and resistance to blackleg and Sclerotinia sclerotiorum (Mithen et al. 1987; Mei et al. 2011). Interspecific hybridizations between B. juncea and B. oleracea could be used to synthesize bridging materials for elucidating genomic homoeology, expanding the genetic variability of Brassica crops and transferring desirable traits between different Brassica species.
In comparison with its black-seeded counterpart, yellow-seeded B. napus has several advantages, including higher oil and protein contents, lower fiber content and higher feed value (Stringam et al. 1974; Slominski et al. 1999; Rakow et al. 2011). Therefore, the development of yellow-seeded varieties has been an ongoing global objective in rapeseed quality breeding. Breeding efforts in the last several decades have led to the extensive development of yellow-seeded B. napus using yellow-seeded forms that exist in the natural germplasm of Brassica species, such as B. rapa (Chen and Heneen 1992, Rahman 2011), B. carinata (Qi et al. 1995), B. juncea (Tang et al.1997), B. juncea and B. carinata (Rashid et al. 1994), B. rapa and B. carinata (Meng et al. 1998; Rahman 2001) and B. rapa and B. juncea (Potapov and Osipova 2003). However, it is still difficult to obtain true-breeding B. napus that consistently yields pure bright yellow seeds (Rahman 2001; Rakow et al. 2011), which may be partly attributed to the lack of truly yellow-seeded B. oleracea as a parent for such crosses (Chen et al.1988; Rakow et al. 1999).
Few reports have yet been published on the occurrence and utilization of genes for yellow seed coat color from B. oleracea. Li et al. (2007) reported the discovery and improvement of several yellow-seeded B. oleracea var. acephala lines. By mating these lines with yellow-seeded B. rapa, we previously obtained stable yellow-seeded B. napus (Wen et al. 2008). The objective of the current study was to develop yellow-seeded B. napus through interspecific crosses between yellow-seeded B. juncea, yellow-seeded B. oleracea and black-seeded B. napus and to provide complementary information on the underlying cytogenetic basis of the production of B. napus through interspecific hybridization. We report herein the characterization of the cytogenetics, fertility and reproduction of the interploid hybrids obtained thereof. To the best of our knowledge, this is the first report rigorously comparing the exclusion efficiency of B-genome chromosomes in the development of B. napus from trigenomic tetraploids and pentaploids and it is also the first attempt to synthesize yellow-seeded B. napus by combining the yellow seed color genes from the A genome of yellow-seeded B. juncea and the C-genome of yellow-seeded B. oleracea var. acephala.
Materials and methods
Plant materials and field crosses
A pure yellow-seeded B. juncea line (breeding accession 6-142; AABB, 2n = 36), a yellow-seeded B. oleracea var. acephala accession (T6; CC, 2n = 18) and a black-seeded double haploid line (DH-6; AACC, 2n = 38) of resynthesized B. napus were selected for the hybridizations. Strain T6 originated from a yellow-seeded mutant of black-seeded B. oleracea var. acephala at XiNan University (Li et al. 2007). DH-6 was a stable line obtained from microspore culture of a resynthesized B. napus, which was produced through crosses between yellow-seeded B. campestris JB2 and yellow-seeded B. oleracea var. acephala T6, but that produced only black seeds in selfing generations. All the materials were planted in the field of Huazhong Agricultural University in Wuhan.
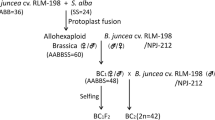
B. juncea 6-142 was used as the maternal parent in crosses with T6 in March 2004. Approximately 16–20 days after pollination, immature embryos were cultured on MS agar medium (Murashige and Skoog 1962). After plantlet formation, sequential culture and chromosome doubling were conducted to generate trigenomic hexaploids (AABBCC) using a method described previously (Wen et al. 2008). Plantlets with roots were transferred to the field in the middle of October. Thereafter, three different crossing approaches were performed in attempt to eliminate the B-genome chromosomes and to develop yellow-seeded B. napus (Fig. 1). In approach 1, the AABBCC plants were used as the maternal parents in a cross with DH-6 to generate trigenomic pentaploid (AABCC) hybrids. Subsequent self-pollination over six flowering seasons resulted in F7 seeds. For approach 2, embryo cultures were used to obtain trigenomic tetraploid hybrids (ABCC) after AABBCC plants were pollinated with the pollen of T6 in March 2005, and the ABCC hybrids were subsequently self-pollinated up to the F7 generation. In approach 3, ABCC plants were pollinated with the pollen of DH-6 in March 2006. The resulting hybrids from this ABCC × AACC cross were advanced to F6 seeds by self-pollination until 2011. To breed yellow-seeded B. napus in each approach, pedigree selection for seed color was applied, and only lighter-colored seeds harvested from the selected plants were sowed and advanced to the next generation.
Cytological methods and pollen stainability
To determine the chromosome numbers of the hybrids and the hybrid progeny, shoot tips, root tips or young ovaries were treated with 2 mM 8-hydroxyquinoline for 3–4 h at 22°C, fixed in a mixture of ethanol and acetic acid (3:1, v:v) for 24 h, and stored at −20°C (Liu and Li 2007). For meiotic analysis, flower buds were fixed in ethanol:acetic acid (3:1, v:v) for 24 h, transferred to a fresh mixture and then stored at −20°C. Mitotic and meiotic observations were made according to the methods of Li et al. (1995). Pollen stainability was determined as the percentage of pollen grains stained with 1% acetocarmine by counting more than 300 pollen grains collected from three flowers of each plant.
DNA extraction, probe labeling and GISH analysis
Three plants each of the AABCC and ABCC hybrids were used for chromosome pairing and separation analysis in pollen mother cells (PMCs). The total genomic DNA of B. nigra cv. Giebra (BB, 2n = 16) was labeled with biotin-11-dUTP (Sino-American Biology Co, Henan, P.R. China) by nick translation and used as a probe. The DNA of B. napus cv. Oro was sheared by boiling for 15 min and used as a blocking agent. Chromosome spreads were prepared according to Zhong et al. (1996). GISH was performed according to Ge and Li (2007). Two-by-two χ2 contingency tests were performed to compare the difference significance of B-genome chromosome pairing and segregation configurations in different interploids.
Pod-setting evaluation
The ratio of siliques to flowers was determined by counting all effective siliques and dividing by the total number of flowers from the main inflorescence or three primary branches. Seed yield in the siliques was assessed by recording the number of seeds per silique obtained from ten effective siliques.
Assessment of seed color in advanced progeny
Hybrid plants arising from the different approaches were self-pollinated in the field up to the F6 or F7 generation, with selection for yellow seed color in each generation. Seed color was assessed visually. Self-pollinated seeds produced on individual plants were classified as yellow, yellow-brown, brown-yellow, brown or black. The term yellow-brown was used when the majority of the seeds (above 60%) were yellow and a small fraction was brown-yellow and/or brown. The term brown-yellow was used when the majority of the seeds (above 60%) were brown and a small fraction was yellow-brown and/or yellow.
Results
Production of hexaploid (AABBCC), pentaploid (AABCC) and tetraploid (ABCC) plants
From 1,024 B. juncea buds pollinated with pollen from B. oleracea, 32 (3.13%) embryos were obtained, and 27 plantlets with intermediate phenotypes were recovered, all having an expected chromosome number of 27 (ABC, Fig. 2a). The addition of colchicine to the medium resulted in 9 (33.33%) chromosome-doubled plants, which exhibited a chromosome number of 2n = 54 at leaf mitotic metaphase (Fig. 2b). Compared with the trigenomic haploids (ABC), the hexaploids (AABBCC) exhibited larger flowers, greatly enhanced pollen fertility, thicker leaves and shorter internodes. GISH analysis of AABBCC plants showed eight labeled bivalents at diakinesis in most of the PMCs observed and an 8:8 segregation of the labeled chromosomes at anaphase I (AI) in all PMCs observed (Fig. 2c, d), revealing that these plants were completely chromosome-doubled. The selfed seeds from AABBCC plants were brown or black.
Chromosome counting and GISH analysis of the interploids derived from B. juncea × B. oleracea var. acephala. a Mitotic metaphase in a trigenomic haploid with a genomic constitution of ABC, 2n = 27. b A somatic cell from the shoot-tip meristem of a trigenomic hexaploid (AABBCC) showing the expected 54 chromosomes at prometaphase. c One diakinesis PMC from a trigenomic hexaploid showing eight labeled B-genome chromosome bivalents (red signals, arrow). d One anaphase I PMC from a trigenomic hexaploid showing an 8:8 segregation of labeled B-genome chromosomes (red signals, arrow). e Mitotic metaphase in a trigenomic pentaploid with a genomic constitution of AABCC, 2n = 46. f Mitotic metaphase in a trigenomic tetraploid with a genomic constitution of ABCC, 2n = 36. Scale bar 10 μm
For approach 1, to generate trigenomic pentaploids (AABCC), female AABBCC plants were crossed with resynthesized B. napus DH-6. Of the 105 hybrids tested, 39 were found to be pentaploids with 2n = 46 (Fig. 2e), whereas the others were aneuploids as a result of dyads or polyads from AABBCC plants. For approaches 2 and 3, crosses between AABBCC plants and B. oleracea var. acephala T6 were conducted to obtain trigenomic tetraploids (ABCC). A total of 153 hybrid plants survived to flowering of which 127 were aneuploids and 26 were trigenomic tetraploids with the expected chromosome number of 36 (Fig. 2f).
Chromosome associations in the PMCs of pentaploid (AABCC) and tetraploid (ABCC) plants
Chromosome pairing was examined at diakinesis or metaphase I(MI) in 99 PMCs from the AABCC hybrids. Except for 26 cells with the expected chromosome associations of 19II + 8I, 73.74% of the 99 PMCs analyzed exhibited variable configurations (Table 1). A maximum of 21II was found in 15 cells (15.15%), indicating two bivalents of B-genomic origin. In addition to the bivalents, multivalents such as trivalents, quadrivalents and even one heptavalent were noted in 38.38% of the PMCs (Fig. 3a). The mean association of the pentaploid hybrids was 6.98I + 18.85II + 0.32III + 0.07IV. The pairing specificities of the ABCC hybrids are summarized in Table 1. Among a total of 56 PMCs studied, 71.39% of the chromosome complement was paired, suggesting the occurrence of considerable intra- or intergenomic chromosome associations. The pairing was mostly observed as bivalency with a frequency of 10.86 per cell. More than nine bivalents occurred in 42 (75%) of the cells (Fig. 3b), with only one (1.79%) cell having the maximum of 13 bivalents. Compared with the AABCC hybrids, a notably higher frequency of multivalents, i.e., trivalents and quadrivalents, was recorded in the ABCC hybrids—more than 1.0 multivalent per PMC at diakinesis. The mean association of the ABCC hybrids was 10.30I + 10.86II + 0.88 III + 0.34 IV.
Cytology and GISH analysis of pentaploid (AABCC) and tetraploid (ABCC) hybrids between B. juncea and B. oleracea var. acephala. Blue and red indicate DAPI fluorescence and signal from the B. nigra probe, respectively. a An AABCC PMC at diakinesis showing a chromosome configuration of 8I (arrowheads) + 12II + 1III (long arrow) + 1IV (the short arrow) +1VII (hollow arrow). b An ABCC PMC at diakinesis showing a chromosome configuration of 11II (arrowheads) + 14I. c An ABCC PMC at diakinesis with two B-genome bivalents (arrows). d An ABCC PMC at diakinesis with one trivalent of B-genomic origin (arrow). e An ABCC PMC at diakinesis with one quadrivalent formed by four labeled B-genome chromosomes (arrow). f An AABCC PMC at early diakinesis with two allosyndetic bivalents formed by two B-genome and two A/C-genome chromosomes. g An ABCC PMC at diakinesis with four B-genome chromosomes associated with A/C-genome chromosomes (arrows). h An ABCC PMC at diakinesis with one trivalent formed by one labeled B-genome chromosome and two A/C-genome chromosomes (arrow). i An AABCC PMC at metaphase showing laggards, several of which are labeled with the B. nigra probe. j An ABCC PMC showing four laggards, which were all labeled by the B. nigra probe at anaphase I (AI). k An AABCC PMC showing 1:7 segregation of B-genome chromosomes at AI. l An AABCC PMC at AI showing the separation of one B-genome chromosome into sister chromatids. m An ABCC PMC at AI showing 10 red signals in the centromeric regions, four of which are smaller or of weaker intensity (arrows), indicating the separation of two B-genome chromosomes into sister chromatids. n An ABCC PMC at AI showing 11 red signals in the centromeric regions, six of which are smaller or weaker in intensity (arrows), indicating the separation of three B-genome chromosomes into sister chromatids (arrows). o An AABCC PMC showing B-genome chromosome laggards at anaphase II (AII). p An ABCC PMC showing B-genome chromosome laggards at AII. a–n share the same scaling, and o–p share another scaling. Scale bar 10 μm (color figure online)
B-genome chromosome pairing in the PMCs of pentaploid (AABCC) and tetraploid (ABCC) plants
GISH analysis of the PMCs from the AABCC and ABCC hybrids indicated that genome composition affected B-genome chromosome behavior at diakinesis (χ2 = 7.57, p < 0.05; Table 2; Fig. 3c–h). In 31 (31.63%) AABCC cells, all eight B-genome chromosomes remained as univalents, whereas in the other cells, one to four B-genome chromosomes were involved in inter- or intragenomic associations (Table 2). In the ABCC plants, the frequency of PMCs with eight B-genome univalents (20.91%) was rather low compared with the AABCC hybrids. Accordingly, in the PMCs analyzed, the percentage of total B-genome univalents was lower in ABCC plants (76.49%) than in AABCC plants (80.36%), but the difference between them was not significant (χ2 = 3.45, p > 0.05). In 17.35 and 12.73% of the PMCs analyzed, a maximum of two bivalents originating from B-genome autosyndesis were encountered in the AABCC and ABCC hybrids, respectively (Fig. 3c). Trivalents of such autosyndetic origin were also observed (Fig. 3d). Strikingly, one quadrivalent formed by B-genome chromosomes was found in two cells from the ABCC hybrids (Fig. 3e). With respect to the frequency of autosyndetic B-genome chromosomes, the difference between these two interploids was not significant (χ2 = 0.03, p > 0.05).
No more than two B-genome chromosomes undergoing allosyndesis in the same cell were observed in AABCC plants (Fig. 3f). However, up to four were observed in ABCC plants (Fig. 3g). A low frequency of allosyndetic multivalents, mainly trivalents and quadrivalents, was found (Fig. 3h). The frequency of B-genome chromosomes involved in allosyndesis was significantly higher in ABCC than in AABCC plants (χ2 = 6.78, p < 0.01). In both interploids, the frequency of B-genome chromosomes involved in autosyndesis was significantly higher than that for allosyndesis (AABCC: χ2 = 13.72, p < 0.01; ABCC: χ2 = 5.11, p < 0.05). The average numbers of unpaired, autosyndetic and allosyndetic B-genome chromosomes were, respectively, 6.43, 1.12, and 0.45 in AABCC plants and 6.12, 1.15, and 0.73 in ABCC plants.
B-genome chromosome segregation in the PMCs of pentaploid (AABCC) and tetraploid (ABCC) plants
Using GISH methods, we further investigated the segregation and elimination of the labeled B-genome chromosomes detected in the PMCs of the AABCC and ABCC plants (Tables 3, 4; Fig. 3i–p). At MI, the majority of chromosomes that orientated away from the equatorial plate were of B-genomic origin (Fig. 3i). At AI, most of the B-genome univalents in the PMCs from AABCC and ABCC, as a rule, failed to orient properly between the poles, either moving to the poles at random or remaining on the equatorial plate as laggards. In no cases were all eight B-genome chromosomes found as laggards at AI. A maximum of 5 and 4 B-genome chromosomes were observed as laggards in the PMCs from the AABCC and ABCC hybrids (Fig. 3j), respectively. The frequency of laggard B-genome chromosomes was significantly higher in the AABCC hybrids (10.27%) than in the ABCC hybrids (4.02%) (χ2 = 20.03, p < 0.01). Among those PMCs that did not exhibit B-genome chromosome irregularities, the most frequent distributions of B-genome chromosomes to the poles were 4:4 in AABCC and 5:3 in ABCC. Additionally, a segregation of 1:7 was scored at a low frequency in both plant types (Fig. 3k). A notable feature at AI was that in some cases, more than eight signals (9–11) were detected in PMCs of AABCC and ABCC hybrids. Some of those signals were smaller in size or weaker in intensity, indicating the occurrence of precocious division of B-genome centromeres (Fig. 3l–n). One to three B-genome chromosomes were assumed to undergo sister chromatid separation at AI, and in most cases, only one labeled chromosome evidenced this kind of abnormality. The frequency of PMCs with precociously divided B-genome chromosomes was slightly higher in the AABCC hybrids than in the ABCC hybrids, but the difference between them was not significant (χ2 = 0.54, p > 0.05). In general, the frequency of B-genome chromosomes exhibiting irregular distributions, such as chromosome lagging and precocious division of univalent centromeres, was significantly higher in the AABCC hybrids than in the ABCC hybrids (χ2 = 20.58, p < 0.001).
At anaphase II, the precociously divided chromosomes either migrated to the poles at random without further division or remained in the center as laggards. In the AABCC and ABCC hybrids, 78.31 and 67.95% of the PMCs showed labeled laggards, respectively (Fig. 3o, p). Occasionally, triads and dyads were observed as a consequence of abnormalities in spindle orientation. All the abnormalities reported herein compromised pollen fertility by generating mini-microspores or multispores. Pollen fertility was estimated at 73.07 ± 8.27% in the AABCC hybrids and 39.32 ± 10.38% in the ABCC hybrids.
Seed set and chromosome number distribution in early generation plants derived from the different approaches
An AABCC plant (F1 in approach 1; see Fig. 1) and an ABCC plant (F1 in approach 2; see Fig. 1), each possessing the expected chromosome number, were selected and assessed for selfed and hybrid seed sets. Consistent with the various meiotic abnormalities displayed in the PMCs and the pollen fertility data, selfed seed set was much lower for ABCC than AABCC plant (Table 5). As expected, the seed set of the ABCC plant was greatly enhanced upon pollination with pollen from DH-6. For the ABCC plant, the number of hybrid seeds per flower was approximately six times higher than the number of selfed seeds per flower. Root tips from 20 selfed and 20 hybrid seeds of the AABCC and ABCC plants were subjected to chromosome counting. Chromosome number varied greatly in the seeds derived from the different approaches. Presumably because of the abortion of most pollen with unbalanced chromosome numbers, the chromosome number distribution in the progeny from approach 2 was quite narrow in contrast with the broadly distributed chromosome number in the progeny from approaches 1 and 3. One seed with 2n = 38 was found among the seeds derived from approach 1, whereas among the seeds from approaches 2 and 3, none had a chromosome number of 38.
In March 2008, a total of 147 (F3), 167 (F3) and 225 (F2) plants obtained through approaches 1, 2 and 3, respectively, were randomly selected and subjected to somatic chromosome counting. As shown in Fig. 4, the chromosome number distribution of the plants from approach 1 was the narrowest: 78.23% (115 out of 147) of the plants examined had 38 or 39 chromosomes. The widest range of chromosome numbers was encountered in plants from approach 2. The numbers in these plants varied from 31 to 52, with 66.47% of the plants between 36 and 40. The frequency of plants with a chromosome number of 38 decreased from the plants of approach 1 (53.74%) to approach 3 (28.00%) to approach 2 (14.37%). A contingency χ2 test showed that significant differences existed in the frequency of plants with 2n = 38 among the different approaches (approaches 1 and 2, χ2 = 53.07; approaches 1 and 3, χ2 = 23.89; approaches 2 and 3, χ2 = 9.53; p < 0.01).
Chromosome number distribution in early generations obtained from the different approaches using B. juncea, B. oleracea var. acephala and artificial B. napus as parents. Approach 1: F3 generation of cross AABBCC × AACC; approach 2: F3 generation of cross AABBCC × CC; approach 3: F2 generation of cross (AABBCC × CC) × AACC. Trigenomic hexaploid AABBCC were obtained by doubling the chromosomes of hybrids from AABB × CC
Identification and characterization of newly derived B. napus lines
After selfing for two more generations, in March 2010, flower buds from partial progeny of plants with 2n = 38 were fixed and stored as described in the “Materials and methods” section. Buds from plants that produced seeds with lighter color were subjected to GISH analysis after harvesting. Ten F5 plants from approach 1 producing yellow-brown and brown-yellow seeds were tested and found to be free of B-genome chromosomes. In the case of approach 2, five F5 plants producing yellow or yellow-brown seeds were selected, and four of the plants, including one derived from a yellow-seeded F4 plant, carried only AC chromosomes. The remaining plant was found to have two labeled chromosomes (Fig. 5a1–a2), indicating the loss of two AC chromosomes and their substitution with two B-genome chromosomes. GISH analysis of one yellow-seeded F4 plant from approach 3 detected no B-genome chromosomes, and all the cells displayed normal chromosome segregation (Fig. 5b1–b2). Another yellow-seeded F4 plant produced only faint B-genome signals, indicating minor cross hybridization (Fig. 5c1–c2). A cytological analysis of the newly derived B. napus lines in the next spring showed that the number of multivalents and univalents was decreasing with advancing generations, and that meiotic stabilization with regular bivalent formation had been achieved (data not shown).
a1–c2 Identification of B-genome chromosomes in selfed progeny of interploid hybrids. a1–a2 A diakinesis PMC of a 38-chromosome derivative from approach 2, showing a configuration of 18 bivalents and 2 labeled univalents (arrows). b1–b2 An AI PMC of a 38-chromosome derivative from approach 3. No intact B-genome chromosomes were detected. c1–c2 A diakinesis PMC of a 38-chromosome derivative from approach 3, showing a configuration of 16II + 1III (arrowhead) +3I (short arrows). Only faint signals were detected (long arrow). Scale bar 10 μm. d–l Seed color of B. juncea, B. oleracea var. acephala and the progeny of interploid hybrids of B. juncea × B. oleracea var. acephala. d Yellow seeds from B. juncea 142. e Yellow seeds from B. oleracea var. acephala T6. f Brown-yellow seeds from an F3 plant from approach 1. g Yellow-brown seeds from an F6 plant from approach 1. h Yellow-brown seeds from an F2 plant from approach 2. i Yellow seeds (F7) from a true-breeding, pure yellow-seeded line from approach 2. j Yellow seeds from an F2 plant from approach 3. k, l Yellow seeds (F6) from two true-breeding, pure yellow-seeded lines from approach 3. Scale bar 1 cm
Having characteristics most similar to natural B. napus, the newly derived B. napus line was readily differentiated from the female B. juncea plant. However, the most conspicuous differences between the new and natural B. napus strains were observed in leaf shape and floral morphology. The radical leaves of the new B. napus lines were found to be elliptical to oblong-ovate, pinnatipartite to pinnatisect, with sinuate-dentate margins, obtuse tips and lobes on the long petioles. Larger and more distinct petals also facilitated the identification of the new B. napus. The newly developed B. napus lines displayed vigorous vegetative growth, high pollen stainability and good seed set. Nine derivative lines were subjected to evaluation for yield traits in June 2011. The range and mean number of seeds per silique in these lines were 15.11–23.83 and 20.59, respectively. The range and mean weight of 1,000 seeds were 2.83–4.48 and 3.74 g, respectively. The seed yield per plant ranged from 12.78 to 17.15 g with an average of 14.48 g (Supplemental Table S1).
Seed color of progeny obtained through the different approaches
In approach 1, the F1 seeds (genome AABCC) were brown or black. Of the 127 F2 plants, six had brown seeds. From the six brown-seeded families, about 180 F3 plants were raised, and 65 produced brown seeds, whereas 26 produced brown-yellow seeds (Supplemental Table S2, Fig. 5f). Seeds of these 91 plants were used to grow the F4 families. No pure yellow-seeded plants occurred in subsequent generations of these plants. However, yellow-brown seeds were observed in the F5 and F6 plants (Fig. 5g).
In approach 2, among 91 F2 plants, three produced yellow-brown seeds (Supplemental Table S2, Fig. 5h), 18 had brown-yellow seeds, and the others segregated for brown and black seeds. Therefore, 210 F3 plants were generated from these 21 F2 plants. All F3 families segregated for seed color. Lighter-colored F4 seeds were sown, and one F4 plant bearing almost all yellow seeds was found among 2,000 plants. Segregation for seed color continued in the following generation, but the yellow-seed-bearing F4 plant bred true for yellow, as confirmed by the F5, F6 and F7 seeds (Fig. 5i).
In approach 3, 21 yellow-brown seeds occurred among the hybrid seeds (F1) of ABCC × AACC. Two hundred and fifty yellow and yellow-brown seeds harvested from these F1 plants were selected to grow the F2 generation. Two F2 plants produced almost all yellow seeds (Supplemental Table S2, Fig. 5j), nine had yellow-brown seeds, and the other plants segregated for brown-yellow, brown and black seeds. Yellow, yellow-brown and brown-yellow seeds were then selected and sown, resulting in 2,113 F3 plants, among which 17 plants producing yellow seeds were found. In the successive generations, two lines raised from two F2 plants produced consistent and uniform yellow seeds from generations F3 to F5 (Fig. 5k–l).
Discussion
Interspecific hybridization between B. juncea and B. oleracea has been used to synthesize bridging materials for transferring desirable traits between Brassica species (Arumugam et al. 1996; Tonguc and Griffiths 2004; Chen et al. 2011). In this study, the AABCC and ABCC hybrids were synthesized as bridges for introducing yellow seed traits from B. juncea and B. oleracea to B. napus. Three different mating approaches were designed to eliminate B-genome chromosomes and synthesize yellow-seeded B. napus, and we compared the efficiency of the approaches. A detailed cytological analysis of the bridges used in each approach and their progeny could track the fate of B-genome chromosomes in the process of B. napus resynthesis, thereby providing new information on homoeologous pairing within the B-genome (autosyndesis) and between the B and A/C genomes (allosyndesis), as well as revealing the cytological basis for B. napus resynthesis in each approach.
It is widely accepted that the three Brassica diploid species, viz., B. rapa, B. nigra and B. oleracea, diverged from a common ancestor and exhibit considerable homology with each other (Lagercrantz and Lydiate 1996; Parkin et al. 2005). Thus, it was not unexpected to observe more than 19 bivalents in the AABCC hybrids and nine bivalents in the ABCC hybrids. In this study, a maximum of 21 and 13 bivalents were encountered in the AABCC and ABCC hybrids, respectively. Li et al. (2004) reported a maximum of 22 bivalents in AABCC hybrids derived from B. carinata × B. rapa. Mason et al. (2010) observed no more than ten bivalents in ABCC hybrids derived from B. napus × B. carinata. The present observations are inconsistent with their findings, which may be due to structural differences between subgenomes originating from different species (such as Bj and Bc, Aj and An). Given that homologous chromosomes can pair well even when the mating partners are of different species (Li et al. 2004; Mason et al. 2010; Navabi et al. 2011), the occurrence of two and four additional bivalents more than was expected in the AABCC and ABCC hybrids may be due to autosyndesis in the B-genome and allosyndesis between the B and A/C genomes. This speculation is strongly supported by our results of GISH analysis. We found that a maximum of two bivalents represented autosyndetic B-genome pairs in both the AABCC and ABCC hybrids, which is in agreement with the findings of Prakash (1973) in haploid B. nigra, Li et al. (2005) and Ge and Li (2007) in ABC hybrids and Mason et al. (2010) in ABCC hybrids. No significant difference was found in the frequency of autosyndetic B-genome chromosomes between the AABCC and ABCC hybrids, suggesting that autosyndesis within the B-genome is unaffected by the potential for allosyndesis between the B and A/C genomes. Mason et al. (2010) also reported that autosyndesis occurs independently of allosyndesis in ABBC hybrids. Thus, the results strongly suggest that the B-genome chromosomes involved in autosyndesis share a closer relationship with each other compared with the A/C genome chromosomes involved in allosyndesis (Quiros 1999). A maximum of two and four B-genome chromosomes were involved in the formation of allosyndetic bivalents in the AABCC and ABCC hybrids, respectively, suggesting that genome composition affects the rate of B-genome allosyndesis, as previously reported by Mason et al. (2010). The frequency of B-genome chromosomes involved in allosyndesis in ABCC plants was significantly higher than that in AABCC plants. The likely explanation for this phenomenon lies in the fact that AABCC hybrids provide conditions for the preferential pairing of the A- and C-genomes because of the presence of homologs whereas in ABCC hybrids, the haploid genome facilitates allosyndetic pairing. In previous investigations, maximum of one to four B-genome chromosomes involved in allosyndesis have been reported in various trigenomic hybrids (Ge and Li 2007; Mason et al. 2010). These inconsistencies imply that structural differentiation of the B-genome in different species and different parental genotypes also influence allosyndesis between the B and A/C genomes in trigenomic hybrids.
Precocious sister centromere separation of univalents at AI is undoubtedly an important mechanism leading to B-genome chromosome elimination in the progeny of the AABCC and ABCC plants. We speculated that this abnormal separation is not unique to B-genome chromosomes because, in several cases, additional unlabeled elements were encountered at AI in PMCs from the ABCC hybrids (Fig. 3n, 40 elements counted). Sister chromatid disjunction of univalent chromosomes at AI is a widely reported meiotic abnormality in wheat, Begonia, Vigna hybrids, Citrus hybrids and other plants, but it has scarcely been mentioned for Brassica species (Aragon-Alcaide et al. 1997; Dewitte et al. 2010; Benavente and Orellana 1986; Del Bosco et al. 1999). This phenomenon may arise spontaneously at a low frequency, or it may be caused by genetic factors (Del Bosco et al. 1999; Dewitte et al. 2010; Clayberg 1959; Chelysheva et al. 2005). However, the possibility that it occurs at random can be excluded in our material because all the plants we tested underwent this abnormality, and the frequency of PMCs exhibiting this phenomenon was over 13%. Studies on mutants have revealed that gene deletions impairing meiosis cohesion complexes, the monopolar attachment of sister kinetochores at meiosis I or the protection of centromeric cohesion throughout meiosis I are responsible for this anomaly (Watanabe 2004). In Arabidopsis, Rec8 and SCC3 are necessary to maintain centromere cohesion and the monopolar kinetochore orientation during meiosis I (Chelysheva et al. 2005). In maize, Hamant et al. (2005) reported the evidence that Sgo1 plays a major role in the protection of centromere cohesion until AII. Moreover, there is considerable evidence that both defects in homolog synapsis and the absence of a homologous partner are associated with an increased frequency of premature separation of sister chromatids at MI or AI in a variety of species (Moore and Orr-weaver 1998). However, the situation observed in the present study is quite different from those mentioned above. At AI, almost all the chromosomes, both from bivalents and univalents, split into sister chromatids in the mutants mentioned above, whereas in our materials, the prematurely dividing chromosomes undoubtedly proceed from the univalents at MI. Aragón-Alcaide et al. (1997) proposed that the presence of the Ph1 and Ph2 genes, which suppress homoeologous pairing in wheat, leads to centromeres with more condensed structures and increased sister chromatid cohesion, thereby preventing the disjunction of univalents at AI. Given that homoeologous pairing in haploid B. napus is under the genetic control of the major gene PrBn, it is reasonable to assume that there are genetic factors responsible for the homoeologous pairing in haploid B. juncea (Jenczewski et al. 2003; Mason et al. 2011). On the basis of these data, together with the prevalence of homoeologous pairing at diakinesis, we speculate that the premature division of univalents at AI is probably due to an afunctional homoeologous pairing control of the alleles from both B. juncea and B. napus parents in the ArAjBjCoCo and AjBjCoCo hybrids.
That genes for yellow seed color exist in both the A and C genomes holds the key to the successful breeding of yellow-seeded B. napus. To retain these yellow seed color genes and even the overall C-genome of B. oleracea in the progeny, crossing programs were performed using artificial B. napus but not natural B. napus. We observed as a general trend that the efficiency of B-genome chromosome elimination and new B. napus reconstruction decreased from approaches 1 to 3 to 2, in that order. This result is in accordance with the cytological observations of the AABCC and ABCC hybrids and their progeny in each approach, suggesting that a higher frequency of B-genome laggards and univalents undergoing precocious division at AI led to a lower frequency of their transition into gametes. The fact that yellow or yellow-brown seeds were produced in each approach indicates that our crossing scheme was effective. In Arabidopsis thaliana, yellow seed color occurs when one or more gene(s) encoding important enzymes are mutated in the pathway for dark-pigment synthesis or accumulation (Rahman 2007), and different Brassica genomes and subgenomes may harbor different but related pathways for pigmentation (Chen and Heneen 1992). Taken together, we conclude that functional complementation took place between the two flavonoid biosynthetic pathways encoded by the Ar and Co genomes, resulting in normal seed pigmentation and black seed color in the resynthesized B. napus ArArCoCo (DH-6), whereas in the newly derived B. napus, the relationship between the Aj and Co genomes is in the opposite situation, resulting in yellow seed color. We expected that plants combining genes for yellow seed color in the Aj and Co genomes would be isolated most efficiently by approach 1. However, yellow seeds were instead produced most abundantly in approach 3, followed by approaches 2 and then 1. Given that the frequency of allosyndetic B-genome chromosomes was significantly higher in ABCC than in AABCC plants, there may be more opportunity for recombination to take place between the B and A/C component genomes in ABCC than in AABCC plants, as also evident from the report by Navabi et al. (2011), and the transfer of yellow seed color genes from the B to A genome may partly account for the more rapid occurrence of yellow seeds in approaches 2 and 3. One of the most important points to consider in generating yellow-seeded B. napus through interspecific hybridization is the genotype of the parents. Studies of three crosses using various B. juncea inbred lines as female parents have indicated an influence of parental genotype on the seed color of newly derived B. napus lines (data not shown), as we found in the hybridization of B. rapa and B. oleracea (Wen et al. 2008). This observation again clearly demonstrates the complexity of the underlying genetic mechanism responsible for seed color. Nevertheless, our schemes represent a step forward in developing more systematic and meaningful procedures for breeding yellow-seeded B. napus.
In summary, this study demonstrated that interspecific hybridization between yellow-seeded B. juncea and yellow-seeded B. oleracea is a feasible strategy for breeding yellow-seeded B. napus. The advantage of using the AABCC and ABCC hybrids as bridge materials is that homoeologous recombination may occur between the A, B and C genome chromosomes in these interploids, thus making it possible to produce more variations. Using molecular marker techniques, it is possible to detect translocations and to differentiate B-genome linkage groups that were retained in the new B. napus line and aneuploids generated incidentally during the breeding process (Navabi et al. 2011). The newly derived B. napus line may become a source of germplasm for broadening the genetic base and expanding yellow seed resources for B. napus. Importantly, by taking advantage of hybrids between the newly derived line and natural B. napus, stronger intersubgenomic heteroses between the subgenomic Aj and An, Ar and An, and Co and Cn components could be studied and utilized (Zou et al. 2010). From a breeding viewpoint, it is imperative that the reproductive efficiency and combining ability of the new B. napus lines, including those studied here, be properly evaluated in field trials before being used for heterosis.
References
Aragón-Alcaide L, Reader S, Miller T, Moore G (1997) Centromeric behaviour in wheat with high and low homoeologous chromosomal pairing. Chromosoma 106:327–333
Arumugam N, Mukhopadhyay A, Gupta V, Pental D, Pradhan AK (1996) Synthesis of hexaploid (AABBCC) somatic hybrid: a bridging material for transfer of ‘tour’ cytoplasmic male sterility to different Brassica species. Theor Appl Genet 92:762–768
Attia T, Röbbelen G (1986) Cytogenetic relationship within cultivated Brassica analyzed in amphihaploids from the three diploid ancestors. Can J Genet Cytol 28:323–329
Attia T, Busso C, Röbbelen G (1987) Digenomic triploids for an assessment of chromosome relationships in the cultivated diploid Brassica species. Genome 29:326–330
Benavente E, Orellana J (1986) Differential anaphase I behavior between wheat and rye univalents in triticale—wheat hybrid plants. Genetics 69:161–166
Busso C, Attia T, Röbbelen G (1987) Trigenomic combinations for the analysis of meiotic control in the cultivated Brassica species. Genome 29:331–333
Chelysheva L, Diallo S, Vezon D, Gendrot G, Vrielynck N, Belcram K, Rocques N, Marquez-Lema A, Bhatt AM, Horlow C, Mercier R, Mezard C, Grelon M (2005) AtREC8 and AtSCC3 are essential to the monopolar orientation of the kinetochores during meiosis. J Cell Sci 118:4621–4632
Chen BY, Heneen WK (1992) Inheritance of seed colour in Brassica campestris L. and breeding for yellow-seeded B. napus L. Euphytica 59:157–163
Chen BY, Heneen WK, Jonsson R (1988) Resynthesis of Brassica napus L. through interspecific hybridization between B. alboglabra Bailey and B. rapa L. with special emphasis on seed colour. Plant Breed 101:52–59
Chen S, Nelson MN, Chèvre AM, Jenczewski E, Li ZY, Mason AS, Meng JL, Plummer JA, Pradhan A, Siddique KHM, Snowdon RJ, Yan GJ, Zhou WJ, Cowling WA (2011) Trigenomic bridges for Brassica improvement. Crit Rev Plant Sci 30:524–547
Choudhary BR, Joshi P, Ramarao S (2002) Cytogenetics of Brassica juncea × Brassica rapa hybrids and patterns of variation in the hybrid derivatives. Plant Breed 121:292–296
Clayberg CD (1959) Cytogenetic studies of precocious meiotic centromere division in Lycopersicon eseulentum Mill. Genetics 44:1335–1346
Del Bosco SF, Tusa N, Conicella C (1999) Microsporogenesis in a Citrus interspecific tetraploid somatic hybrid and its fusion parents. Heredity 83:373–377
Dewitte A, Eeckhaut T, Van Huylenbroeck J, Van Bockstaele E (2010) Meiotic aberrations during 2n pollen formation in Begonia. Heredity 104:215–223
Ge XH, Li ZY (2007) Intra- and intergenomic homology of B-genome chromosomes in trigenomic combinations of the cultivated Brassica species revealed by GISH analysis. Chromosome Res 15:849–861
Getinet A, Rakow G, Downey RK (1996) Agronomic performance and seed quality of Ethiopian mustard in Saskatchewan. Can J Plant Sci 76:387–392
Gladis T, Hammer K (2001) Nomenclatural note on the Brassica oleracea-group. Genet Resour Crop Evol 48:7–11
Hamant O, Golubovskaya I, Meeley R, Fiume E, Timofejeva L, Schleiffer A, Nasmyth K, Cande WZ (2005) A REC8-dependent plant shugoshin is required for maintenance of centromeric cohesion during meiosis and has no mitotic functions. Curr Biol 15:948–954
Jenczewski E, Eber F, Grimaud A, Huetb S, Lucasa MO, Monodb H, Chèvrea AM (2003) PrBn, a major gene controlling homeologous pairing in oilseed rape (Brassica napus) haploids. Genetics 164:645–653
Jiang CC, Ramchiary N, Ma YB, Jin M, Feng J, Li RY, Wang H, Long Y, Choi SR, Zhang CY, Cowling WA, Park BS, Lim YP, Meng JL (2011) Structural and functional comparative mapping between the Brassica A genomes in allotetraploid Brassica napus and diploid Brassica rapa. Theor Appl Genet 123:927–941
Lagercrantz U, Lydiate DJ (1996) Comparative genome mapping in Brassica. Genetics 144:1903–1910
Li Z, Liu HL, Luo P (1995) Production and cytogenetics of intergeneric hybrids between Brassica napus and Orychophragmus violaceus. Theor Appl Genet 91:131–136
Li MT, Qian W, Meng J, Li Z (2004) Construction of novel Brassica napus genotypes through chromosomal substitution and elimination using interploid species hybridization. Chromosome Res 12:417–426
Li MT, Li ZY, Zhang CY, Qian W, Meng JL (2005) Reproduction and cytogenetic characterization of interspecific hybrids derived from crosses between Brassica carinata and B. rapa. Theor Appl Genet 110:1284–1289
Li JN, Yin JM, Zhou QY, Lin N, Chen L, Chen SZ (2007) Agronomic performance and seed quality of a new source of yellow-seeded Brassica oleracea. In: Proceedings of 12th International Rapeseed Congress, Wuhan, China, 26–30 March, pp 114–116
Liu M, Li ZY (2007) Genome doubling and chromosome elimination with fragment recombination leading to the formation of Brassica rapa-type plants with genomic alterations in crosses with Orychophragmus violaceus. Genome 50:985–993
Lysak MA, Cheung K, Kitschke M, Bures P (2007) Ancestral chromosomal blocks are triplicated in Brassiceae species with varying chromosome number and genome size. Plant Physiol 145:402–410
Mason A, Huteau V, Eber F, Coriton O, Yan G, Nelson MN, Cowling WA, Che`vre AM (2010) Genome structure affects the rate of autosyndesis and allosyndesis in AABC, BBAC and CCAB Brassica interspecific hybrids. Chromosome Res 18:655–666
Mason A, Nelson MN, Castello MC, Yan GJ, Cowling WA (2011) Genotypic effects on the frequency of homoeologous and homologous recombination in Brassica napus × B. carinata hybrids. Theor Appl Genet 122:543–553
Mei J, Qian L, Disi JO, Yang X, Li Q, Li J, Frauen M, Cai D, Qian W (2011) Identification of resistant sources against Sclerotinia sclerotiorum in Brassica species with emphasis on B. oleracea. Euphytica 177:393–399
Meng JL, Shi SW, Gan L, Li ZY, Qu XS (1998) The production of yellow-seeded Brassica napus (AACC) through crossing interspecific hybrids of B. campestris (AA) and B. carinata (BBCC) with B. napus. Euphytica 103:329–333
Mithen RF, Lewis BG, Heaney RK, Fenwick GR (1987) Resistance of leaves of Brassica species to Leptosphaeria maculans. Trans Br Mycol Soc 88:525–531
Moore DP, Orr-Weaver TL (1998) Chromosome segregation during meiosis: building an unambivalent bivalent. Curr Top Dev Biol 37:263–299
Murashige T, Skoog F (1962) A revised medium for rapid growth and bioassays with tobacco tissue cultures. Plant Physiol 15:473–497
Navabi ZK, Stead KE, Pires JC, Xiong ZY, Sharpe AG, Parkin IAP, Rahman MH, Good AG (2011) Analysis of B-genome chromosome introgression in interspecific hybrids of Brassica napus × B. carinata. Genetics 187:659–673
Nelson MN, Mason AS, Castello MC, Thomson L, Yan G, Cowling WA (2009) Microspore culture preferentially selects unreduced (2n) gametes from an interspecific hybrid of Brassica napus L. × Brassica carinata Braun. Theor Appl Genet 119:497–505
Parkin I, Gulden S, Sharpe A, Lukens L, Trick M, Osborn T, Lydiate D (2005) Segmental structure of the Brassica napus genome based on comparative analysis with Arabidopsis thaliana. Genetics 171:765–781
Potapov DA, Osipova GM (2003) Development of yellow seeded Brassica napus in Siberia. In: Proceedings of 11th International Rapeseed Congress, vol 1, Copenhagen, 6–10 July, pp 250–252
Prakash S (1973) Haploidy in Brassica nigra Koch. Euphytica 22:613–614
Prakash S, Chopra VL (1988) Introgression of resistance to shattering in Brassica napus from Brassica juncea through non-homologous recombination. Plant Breeding 101:167–168
Qi CK, Fu SZ, Pu HM (1995) A successful transfer of yellow-seeded trait from Brassica carinata to B. napus. In: Proceedings of 8th International Rapeseed Congress, vol 4, Cambridge, 4–7 July, pp 1137–1140
Quiros CF (1999) Genome structure and mapping. In: Go′mez-Campo C (ed) Biology of Brassica Coenospecies. Elsevier, Amsterdam, pp 217–246
Rahman MH (2001) Production of yellow-seeded Brassica napus through interspecific crosses. Plant Breed 120:463–472
Rahman M (2007) Development of molecular markers for marker assisted selection for seed quality traits in oilseed rape. PhD Dissertation, University of Manitoba
Rahman H (2011) Untraditional approaches of interspecific hybridization in Brassica for introgression of traits from allied species. In: Proceedings of 13th International Rapeseed Congress, Prague, 5–9 June, pp 867–870
Rakow G, Raney JP, Relf-Eckstein J (1999) Agronmic performance and seed quality of a new source of yellow seeded Brasscia napus. In: Proceedings of the 10th International Rapeseed Congress Canberra, 26–29 September
Rakow G, Relf-Eckstein JA, Olson T (2011) Review and update on the development of yellow seed Brassica napus canola. p. 55. In: Abstract Book of 13th International Rapeseed Congress, Prague, 5–9 June
Rashid G, Rakow RK, Downey (1994) Development of yellow seeded Brassica napus through interspecific crosses. Plant Breed 112:127–134
Röbbelen G (1960) Beiträge zur Analyse des Brassica-Genomes. Chromosoma 11:205–228
Roy NN (1984) Interspecific transfer of Brassica juncea-type high blackleg resistance to Brassica napus. Euphytica 33:295–303
Slominski BA, Simbaya J, Campbell LD, Rakow G, Guenter W (1999) Nutritive value for broilers of meals derived from newly developed varieties of yellow-seeded canola. Anim Feed Sci Technol 78:249–262
Stringam GR, Mcgregor DI, Pawlowski SH (1974) Chemical and morphological characteristics associated with seed coat colour in rapeseed. In: Proceedings of 4th International Rapeseed Congress Giessen, 4–8 June, pp 9–108
Tang ZL, Li JN, Zhang XK, Chen L, Wang R (1997) Genetic variation of yellow-seeded rapeseed lines (Brassica napus L.) from different genetic sources. Plant Breed 116:471–474
Tonguc M, Griffiths PD (2004) Development of black rot resistant interspecific hybrids between Brassica oleracea L. cultivars and Brassica accession A 19182, using embryo rescue. Euphytica 136:313–318
U N (1935) Genome analysis on Brassica with special reference to the experimental formation of B. napus and peculiar mode of fertilization. Jpn J Bot 7:389–452
Watanabe Y (2004) Modifying sister chromatid cohesion for meiosis. J Cell Sci 117:4017–4023
Wen J, Tu JX, Li ZY, Fu TD, Ma CZ, Shen JX (2008) Improving ovary and embryo culture techniques for efficient resynthesis of Brassica napus from reciprocal crosses between yellow-seeded diploids B. rapa and B. oleracea. Euphytica 162:81–89
Woods DL, Capraca JJ, Downey RK (1991) The potential of mustard (Brassica juncea (L.) Coss.) as an edible oil crop on the Canadian prairies. Can J Plant Sci 71:195–198
Zhong XB, de Hans JJ, Zabel P (1996) Preparation of tomato meiotic pachytene and mitotic metaphase chromosomes suitable for fluorescence in situ hybridization (FISH). Chromosome Res 4:24–28
Zou J, Zhu JL, Huang SM, Tian ET, Xiao Y, Fu DH, Tu JX, Fu TD, Meng JL (2010) Broadening the avenue of intersubgenomic heterosis in oilseed Brassica. Theor Appl Genet 120:283–290
Acknowledgments
This research was financed by the High-Tech program ‘‘863’’ (2011AA10A104), the Program for Modern Agricultural Industrial Technology System (nycytx-00501) and the National Natural Science Foundation of China (31000721).
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by I. Rajcan.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Wen, J., Zhu, L., Qi, L. et al. Characterization of interploid hybrids from crosses between Brassica juncea and B. oleracea and the production of yellow-seeded B. napus . Theor Appl Genet 125, 19–32 (2012). https://doi.org/10.1007/s00122-012-1813-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00122-012-1813-y