Abstract
Improved knowledge of genome composition, especially of its repetitive component, generates important informations in both theoretical and applied research. In this study, we provide the first insight into the local organization of the sunflower genome by sequencing and annotating 349,380 bp from 3 BAC clones, each including one single-copy gene. These analyses resulted in the identification of 11 putative gene sequences, 18 full-length LTR retrotransposons, 6 incomplete LTR retrotransposons, 2 non-autonomous LTR-retroelements (LINEs), 2 putative DNA transposons fragments and one putative helitron. Among LTR-retrotransposons, non-autonomous elements (the so-called LARDs), which do not carry any protein-encoding sequence, were discovered for the first time in the sunflower. The insertion time of intact retroelements was measured, based on sister LTRs divergence. All isolated elements were inserted relatively recently, especially those belonging to the Gypsy superfamily. Retrotransposon families related to those identified in the BAC clones are present also in other species of Helianthus, both annual and perennial, and even in other Asteraceae. In one of the three BAC clones, we found five copies of a lipid transfer protein (LTP) encoding gene within less than 100,000 bp, four of which are potentially functional. Two of these are interrupted by LTR retrotransposons, in the intron and in the coding sequence, respectively. The divergence between sister LTRs of the retrotransposons inserted within the genes indicates that LTP gene duplication started earlier than 1.749 MYRS ago. On the whole, the results reported in this study confirm that the sunflower is an excellent system to study transposons dynamics and evolution.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Improved knowledge of genome composition, especially of its repetitive component, generates important information in both theoretical and applied research, for example to improve strategies for genetic and physical mapping of genomes and for the discovery and development of molecular markers. Moreover, knowledge of genome composition is a prerequisite for the annotation steps in sequencing projects both of ESTs (Expressed Sequence Tags) and of genomic regions.
To date, substantial progress has been made in unveiling the structure and organization of plant genomes. In the emerging view of plant evolution, it is well established that angiosperm species radiation has been accompanied, if not promoted, by polyploidization events and differential amplification of a repetitive component of their genomes represented by the long-terminal repeat (LTR) retrotransposons (REs) (Grover et al. 2008; Soltis and Soltis 1999). LTR-retrotransposons (LTR-REs) are capable of replicating through a copy and paste mechanism and have the potential to increase the genome size of their host in a very short time span (Hawkins et al. 2006; Neumann et al. 2006; Piegu et al. 2006).
Sequencing of several plant genomes have revealed that the large degree of genomic variation and the occurrence of non-shared genomic sequences in closely allied grass species can be ascribed to the very young age of their extant LTR-REs complement.
The replicative mechanism of LTR-REs, coupled with the error-prone nature of transcription and reverse transcription, determines the generation of different RE families, characterized by sequence variability in both the coding, transcribed portion and in the LTRs (Beguiristain et al. 2001). RE families have been reported to amplify differentially in different lineages within single plant groups or even within a single species (e.g., in maize) over a time span of less than 1 million years (Brunner et al. 2005; Wang and Dooner 2006). Similar events have taken place in several cereal species (Scherrer et al. 2005; Piegu et al. 2006; Vitte and Bennetzen 2006; Paterson et al. 2009) and in some dicots as well, even though to a less dramatic extent (Hawkins et al. 2006; Holligan et al. 2006; Neumann et al. 2006; Ungerer et al. 2006). In the recently sequenced sorghum genome, for example, the concomitant action of transposable element insertion and removal by illegitimate recombination or by DNA loss resulted in an average insertion age of 0.8 million years and in 50% of the detected elements having inserted within the last 500,000 years (Paterson et al. 2009).
Among species with large genomes, grasses such as maize, barley and wheat are by far the group of plants for which most information on retrotransposon-related genome structure has been collected. Apart from Gossypium species, relatively little attention has been given to large genome-sized dicotyledons, despite their great economic importance. For example, studies on the genome composition and organization in the Asteraceae family, which is very large and includes very important crop species such as sunflower, are at their very beginning (Cavallini et al. 2010).
Sunflower (Helianthus annuus L.) is the most important species belonging to the genus Helianthus, the relatively recent origin of which ranges between 4.75 and 22.7 million years. Based on the geographic distributions of its closest relatives, the genus Helianthus likely originated in Mexico, with subsequent migration through North America (Schilling et al. 1998). Sunflower haploid genome size is around 3,000 Mb. New Helianthus species have arisen by interspecific hybridization, some of which have been extensively studied (Rieseberg et al. 2003; Gross et al. 2007).
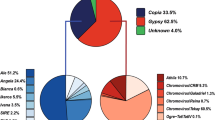
Sample sequencing of a small-insert genomic library from sunflower provided a set of sequences that were used to analyze the composition of the sunflower genome in terms of types and abundance of repetitive elements (Cavallini et al. 2010). The fraction of repetitive sequences amounted to 62% of the sequences, while the putative functional genes accounted for 4%; the largest component of the repetitive fraction of the sunflower genome was represented by LTR-REs, especially of the Gypsy superfamily. Class II elements were barely represented in the library.
The identification of transposable elements was, however, difficult in sunflower because of the paucity of sequences of previously described and annotated elements. While a fraction of the coding portions of the elements were recognized through the BlastX homology searches, the non-coding portions (e.g., the long-terminal repeat regions of LTR-REs) were much more difficult to detect due to the high rate of sequence evolution of transposable elements between species (Ma and Bennetzen 2004). Sequencing of large genome regions appears to be more effective for identifying and characterizing repetitive sequences than BLAST homology searches of relatively short sequences. For example, a more accurate dating of amplification events of the LTR-RE component requires a comparison of the two LTR sequences from single elements that can be obtained from the sequencing of large genomic regions (SanMiguel et al. 1996).
For these reasons, we sequenced and annotated three clones from a sunflower BAC library, for a total of 349,380 bp. By this analysis, we provide the first insight into the local organization of the sunflower genome showing nests of REs inserted into each other and allowing the estimation of retroelement insertion ages. Different waves of retroelement mobilization during the evolution of this species and the occurrence of very recent retrotransposition events are suggested.
Materials and methods
BAC library screening
A bacterial artificial chromosome (BAC) library from sunflower inbred Ha383 was available from the CUGI (USA). We chose three genes for which bibliographic information and experimental evidences were suggested to be in single copy: a Lipid Transfer Protein-Encoding Gene (LTP), a Dehydrin-Encoding Gene (DHN) and a Z-Carotene Desaturase-Encoding Gene (DES).
The three selected genes were used to develop three probes to screen the BAC library. For each gene, we performed PCR, using specific primers: 5′-TGGCAAAGATGGCAATGATG-3′ and 5′-ATCAAAGACACATACACATCCATA-3′for LTP; 5′- GCAGCATATGGCAAACTACCGAGGAGATAA-3′ and 5′-CGAATTCGTGAAACCACATACAAAACAAAA-3′ for DHN; 5′-GGCAAGCTGCAGGGTTGG-3′ and 5′-AGACTCAGCTCATCAACT-3′ for DES. Sequences were amplified using 100 ng of genomic DNA as a template; thermocycling was performed at 94°C for 30 s, 60°C for 30 s and 72°C for 60 s, for 30 cycles, using Taq-DNA polymerase (Promega). PCR products were then used as templates for probes construction.
Radioactive 32P probes were prepared with [α-32P]dCTP by a random-primed synthesis with Klenow fragments (Roche) using 25 ng of each PCR product. Probes were purified using ProbeQuant G-50 Micro Columns (GE Healthcare). BAC library hybridizations with the three probes were carried out in 5× SSC, 5× Denhardt solution, 0.5% SDS and 100 μg/ml salmon sperm DNA for 16 h at 65°C and the nylon filters were washed with 0.3 × SSC and 0.1% SDS at 65°C. Filters were exposed for 2 days to a multipurpose phosphor storage screen (Cyclone Storage Phosphor System, Packard, CT, USA) to obtain a digital image of the radioactivity distribution. The obtained digital images were then analyzed using a phosphoimager (Cyclone Storage Phosphor system, Packard).
To avoid false-positive results, hybridization-positive clones were submitted to a PCR amplification using the specific primers reported above: by this way we could verify if the selected gene was actually included in the clone.
Among the hybridization-positive, PCR-positive BAC clones, we selected one clone per gene to be sequenced and analyzed (DES: clone 0516 M24; DHN: clone 0340 D07; LTP: clone 0148 M20).
BAC clones sequencing
The three selected BAC clones were sequenced with a shotgun strategy (Tarchini et al. 2000) using a standard protocol at 11–12× redundancy (considering only bases of Phred quality ≥ 20). As much as 10 μg of DNA was extracted by two subsequent maxipreps from each of three Helianthus annuus genomic BAC clones. BAC DNAs were treated with Plasmid-Safe™ ATP-Dependent DNase (Epicentre) to remove contaminating bacterial chromosomal DNA.
DNA was sheared by Hydroshear (Genomics Solution) at the following setting parameters: DNA volume: 200 μl, # of cycles = 15, speed code = 13.
DNA was purified and concentrated by using filter columns (QIAquick PCR Purification Kit, QIAGEN™) and resuspended in 40 μL of double-distilled water. Uncompleted ends were repaired in a 50-μL reaction mix using the End-It™ DNA End-Repair Kit (Epicentre™), following the indications of the manufacturer. End-repaired DNA was run on a 1% agarose gel. Fragments in the size range of 2.5–4.0 kb were selected and DNA was purified from the gel using the QIAquick Gel Extraction Kit (QIAGEN™) and ligated into pSmart-LC plasmid using the CloneSmart LCAmp Blunt Cloning Kit (Lucigen™) according to the manifacturer’s protocol; 1 μL of this ligation mix was then used to transform E. coli strain DH10β using the OF10G Supreme™ Electrocompetent Cells (Lucigen™) and a Bio-Rad Gene Pulser II electroporator. Recombinants were selected on Luria–Bertani plates with ampicillin.
Mate-paired reads were produced by sequencing with BigDye Terminator Cycle Sequencing Kit (Applied Biosystems™) and the SL1 and SR2 primers. The samples were purified by ethanol precipitation and were subsequently run on an ABI 3730xl capillary sequencer, starting from minipreps prepared with the MultiScreen Plasmid384 system (Millipore). The total number of sequences (1,536 mate-paired per clone, 700 bp read length on average) was then trimmed using PHRED and assembled using PHRAP (http://www.phrap.org) and PCAP. PCR primers were designed to walk across the sequence gaps by extracting the non-repetitive ends of the relevant contig sequences and importing them together into the Primer 3.0 program (Rozen and Skaletsky 2000). Subcontigs robustly connected by clone mates were merged manually where the sequencing failed. Merged sequences were further confirmed by PCR on genomic DNA.
Sequences are deposited at EMBL database under the accession numbers JN021934-36, and at the Department of Crop Plant Biology of Pisa University repository Web site (http://www.agr.unipi.it/Sequence-Repository.358.0.html).
Sequence analysis
The method used for BAC sequence annotation and transposable elements identification was partially based on an automatic pipeline for BLAST searches. Customized PERL scripts were utilized to fragment the complete sequences of both BAC clones into several partially overlapping 2,500 bp-long regions, which were subsequently analyzed by automatic BLASTX and BLASTN searches with MPI BLAST software (http://mpiblast.lanl.gov) against public non-redundant databases at GenBank. BLAST results for each fragment were later recombined into a single file after automatic correction of nucleotide coordinates. Since the number of BLAST hits that can be provided in a single search is limited and highly conserved motifs are redundant, this procedure increased the number of matches along the whole BAC sequences by allowing for detection of additional weaker, but still significant homologies. To limit false-positive detection, we used a fixed E-value threshold of E < 10−5 for BLASTN and E < 10−10 for BLASTX.
Repetitive DNA content of each BAC clone was estimated by masking sequences using BLAST software against the RepBase (Jurka 2000) and the sunflower small-insert genomic library (Cavallini et al. 2010).
To identify homologies to conserved features of already known retroelements, the complete sequences from each of the three BAC clones were used to conduct BLASTX and BLASTN searches against non-redundant databases at GenBank and screened for similarity matches to either REs gag-pol polyprotein or transposase, or other characterized gene products typically encoded by transposable elements. LTR retroelements were also identified using LTR FINDER (Xu and Wang 2007) and DOTTER softwares (Sonnhammer and Durbin 1995). LTR-FINDER uses a suffix array-based algorithm to construct all exact match pairs that are extended to long highly similar pairs. Alignment boundaries are obtained adjusting the ends of LTR pair candidates using the Smith–Waterman algorithm. These boundaries are re-adjusted based on the occurrence of typical LTR-RE features, such as being flanked by the dinucleotides TG and CA, at the 5′ and 3′ ends, respectively, and the presence of a target-site duplication (TSD) of 4–6 bp of a putative 20–25 bp-long primer binding site (PBS), complementary to a tRNA at the end of putative 5′-LTR, and of a 20–25 bp-long polypurine tract (PPT) just upstream of the 5′ end of the 3′ LTR.
For LTP gene copies analysis, sequences were aligned using ClustalW (Thompson et al. 1994), then genetic similarity between each sequence was measured using the DNAdist program of the PHYLYP package (Felsenstein 1989). The triangular matrix was imported into NTSYS-pc version 2.01 h package (Rohlf 1998) to construct dendrograms using the UPGMA in the SAHN routine for cluster analysis. The number of synonymous substitutions per site between LTP genes was calculated using DnaSP (Rozas and Rozas 1999).
Insertion age calculation of full-length retroelements
Retrotransposon insertion age was estimated comparing the 5′- and 3′-LTRs of each putative RE. The two LTRs of a single RE are identical at the time of insertion because they are mostly copied from the same template. The two LTRs were aligned with ClustalW software, indels were eliminated and the number of nucleotide substitutions per site were calculated using DnaSP (Rozas and Rozas 1999).
Insertion time estimates are based on the occurrence of nucleotide substitutions between LTRs using a nucleotide substitution rate of 2.0 × 10−8 synonymous substitutions per site per year proposed for sunflower REs by Ungerer et al. (2009). According to this rate, insertion time for each intact RE was estimated.
DNA isolation and hybridization
Seeds of the sunflower HCM line were washed in tap water and germinated on moist paper in Petri dishes and plants were grown in the open air. Young leaves were collected and DNA purification was carried out according to Cavallini et al. (2010). A sunflower small-insert library (Cavallini et al. 2010) was used for relative quantification of the transposons identified in the BAC clones. As much as 40 µl of plasmid DNA from each of the clones of the sunflower small-insert library was first linearized by overnight digestion with EcoRI (4 units) in a total volume of 50 μl. DNA was then denatured for 10 min at 91°C and gridded at moderate density (4 × 4) in duplicate using a Beckman Biomek 2000 replicator tool onto nylon membranes that had been presoaked in denaturation buffer. Filters were then denatured for 3 min in 1.5 M NaCl and 0.5 M NaOH, neutralized for 15 min in 1.5 M NaCl and 0.5 Tris HCl, pH8, and rinsed in 5× SSC. Filters were then exposed to UV light for 2.5 min. The clones arrayed on the membranes were probed using total labeled genomic DNA from Helianthus annuus, H. petiolaris, H. argophyllus, H. debilis, H. ciliaris, H. pumilus, H. atrorubens, H. giganteus, H. simulans, H. tuberosus, Viguiera multiflora, Tithonia rotundifolia, and other Asteraceae (Xanthium strumarium, Calendula officinalis, Senecio vulgaris, Tagetes erecta, Achillea spp., Bellis perennis, Gerbera spp., Leontopodium spp., Taraxacum officinalis and Cynara scolymus). Total genomic DNA from each species was isolated from young leaves and digoxigenin labeled by the random-primed DNA labeling technique using a DIG DNA Labeling Kit (Roche) according to the manufacturer’s recommendations. Hybridization and detection were performed as described by Cavallini et al. (2010). Labeled lambda DNA was also used as control probe. The relative hybridization intensity for each spot in macroarrays was analyzed by eye and quantified in arbitrary units in the range 0–3, where 0 is for not labeled, 1 for slightly labeled, 2 for labeled and 3 is for heavily labeled. For each transposons identified in BAC clones, the hybridization intensity was calculated as the mean of intensity of each corresponding clone.
Whole genome shotgun sequencing by Illumina’s Sequencing-By-Synthesis (SBS) technology.
A genomic library was prepared from 5 μg of genomic DNA from the same line of H. annuus using the Illumina PE DNA Sample Prep kit according to the manufacturer. After spin column extraction and quantification, the library was loaded on Cluster Station to create CSMA (clonal single molecular array) and sequenced at ultra-high throughput on the Illumina’s Genome Analyzer IIx platform to produce 75-bp paired-end reads. Then, alignments to BAC sequences were performed at 1,000-bp intervals using the program Genomics Workbench 3.0 (CLC Bio) and the number of Illumina hits was calculated along the BAC sequences.
Results
BAC sequencing and annotation
We chose three genes that bibliographic information and experimental evidences suggested to be in single copy: a Lipid Transfer Protein-Encoding Gene (LTP), a Dehydrin-Encoding Gene (DHN) and a Z-Carotene Desaturase-Encoding Gene (DES). The three selected genes were used as probes to screen a BAC library. Three selected BAC clones were sequenced, yielding the nucleotide sequence of three large genomic regions of 135,613 bp (LTP clone), 110,201 bp (DES clone) and 103,566 bp (DHN clone). Sequencing of 3 BAC clones provides significant new insights into sunflower genomic organization (Table 1). BLASTX and BLASTN searches against non-redundant databases at GenBank identified besides LTP, DES and DHN genes, other eight protein-encoding genes (Table 2). The BAC clone carrying the LTP gene revealed that this gene is present in five copies of different length and sequence (see below).
The pairwise comparison between the three BAC clones resulted in a low percentage of significant homology, ranging from 2.9 to 12.2% of each clone sequence, indicating no excessive redundancy between the three regions.
Eleven gene sequences (accounting for 21,525 bp, Table 2) were found in the three BAC sequences (accounting for 349,380 bp), i.e., gene sequences account for 6.16% of the BAC sequences. For comparison, it may be observed that in the sunflower small-insert library (Cavallini et al. 2010), identified gene sequences (700 bp long, on average) were 64 over 1,638 of the whole library, i.e., 3.91%. Consequently, gene sequence content appears overestimated in the BAC clones selected for sequencing, as expected because clones that contain genes (therefore probably corresponding to genic regions) were specifically chosen.
Performing BLASTX, JDOTTER and LTR-FINDER analyses resulted in the identification of 18 full-length LTR-REs, namely with intact ends, irrespective of whether these elements were potentially functional or contained inactivating mutations in their internal sequence (Tables 1, 3). Seven of them belong to the Gypsy superfamily, five to the Copia superfamily and six are putative LARDs, i.e., non-autonomous retroelements. We also found eight incomplete REs (5 Gypsy, 1 LARD and 2 LINEs) that exhibited ill-defined or truncated boundaries. Moreover, two putative DNA transposons fragments, and a putative helitron, interrupted by two LTR-REs, were present.
The arrangement of REs denoted extensive transposition activity in the regions and, similar to that observed in maize (SanMiguel et al. 1996), in many cases elements inserted into others; in one case, two different retroelements were inserted in a single element. On the whole, 15 out of 29 transposons found in the BAC sequences were single, namely adjacent to sequences of the host genome.
All the putatively intact LTR-REs are annotated in Table 4. Of the 29 transposons identified in the BAC clones, 21 were also detected in the small-insert library by homology searches (BLAST E-value smaller or equal to 1 × 10−10). The annotated map of DES, DHN and LTP BAC clones are reported in Fig. 1.
Annotation of DES, DHN and LTP BAC clones and number of Illumina hits matching to BAC sequences. Transposon sequences are indicated according to Wicker et al. (2007). Incomplete LTR-REs are indicated with the letter f in their code
To improve BAC annotation, 55 millions of 75-mers obtained by Illumina SBS were aligned to BAC sequences (Fig. 1). Peaks of Illumina 75-mers occurred in regions corresponding to LTR-REs, especially Gypsy elements and LARDs, while Copia elements were less represented. However, extensive variation in redundancy, as determined by Illumina library alignment, can be observed within superfamilies. For example, DESRLG1f, DESRLG2, DESRLG2f, DESRLX3f, DHNRLG2 and LTPRLG1 show the largest redundancy, with 40,000 Illumina hits or more.
Only a few regions (at the 5′-end of the DHN clone and at the 3′-end of the DES clone) showed high Illumina redundancy and could not be annotated by BLAST analysis, confirming that most of the repetitive component of the sunflower genome were represented by retrotransposons. Interestingly, at the 3′-end of DES clone the highest peak of Illumina hits is found, with more than 160,000 hits. This region corresponds to the sunflower most repetitive family (named Contig 61, Cavallini et al. 2010), whose nature was unknown. Unfortunately, not even the present analyses allowed establishing the nature of this repeat, which therefore remains unknown.
It is also to be noted that, in nested elements, inserted elements are often differently redundant than host elements. For example, in the LTP clone the Gypsy element LTPRLG1, interrupted by another Gypsy element (LTPRLG2), is highly redundant, contrary to the nested element. The opposite trend is observed for DHNRLG2 inserted into DHNRLC1 (Fig. 1).
Transposon dynamics
Of the 29 transposons identified in the three BAC clones, 26 are retroelements; there were 24 LTR-REs (Gypsy, Copia, and LARDs). In many cases (18 REs), they were complete elements (Table 3).
A complete element can be defined as one that shows two relatively intact LTRs and identified PPT and PBS sites, and is also flanked by TSDs. They were first classified as belonging to Gypsy (RLG, Wicker et al. 2007), Copia (RLC) or LARD (RLX) superfamilies according to BLAST similarity of their internal (i.e., between LTRs) portion to NCBI and REPBASE (Jurka 2000) databases. The coordinates and the characteristics of the complete LTR-REs are reported in Table 4.
The time of insertion of intact retroelements was estimated, based on sister LTR divergence. Indeed, at the time an element inserts into the genome, the LTRs are usually 100% identical since the retroelement transcription starts from the R region in 5′ LTR and terminates at the end of the R region in 3′ LTR, thus including only one copy of each U5 and U3 regions. Combination of single-copy U5 and U3 regions with a hybrid R region during reverse transcription into cDNA yields two identical LTRs at both termini of retroelements prior to integration (Kumar and Bennetzen 1999). As time passes, mutations occur within the LTRs at a rate that has been proposed to be higher than that of single-copy regions, at least in rice (Ma and Bennetzen 2004). Hence, LTR retroelements have a built-in clock that can be used to estimate the insertion age (SanMiguel and Bennetzen 1998).
It is to be recalled that the estimation of insertion time by the number of mutations in sister LTRs is subject to error, because it assumes the same mutation rates in all retroelements and chromosome positions (Cossu et al., in preparation). Anyway, this method appears as the most suitable to study RE dynamics.
Eighteen LTR pairs, logically identified in full-length elements by JDOTTER and homology analyses, were aligned and nucleotide distance was assessed. The same analysis was performed to four complete LTR-REs (one Copia, two Gypsy, and one LARD) found in the sequence of two other BAC clones available in GenBank (FJ269356 and GU074383). Insertion age was calculated using the substitution rate of 2.0 × 10−8 reported for sunflower REs by Ungerer et al. (2009) according to a personal communication by M. Barker and L. Rieseberg, University of British Columbia. Insertion time estimates based on LTR divergence were consistent with the relative layering of nested REs.
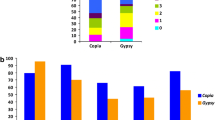
We observed a peak of elements with LTR divergence between 1.0 and 1.2 MYRS (Fig. 2); another peak is observed within the last 200,000 years, and a Copia RE does not show variations in its LTRs, suggesting that its insertion should have occurred between 0 and 54.000 years, i.e., the retrotransposition process could be still active.
The three superfamilies showed different time span activity that overlapped only partially. Gypsy elements are by far the most recently inserted, followed by LARDs; Copia elements transposition is scattered, from relatively ancient to very recent (Fig. 2).
Genome expansion related to the amplification of Gypsy and Copia retroelements has been shown to occur in the evolution of three Helianthus hybrid species adapted to extreme environments (Ungerer et al. 2009). In agreement with the results reported by Ungerer et al. (2009), our data show that mobilization waves of REs in sunflower are very recent, compared to other species (see for example Baucom et al. 2009; Bennetzen 2007; Ma et al. 2004).
To analyze the conservation of transposons (complete and fragmented) contained in the BACs within the genus Helianthus and other Asteraceae, we hybridized genomic DNA from four annual and six perennial Helianthus species, from Viguiera multiflora and Tithonia rotundifolia (two Helianthus related species), and from another ten Asteraceae species (see “Materials and methods”) to a panel of 1,344 clones from a small-insert library of sunflower spotted on nylon membranes (Cavallini et al. 2010) and analyzed clones sharing their sequence with REs identified in the BAC clones.
The signals detected in many spots indicated that the repetitive sequences occurring in the BAC clones are present in high copy number in H. annuus and conserved enough in sequence to be detected by hybridization in the other species (Fig. 3). The conservation of transposon families is clearly evident not only within Helianthus, but also in other Asteraceae, despite their estimated evolutionary distance.
Mean hybridization intensity of clones from a small-insert library and with sequence similarity to 21 transposons identified in three BACs, spotted on nylon membrane and hybridized with labeled genomic DNAs of H. annuus, four annual and six perennial Helianthus species, Viguiera multiflora, Tithonia rotundifolia and other ten Asteraceae species. Hybridization signal intensity of each clone was evaluated in arbitrary units: 0, lack of signal; 1, low-intensity signal; 2, medium intensity signal; and 3, strong-intensity signal. For each transposons is reported the mean of labeling intensities of small-insert clones corresponding to that transposon
The three superfamilies show different patterns of hybridization in different groups of species of Helianthus (Fig. 3): Copia elements are equally redundant in annuals and perennials, while Gypsy REs are generally much more frequent in annual species than in perennials. Interestingly, LARDs are generally much more redundant in perennial species than in annuals, despite being identified in H. annuus (i.e., an annual species).
These different redundancy patterns suggest that the REs identified in the three BAC clones occurred in the progenitor of the genus before splitting of annuals and perennials; however, LARDs have increased their number especially in perennials, and Gypsy elements especially in annuals. This is consistent with the recent burst of transposition observed for Gypsy elements in the sequenced BACs.
Concerning DNA transposons, those containing a transposase gene are fragmented, indicating that they were subjected to large mutations and/or deletions. The third is probably a helitron, because of the occurrence of putative diagnostic features (Du et al. 2008). Such features include: (i) a putative helicase encoding sequence; (ii) many ATC trinucleotides in the 5′ helicase flanking region; (iii) two CTRRT sequences, preceded (at −11 nucleotides) by putative hairpin sequences in the 3′ helicase flanking region. The helicase gene resulted interrupted by the insertion of a Gypsy element, on its turn interrupted by a LARD. This putative helitron sequence is the first to be described in sunflower. The insertion of the Gypsy element into the helitron can be dated to 1.14 MYRS ago; accordingly, the putative helitron was inserted before that date.
The LTP locus
Sequencing of the BAC clone highlighted that the LTP locus comprises five copies of the LTP gene, named LTP1 to LTP5; three of these LTP gene copies are forward oriented (LTP2, 4 and 5) and two are reverse oriented (LTP1 and 3; Fig. 4). All copies show two exons and one intron. LTP1 is interrupted by a non-autonomous RE in its coding region and it is presumably inactivated. Also LTP4 is interrupted by an LTR-RE (of the Copia superfamily); in this case, however, the retroelement is inserted into the intron, and therefore the functionality of LTP4 cannot be ruled out. In fact, the coding regions of LTP4, as also those of LTP2, 3 and 5, do not show stop codons, indicating the possibility that all these gene copies encode functional protein sequences.
Schematic representation of five copies of the LTP-encoding gene in the LTP BAC clone. Boxes indicate the coding portion of the genes; inner boxes represent introns. REs interrupting or strictly adjacent to LTP genes are represented as triangles. Numbers indicate the coordinates of each gene in the LTP–BAC sequence
Considering LTP gene copies without inserted REs, the coding portion is always 351 bp; intron length is more variable, ranging from 121 to 627 bp. LTP1 and LTP4 have an RE inserted in their coding portion and intron, respectively; excluding inserted REs from their sequence, LTP1 coding portion is 357 bp long and LTP4 intron is 6627 bp long.
Dot plot analysis shows that only coding portions are repeated, while regions adjacent to each gene copy seem to be specific to each gene. In fact, extensive variability is found in the putative proximal promoter regions; a 2,000-pb region, upstream of each gene, was scanned for regulatory cis-elements against the PLACE database (Higo et al. 1999): a number of putative regulatory elements were found, of different types and in different number for the different gene copies. The number of some cis-elements, selected especially among those responsive to environmental changes, show large variability (see Supplementary Materials), suggesting that each gene follows a specific expression pattern. On the contrary, at protein sequence levels, only minor variations are observed, which probably do not affect LTP function. Actually, K a (the number of non-synonymous substitutions per site) ranges from 0.01 to 0.04. Such values are very low compared to K s (the number of synonymous substitutions per site), ranging from 0.1 to 0.3, i.e., tenfold the K a. This suggests conservative selection for LTP gene sequences.
A phylogenetic analysis, by the neighbor-joining method of the 5 LTP gene copies was performed using an LTP encoding sequence of Lactuca sativa (GenBank accession number EF101532) as outgroup (Fig. 5). The dendrogram allows deducing a first duplication originating two ancestor sequences that on their turn duplicated once and twice, respectively. The occurrence of intact REs within LTP1 and LTP4 allows at least partial elucidation of the time course of LTP gene duplications. According to divergence between sister LTRs, the Copia element interrupting LTP4 inserted recently, because no nucleotide substitutions were observed between LTRs. On the contrary, insertion date of the LARD nested into LTP1 amounts to 1.749 MYRS. Therefore, it can be concluded that gene duplication started before 1.749 MYRS ago.
Dendrogram obtained from UPGMA cluster analysis of five LTP gene copies in the LTP BAC clone. LTP Ls indicates an LTP coding sequence of Lactuca sativa used as the outgroup. For sunflower sequences, the putative time interval of duplication (in MYRS) is indicated at each node, based on a synonymous substitution rate per year of 1 × 10−8
Actually, hypothesizing that duplicated LTP genes originated from a unique ancestor, the number of synonymous substitutions per site between LTP gene copies should allow to date each duplication event. Based on the synonymous substitution rate of 1.0 × 10−8 proposed for sunflower genes by Barker and Rieseberg (see above), we have calculated the putative dates of duplication events (Fig. 5). It can be supposed that duplications started between 31.5 and 34.5 MYRS ago, and that the last duplication (involving LTP1 and LTP3 genes) occurred between 7.8 and 10.0 MYRS ago, i.e., before the insertion of REs within two of LTP genes, as expected.
Discussion
Sequencing large genomic regions allowed improving the characterization of the sunflower genome, beyond available biochemical, cytological and molecular data.
The repetitive component of the H. annuus genome amounts to more than 60% (Cavallini et al. 2010). LTR-RE redundancy is very large and has been described in a number of studies (Santini et al. 2002; Natali et al. 2006; Ungerer et al. 2009; Cavallini et al. 2010). As in the genome of other plant species, LTR-REs are the vast majority, with large prevalence of Gypsy over Copia elements. In each of the three selected BAC clones, we could find nested REs, suggesting that transposition is pervasive of the whole genome.
In the three BAC clones, we have isolated and characterized a number of complete retroelements, adding numerous sequences to the only complete retroelement till now described in the sunflower, HACRE1 (Buti et al. 2009). Both the number of retroelements in the sequenced BAC clones and Illumina data confirm that Gypsy elements are prevalent over Copia ones in the sunflower genome (see Cavallini et al. 2010), similar to other plant species. For example, in angiosperms, Gypsy superfamily is more represented than Copia superfamily in the genomes of papaya, (with respective ratio of 5:1, Ming et al. 2008), Sorghum (4:1, Paterson et al. 2009), rice (3:1, The International Rice Genome Sequencing Project 2005) and poplar (Tuskan et al. 2006). On the contrary, Copia elements are prevalent over Gypsy ones in grapevine (2:1, The French-Italian Public Consortium for Grape Genome Characterization 2007). Maize genome shows a similar abundance of the two classes (Meyers et al. 2001), with Gypsy elements especially concentrated in gene-poor regions and Copia REs overrepresented in gene-rich ones (Baucom et al. 2009; Schnable et al. 2009). Similar data are reported for other cereal species with large genomes such as wheat and barley (Vicient et al. 2005; Paux et al. 2006). Species of the Gossypium genus show a variable proportion of Gypsy versus Copia elements with Gypsy elements prevailing in species with larger genome sizes (Hawkins et al. 2006). Such a comparison, though referred to superfamilies, confirms that the dynamics of retrotransposons are different in different species. Further data would be necessary to evaluate if different RE families have undergone different transposition waves, as for example observed in poplar (Cossu et al., submitted).
It is worth noting that, for the first time, putatively complete non-autonomous elements (the so-called LARDs, Kalendar et al. 2004) have been identified in the sunflower; in fact, this class of REs can be identified only when their complete sequence is available, allowing to recognize the occurrence of LTRs. The number of intact LARDs is the same as the intact Copia elements, suggesting that the redundancy of LARD superfamily is similar to that of the Copia.
Most of the identified REs appear to be specific to Helianthus, as already suggested by previous studies (Natali et al. 2006). The redundancy of each element was estimated using an Illumina library of the same sunflower line. Illumina 75mers were aligned to the three BAC sequences and showed a strict correspondence to the annotation: peaks of redundancy are observed in the regions containing REs; moreover, differences can be found among different elements confirming the possibility of using SBS technologies for relative quantification purposes, as reported by Swaminathan et al. (2007).
Concerning retrotransposon dynamics, the identification of sister LTRs allowed for the first time to date the insertion of retroelement in the sunflower genome using this method, established by Ma et al. (2004) in maize or barley. An analysis of insertion age based on comparison of RT-coding sequences of sunflower was carried out by Ungerer et al. (2009) that reported large and recent activity of elements in Helianthus species derived from interspecific hybridization between H. annuus and H. petiolaris. All the REs identified in the three BAC clones show a relatively recent insertion time, in a time span of 0 to 2.6 MYRS. These data indicate that in the sunflower, as in maize (Brunner et al. 2005; Wang and Dooner 2006), retrotransposon burst is very recent and probably still occurring, as already suggested by Cavallini et al. (2010), Ungerer et al. (2009), and Vukich et al. (2009a). On the other hand, it has been recently demonstrated that many sunflower elements are transcribed even in the absence of environmental stimuli (Buti et al. 2009; Vukich et al. 2009b). Vukich et al. (2009b) also showed that, even at a very low rate, transcription of retroelement is followed by insertion in another chromosomal site, i.e., it results in an increase of retrotransposon number.
With regard to LTR-RE superfamilies, some differences can be observed in the insertion time between Copia and Gypsy elements. Also in other species, LTR-RE superfamilies are subjected to different amplification histories during the evolution of the host; for instance, in wheat, Copia and Gypsy superfamilies are differently represented in the A and B genome (Charles et al. 2008). An example of different amplification histories among RE families was reported for Copia elements of Vitis vinifera (Moisy et al. 2008) and Populus trichocarpa (Cossu et al., submitted).
It has been suggested that the capacity to transpose of an LTR-RE is related to its redundancy, i.e., low redundant REs are more active than high redundant ones because these are more commonly subject to inactivation by small RNAs. In this sense, the few elements in plants for which new insertion events were shown are three Copia-like elements, Tnt1, Tto1 and Tos17, present in a relatively low copy number (<1,000) per haploid genome (see Yamazaki et al. 2001) and a low redundant Gypsy element of sunflower (Vukich et al. 2009b). Interestingly, in the BAC clones sequenced here, Illumina analysis shows two cases in which the inserted elements are much less redundant than the interrupted ones. However, in other cases, especially when Copia REs are interrupted by Gypsy ones, these are more redundant, suggesting that the negative correlation between RE transposition and redundancy is not a general rule.
Beside recent retrotransposon activity, occurrence of past activity is indicated by the hybridization of genomic DNA of annual and perennial species of Helianthus to clones of the sunflower small-insert library described by Cavallini et al. (2010). Clones homologous to sequences of the REs identified in the BACs show hybridization signals in both Helianthus sections, indicating that such retroelements were already present in the Helianthus ancestor before splitting between annuals and perennials. Then, variations (either increases or decreases) had occurred in the extant species. It is known that the rates of both genome expansion and genome contraction processes appear to vary between species (Bennetzen et al. 2005; Vitte and Bennetzen 2006), allowing some genomes to shrink, while others expand. Rearrangements, and illegitimate and unequal homologous recombination are the processes that drive DNA removal in plants by multiple mechanisms, including repair of double-strand breaks (non-homologous end-joining) and slipstrand mispairing (Ma and Bennetzen 2004). Therefore, as in other genera, retrotransposon activity seems to be a major force acting in the diversification of species (Ungerer et al. 2006, 2009).
Regarding the structure of sequenced loci, the LTP locus appears to be the most interesting, with five copies of the LTP gene within less than 100,000 bp, four of which are potentially functional, and LTP1 probably inactivated by a retroelement insertion. Sequence analysis of the proximal putative promoter sequence suggests the mode by which the plant uses gene redundancy: the promoter sequences of sunflower LTP genes are very different and should insure large differences in the regulation pattern of each copy. Such differences have been observed in other species such as grapevine (Falginella et al. 2010). On the other hand, only minor differences may be observed as to the proteins encoded by the four LTP putatively functional genes. It can be concluded that the major specificities of the five LTP genes (or at least of the four putatively functional ones) stand in their regulation pattern rather than in their biochemical function.
Finally, it can be observed that LTP1 inactivation by the Copia retroelement has occurred very recently (as indicated by complete similarity between sister LTRs), further suggesting that sunflower is still evolving at a high rate.
Actually, a relative incompleteness of species differentiation within Helianthus is indicated by cross-compatibility between H. annuus and annual Helianthus species and sometimes also between H. annuus and perennial species (Whelan 1978). On the whole, the results reported in this study confirm that the sunflower is an excellent system to study plant genome evolution.
References
Baucom RS, Estill JC, Leebens-Mack J, Bennetzen JL (2009) Natural selection on gene function drives the evolution of LTR retrotransposon families in the rice genome. Genome Res 19:243–254
Beguiristain T, Grandbastien MA, Puigdomenech P, Casacuberta JM (2001) Three Tnt1 subfamilies show different stress-associated patterns of expression in tobacco. Consequences for retrotransposon control and evolution in plants. Plant Physiol 127:212–221
Bennetzen JL (2007) Patterns in grass genome evolution. Curr Opin Plant Biol 10:176–181
Bennetzen JL, Ma J, Devos KM (2005) Mechanisms of recent genome size variation in flowering plants. Ann Bot 95:127–132
Brunner S, Fengler K, Morgante M, Tingey S, Rafalski A (2005) Evolution of DNA sequence nonhomologies among maize inbreds. Plant Cell 17:343–360
Buti M, Giordani T, Vukich M, Gentzbittel L, Pistelli L, Cattonaro F, Morgante M, Cavallini A, Natali L (2009) HACRE1, a recently inserted copia-like retrotransposon of sunflower (Helianthus annuus L.). Genome 11:904–911
Cavallini A, Natali L, Zuccolo A, Giordani T, Jurman I, Ferrillo V, Vitacolonna N, Sarri V, Cattonaro F, Ceccarelli M, Cionini PG, Morgante M (2010) Analysis of transposons and repeat composition of the sunflower (Helianthus annuus L.) genome. Theor Appl Genet 120:491–508
Charles M, Belcram H, Just J, Huneau C, Viollet A, Couloux A, Segurens B, Carter M, Huteau V, Coriton O, Appels R, Samain S, Chalhoub B (2008) Dynamics and differential proliferation of transposable elements during the evolution of the B and A genomes of wheat. Genetics 180:1071–1086
Du C, Caronna J, He L, Dooner HK (2008) Computational prediction and molecular confirmation of Helitron transposons in the maize genome. BMC Genomics 9:51
Falginella L, Castellarin SD, Testolin R, Gambetta GA, Morgante M, Di Gaspero G (2010) Expansion and subfunctionalisation of flavonoid 3′, 5′-hydroxylases in the grapevine lineage. BMC Genomics 11:562
Felsenstein J (1989) PHYLIP—Phylogeny Inference Package (Version 3.2). Cladistics 5:164–166
Gross BL, Turner KG, Rieseberg LH (2007) Selective sweeps in the homoploid hybrid species Helianthus deserticola: evolution in concert across populations and across origins. Mol Ecol 16:5246–5258
Grover C, Hawkins J, Wendel J (2008) Phylogenetic insights into the pace and pattern of plant genome size evolution. In: Volff J-N (ed) Plant Genomes. Genome Dynamics. Vol. 4. Karger, Basel (Switzerland), pp 57–68
Hawkins JS, Kim HR, Nason JD, Wing RA, Wendel JF (2006) Differential lineage-specific amplification of transposable elements is responsible for genome size variation in Gossypium. Genome Res 16:1252–1261
Higo K, Ugawa Y, Iwamoto M, Korenaga T (1999) Plant cis-acting regulatory DNA elements (PLACE) database. Nucl Acids Res 27:297–300
Holligan D, Zhang X, Jiang N, Pritham EJ, Wessler SR (2006) The transposable element landscape of the model legume Lotus japonicus. Genetics 174:2215–2228
Jurka J (2000) Repbase update: a database and an electronic journal of repetitive elements. Trends Genet 16:418–420
Kalendar R, Vicient CM, Peleg O, Anamthawat-Jonsson K, Bolshoy A, Schulman AH (2004) Large retrotransposon derivatives: abundant, conserved but nonautonomous retroelements of barley and related genomes. Genetics 166:1437–1450
Kumar A, Bennetzen JB (1999) Plant retrotransposons. Ann Rev Genet 33:479–532
Ma J, Bennetzen JL (2004) Rapid recent growth and divergence of rice nuclear genomes. Proc Natl Acad Sci USA 101:12404–12410
Ma J, Devos KM, Bennetzen JL (2004) Analyses of LTR-retrotransposon structures reveal recent and rapid genomic DNA loss in rice. Genome Res 14:860–869
Meyers BC, Tingey SV, Morgante M (2001) Abundance, distribution, and transcriptional activity of repetitive elements in the maize genome. Genome Res 11:1660–1676
Ming R, Hou S, Feng Y, Yu Q, Dionne-Laporte A, Albert H, Suzuki JY, Tripathi S, Moore PH, Gonsalves D (2008) The draft genome of the transgenic tropical fruit tree papaya (Carica papaya Linnaeus). Nature 452:991–997
Moisy C, Garrison KE, Meredith CP, Pelsy F (2008) Characterization of ten novel Ty1/Copia-like retrotransposon families of the grapevine genome. BMC Genomics 9:469
Natali L, Santini S, Giordani T, Minelli S, Maestrini P, Cionini PG, Cavallini A (2006) Distribution of Ty3-Gypsy- and Ty1-Copia-like DNA sequences in the genus Helianthus and other Asteraceae. Genome 49:64–72
Neumann P, Koblizkova A, Navratilova A, Macas J (2006) Significant expansion of Vicia pannonica genome size mediated by amplification of a single type of giant retroelement. Genetics 173:1047–1056
Paterson AH, Bowers JE, Bruggmann R, Dubchak I, Grimwood J, Gundlach H, Haberer G, Hellsten U, Mitros T, Poliakov A, Schmutz J, Spannagl M, Tang H, Wang X, Wicker T, Bharti AK, Chapman J, Feltus FA, Gowik U, Grigoriev I, Lyons E, Maher CA, Martis M, Narechania A, Otillar RP, Penning BW, Salamov AA, Wang Y, Zhang L, Carpita NC, Freeling M, Gingle AR, Hash CT, Keller B, Klein P, Kresovich S, McCann MC, Ming R, Peterson DG, Rahman M, Ware D, Westhoff P, Mayer KFX, Messing M, Rokhsar DS (2009) The Sorghum bicolor genome and the diversification of grasses. Nature 457:551–556
Paux E, Roger D, Badaeva E, Gay G, Bernard M, Sourdille P, Feuillet C (2006) Characterizing the composition and evolution of homoeologous genomes in hexaploid wheat through BAC-end sequencing on chromosome 3B. Plant J 48:463–474
Piegu B, Guyot R, Picault N, Roulin A, Saniyal A, Kim H, Collura K, Brar DS, Jackson S, Wing RA, Panaud O (2006) Doubling genome size without polyploidization: dynamics of retrotransposition-driven genomic expansions in Oryza australiensis, a wild relative of rice. Genome Res 16:1262–1269
Rieseberg LH, Raymond O, Rosenthal DM, Lai Z, Livingstone K, Nakazato T, Durphy JL, Schwarzbach AE, Donovan LA, Lexer C (2003) Major ecological transitions in wild sunflowers facilitated by hybridization. Science 301:1211–1216
Rohlf FJ (2008) NTSYSpc: Numerical Taxonomy System, ver. 2.00. Exeter Publishing Ltd, Setauket
Rozas J, Rozas R (1999) DnaSP version 3: an integrated program for molecular population genetics and molecular evolution analysis. Bioinformatics 15:174–175
Rozen S, Skaletsky H (2000) Primer3 on the WWW for general users and for biologist programmers. In: Krawetz S, Misener S (eds) Bioinformatics methods and protocols: methods in molecular biology. Humana Press, Totowa, pp 365–386
SanMiguel P, Bennetzen JL (1998) Evidence that a recent increase in maize genome size was caused by the massive amplification of intergene retrotransposons. Ann Bot 82:37–44
SanMiguel P, Tikhonov A, Jin YK, Motchoulskaia N, Zakharov D, Melake-Berhan A, Springer PS, Edwards KJ, Lee M, Avramova Z (1996) Nested retrotransposons in the intergenic regions of the maize genome. Science 274:765–768
Santini S, Cavallini A, Natali L, Minelli S, Maggini F, Cionini PG (2002) Ty1/Copia- and Ty3/Gypsy-like DNA sequences in Helianthus species. Chromosoma 111:192–200
Scherrer B, Isidore E, Klein P, Kim JS, Bellec A, Chalhoub B, Keller B, Feuillet C (2005) Large intraspecific haplotype variability at the Rph7 locus results from rapid and recent divergence in the barley genome. Plant Cell 17:361–374
Schilling EE, Linder CR, Noyes RD, Rieseberg LH (1998) Phylogenetic relationships in Helianthus (Asteraceae) based on nuclear ribosomal DNA internal transcribed spacer region sequence data. Syst Bot 23:177–187
Schnable PS, Ware D, Fulton RS, Stein JC, Wei F, Pasternak S, Liang C, Zhang J, Fulton L, Graves TA, Minx P, Reily AD, Courtney L, Kruchowski SS, Tomlinson C, Strong C, Delehaunty K, Fronick C, Courtney B, Rock SM, Belter E, Du F, Kim K, Abbott RM, Cotton M, Levy A, Marchetto P, Ochoa K, Jackson SM, Gillam B, Chen W, Yan L, Higginbotham J, Cardenas M, Waligorski J, Applebaum E, Phelps L, Falcone J, Kanchi K, Thane T, Scimone A, Thane N, Henke J, Wang T, Ruppert J, Shah N, Rotter K, Hodges J, Ingenthron E, Cordes M et al (2009) The B73 maize genome: complexity, diversity, and dynamics. Science 326:1112–1115
Soltis DE, Soltis PS (1999) Polyploidy: recurrent formation and genome evolution. Trends Ecol Evol 9:348–352
Sonnhammer EL, Durbin R (1995) A dot-matrix program with dynamic threshold control suited for genomic DNA and protein sequence analysis. Gene 167:GC1–GC10
Swaminathan K, Varala K, Hudson ME (2007) Global repeat discovery and estimation of genomic copy number in a large, complex genome using a high-throughput 454 sequence survey. BMC Genomics 8:132
Tarchini R, Biddle P, Wineland R, Tingey S, Rafalski A (2000) The complete sequence of 340 kb of DNA around the rice Adh1-adh2 region reveals interrupted colinearity with maize chromosome 4. Plant Cell 12:381–391
The French-Italian Public Consortium for Grape Genome Characterization (2007) The grapevine genome sequence suggests ancestral hexaploidization in major angiosperm phyla. Nature 449:463–467
The International Rice Genome Sequencing Project (2005) The map-based sequence of the rice genome. Nature 436:793–800
Thompson JD, Desmond G, Gibson H, Gibson TJ (1994) CLUSTALW: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucl Acids Res 22:4673–4680
Tuskan GA, DiFazio S, Jansson S, Bohlmann J, Grigoriev I, Hellsten U, Putnam N, Ralph S, Rombauts S, Salamov A, Schein J, Sterck L, Aerts A, Bhalerao RR, Bhalerao RP, Blaudez D, Boerjan W, Brun A, Brunner A, Busov V, Campbell M, Carlson J, Chalot M, Chapman J, Chen GL, Cooper D, Coutinho PM, Couturier J, Covert S, Cronk Q, Cunningham R, Davis J, Degroeve S, Déjardin A, de Pamphilis C, Detter J, Dirks B, Dubchak I, Duplessis S, Ehlting J, Ellis B, Gendler K, Goodstein D, Gribskov M, Grimwood J, Groover A, Gunter L, Hamberger B, Heinze B, Helariutta Y et al (2006) The genome of black cottonwood, Populus trichocarpa (Torr. & Gray). Science 313:1596–1604
Ungerer MC, Strakosh SC, Zhen Y (2006) Genome expansion in three hybrid sunflower species is associated with retrotransposon proliferation. Curr Biol 16:R872–R873
Ungerer MC, Strakosh SC, Stimpson KM (2009) Proliferation of Ty3/gypsy-like retrotransposons in hybrid sunflower taxa inferred from phylogenetic data. BMC Biol 7:40
Vicient CM, Kalendar R, Schulman AH (2005) Variability, recombination, and mosaic evolution of the barley BARE-1 retrotransposon. J Mol Evol 61:275–291
Vitte C, Bennetzen JL (2006) Analysis of retrotransposon structural diversity uncovers properties and propensities in angiosperm genome evolution. Proc Natl Acad Sci USA 103:17638–17643
Vukich M, Schulman AH, Giordani T, Natali L, Kalendar R, Cavallini A (2009a) Genetic variability in sunflower (Helianthus annuus L.) and in the Helianthus genus as assessed by retrotransposon-based molecular markers. Theor Appl Genet 119:1027–1038
Vukich M, Schulman AH, Giordani T, Natali L, Kalendar R, Cavallini A (2009b) Copia and Gypsy retrotransposons activity in sunflower (Helianthus annuus L.). BMC Plant Biol 9:150
Wang Q, Dooner HK (2006) Remarkable variation in maize genome structure inferred from haplotype diversity at the bz locus. Proc Natl Acad Sci USA 103:17644–17649
Whelan EDP (1978) Cytology and interspecific hybridization. In: Carter JF (ed), Sunflower Science and Technology. Am Soc Agronomy, pp 339–370
Wicker T, Sabot F, Hua-Van A, Bennetzen JL, Capy P, Chalhoub B, Flavell A, Leroy P, Morgante M, Panaud O, Paux E, SanMiguel P, Schulman AH (2007) A unified classification system for eukaryotic transposable elements. Nature Rev Genet 8:973–982
Wilson RK, Mardis ER (1997) Shotgun sequencing. In: Birren B, Green ED, Klapholtz S, Myers RM, Roskams J (eds) Genome analysis: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor
Xu Z, Wang H (2007) LTR_FINDER: an efficient tool for the prediction of full-length LTR retrotransposons. Nucl Acids Res 35:W265–W268
Yamazaki M, Tsugawa H, Miyao A, Yano M, Wu J, Yamamoto S, Matsumoto T, Sasaki T, Hirochika H (2001) The rice retrotransposon Tos17 prefers low-copy-number sequences as integration targets. Mol Genet Genom 265:336–344
Acknowledgments
The research work was supported by PRIN-MIUR, Italy, Project “Variabilità di sequenza ed eterosi in piante coltivate”.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by A. Bervillé.
Electronic supplementary material
Below is the link to the electronic supplementary material.
122_2011_1626_MOESM1_ESM.tif
Number of putative cis-regulatory elements in 2,000-bp upstream of each copy of the LTP gene resulting from sequence analysis at the PLACE Web site. Regulatory element codes as in Higo et al. (1999). (TIFF 119 kb)
Rights and permissions
About this article
Cite this article
Buti, M., Giordani, T., Cattonaro, F. et al. Temporal dynamics in the evolution of the sunflower genome as revealed by sequencing and annotation of three large genomic regions. Theor Appl Genet 123, 779–791 (2011). https://doi.org/10.1007/s00122-011-1626-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00122-011-1626-4