Abstract
A relationship between pepper trichome and pepper mottle virus (PepMoV) resistance was examined. In an intraspecific F2 mapping population from the cross between Capsicum annuum CM334 (trichome-bearing and PepMoV resistant) and Chilsungcho (glabrous and PepMoV susceptible), major QTLs for both traits were identified by composite interval mapping in linkage group (LG) 24 corresponding a telomere region on pepper chromosome 10. Ptel1 of putative trichome enhancing locus was a common major QTL for trichome density on the main stem and calyx. Ptel1 apart from HpmsE031 at a 1.03 cM interval was specifically associated to the trichome density on the main stem, whereas Ptel2 near m104 marker on LG2 was specific for the calyx trichome. Epistatic analysis indicated that Ptel1 engaged in controlling the trichome density by mutual interactions with the organ-specific QTLs. For PepMoV resistance, two QTLs (Pep1 and Pep2) were identified on the LG 24. Pep1 was located with Ptel1 in the R-gene cluster (RGC) for potyvirus resistance including Pvr4 with broad spectrum resistance to potyviruses. Pep1 flanking TG420 marker seemed to be the major factors determining correlation with PepMoV resistance. These results indicate that the level of trichome density on pepper main stem can be used as a morphological marker for Pvr4 in pepper breeding.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Trichomes are hair-like structure on epidermal cells of many plant species, which can largely be classified into glandular or non-glandular forms. This organ has generally been regarded as a trait of little worth in agriculture due to its negative aspects such as a causing agent for allergy and the lower preference of farmers. However, the trichomes have been demonstrated to be directly or indirectly engaged in the plant protection from ultraviolet radiation, drought, high salinity (Espigares and Peco 1995; Skaltsa et al. 1994), heavy metals (Ager et al. 2003), herbivores, and pathogen attack (Elle et al. 1999; Johnson 1975; Levin 1973). Glandular trichomes in Solanaceae species give the high level of resistance against a number of phytophagous arthropods by producing exudate containing toxic acylsugars such as at the tips of tomato type VI trichomes (Kennedy 2003).
The most well-known non-glandular trichome is cotton fiber that is the complex of extremely elongated single cells (Wilkins et al. 2000). This type of trichome in willow inhibits grazing of adult leaf beetle (Phratora vulgatissima) by increasing trichome density on the new developing leaves (Dalin and Bjorkman 2003).
The high-dense trichomes can be frequently observed in a number of tomato, eggplant, and potato cultivars geared worldwide, while a few or no trichomes on the main stem of all pepper cultivars. Comparative study in the previous literatures showed that Ptl1 (Kim et al. 2010) may be located in the same region as the H (hair absence) locus in tomato and ovh 10.1 (ovary hair) in eggplant (Tanksley et al. 1992). Those literatures may indicated that the genetic evidence on the conservation of trichome loci in Solanaceae family. The potyvirus resistance loci, has also been reported in Capsicum annuum Criollo de Morelos 334 (CM334) (Dogimont et al. 1996) and some of C. chinense accessions (Grube et al. 2000; Murphy et al. 1998). Pvr4 from CM334 has broad spectrum resistance to Potato virus Y pathotypes including Pepper mottle virus (PepMoV), which has been known to closely linked to Pvr7 on pepper chromosome 10 (Caranta et al. 1999; Grube et al. 2000). A recent study in our laboratory suggested that the resistance of PepMoV would be an obvious continuity with the trichome presence on the main stem of F2 individuals from the cross of CM334 (high-dense trichomes) and Chilsungcho (glabrous) peppers (Kim et al. 2010). Since, many loci in this genomic region have been reported to be associated with resistance to various plant pathogens; it would be useful if we figure out the association of trichome-bearing with disease resistance traits for phenotypic selection of Potyvirus resistance in breeding program.
In this study, linkage relationships between trichomes on the main stem and PepMoV resistance were analyzed using composite interval mapping. Here, we report the close association of QTL for PepMoV resistance and trichome density in pepper clustering at a narrow region near TG420 marker on chromosome 10.
Materials and methods
Plant materials
Capsicum annuum CM334 is bearing trichomes and resistant to PepMoV. C. annuum Chilsungcho is a domestic, glabrous in the aerial parts, and susceptible to the virus. Both parents, their self-pollinated progenies, F1, BC1F1, and F2 were used for genetic analysis of trichome inheritance and PepMoV resistance. In addition, C. annuum ECW123R was used as a susceptible parent for further analysis of PepMoV resistance. An intraspecific mapping population consists of 100 F2 individuals from the CM334 and Chilsungcho cross (Min et al. 2008), which used for QTL mapping of trichome and PepMoV resistance. Phenotype data were obtained from each F2 (trichome) of the mapping population and their F3 families (trichome and PepMoV resistance). All plants were grown in a greenhouse at Seoul National University, Korea.
Morphology observation and scoring of trichome density
Morphological characters of trichomes on the main stem were scanned with an electron microscope (JSM 5410LV, Jeol Ltd., Tokyo, Japan) at The National Instrumentation Center for Environmental Management at Seoul National University.
On the main stem, the presence or absence of trichomes was visually evaluated and the trichome density was indexed by a scale ranging from 1 (glabrous) to 5 (very high, the whole surface of the stem covered with trichomes) (Doganlar et al. 2002) at the stage having 10–11 fully expanded true leaves. The trichomes on the sepal located between the second and third calyx on the left side of flowers were directly counted under a dissecting microscope (S-700, Carl Zeiss Inc., Göttingen, Germany). For phenotyping, the trichome densities on the main stem and calyx, total 100 F2 individuals in the mapping population, 20 flowers per each F2 plant were randomly picked at full-blooming stage. Twenty flowers of each F3 families in the mapping population were used for counting trichome number on the calyx. Forty progenies of each F3 families were used for screening trichome density on the main stem. These experiments were triplicated and the mean values were taken for analysis.
Virus inoculation
PepMoV-SNU1 (Han et al. 2006) was propagated in Nicotiana tabaccum Xanthi-nc. Infected leaves were harvested from ten individuals showing severe mosaic symptoms with stem necrosis at 14 days after inoculation and stored at −80°C prior to use. The virus infected leaves were macerated with a mortar and pestle in five times volume of 0.1 M phosphate buffer (pH 7.4) with 1% Na2SO3. The homogenate was centrifuged at 8,000 rpm for 10 min. The resulting supernatant was used as an inoculum. Inoculation was performed at the fourth true leaf stage and again at 14 days after the first inoculation. Two leaves per each plant were dusted thoroughly with Caborundum (600 mesh), gently rubbed with the cotton wool wetted with the inoculum, and washed immediately with tap water. The inoculated plants were grown in a greenhouse at Seoul National University, Korea.
Resistance evaluation
Virus infection was checked by Double Antibody Sandwich (DAS)-ELISA for the genetic analysis of PepMoV resistance. DAS-ELISA was performed according to the manufacturer’s protocol (Agdia Inc., Elkhart, IN, USA). For QTL mapping of PepMoV resistance, virus infection was examined 30 days after the inoculation by ring-blot immunobinding assay (Han et al. 2007). The evaluations were triplicated on 40 F3 progenies per each F2 individual from the same mapping population for trichome density. Viral resistance was accessed by scoring percent infection. Each test was analyzed for Pearson correlation coefficient with the statistical analysis system (SAS) (Hanrahan et al. 1990). The mean value of the first and third tests that showing high correlation coefficient (r = 0.93) at P < 0.05 was used in QTL mapping.
QTL analysis and epistatic interactions
With a total of 389 markers including eight RAMP (randomly amplified microsatellite polymorphism) markers (Min et al. 2008) that developed from the candidate genes of Arabidopsis trichome initiation and those of the SNU5 map (Lee et al. 2009), linkage analysis was performed using the Kosambi function of the Carthagene program (De Givry et al. 2005) at the threshold value of logarithm of odds (LOD) 4.0 and maximum distance of 30 cM. QTL were analyzed using a composite interval mapping (CIM) function of QTL Cartographer V2.0 (Zeng 1994). At P < 0.05, the critical values for significance thresholds were determined by 1,000 permutation tests and the 951st value was taken for QTL analysis (Zeng 1994). Epistatic interactions between pairs of markers on the linkage map were detected using a fixed interaction model of two-factor analysis of variance (ANOVA) at a significance of P < 5 × 10−3 (Caranta et al. 2002). Chromosomes carrying QTL were visualized using the MapChart software (Voorrips 2002).
Results
Trichome morphology, distribution, and PepMoV resistance
Based on the observations using a dissecting microscope and a scanning electron microscope, CM334 plants had a number of non-glandular unbranched trichomes, while no trichomes (glabrous) were observed in the aerial parts of Chilsungcho plants (Fig. 1). For PepMoV-SNU1 resistance, CM334 was immune, showing in no symptom, whereas Chilsungcho was systemically infected and presented typical mottle symptom (Fig. 1e, j). The resistance in CM334 was dominantly inherited. However, the ratio of resistant to susceptible was 59:7 for F2 and 21:9 for BC 1F1 (Table 1). This result was not accordance with the previous reports describing one dominant gene for PepMoV resistance in an F2 segregating population from Yolo Wonder and CM334 cross. To further confirm the mode of resistance inheritance in CM334, a segregating population was newly generated by changing Chilsungcho to C. annuum ECW123R highly susceptible to PepMoV-SNU1. The resistance in the new population was also quantitatively inherited (Table 1). Furthermore, the ratio of resistant to susceptible in F3 families from the first mapping population could not be classified as a single dominant gene model (Fig. 2e).
Phenotype of trichome and pepper mottle virus (PepMoV) resistance in C. annuum CM334 and Chilsungcho. Photographs were taken using a digital camera (a, b, c, d, f, g, h) and a scanning electron microscope (d, i) at ×50. The red ovals (c, h) on the calyx are the areas for counting trichomes. At 14 days after inoculation with PepMoV-SNU1, CM334 was not infected (e) whereas Chilsungcho showed severe mottle symptom on upper leaves (j)
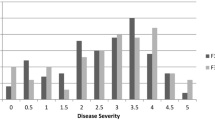
Frequency distribution of trichome density on the main stem (a, b) and calyx (c, d) in F2 and F3 populations from the cross between CM334 and Chilsungcho. Arrows indicated trichome density of CM334, Chilsungcho, and F1 population. Frequency distribution of arcsine value for the resistance to pepper mottle virus-SNU1 (PepMoV-SNU1) in 100 F3 families (e). F3 families were from a cross between Capsicum annuum CM334 and C. annuum Chilsungcho, which were used a mapping population in this study. The resistance level in F2 was deduced by percent infection of 40 F3 families per each F2 individuals in which the value was transformed into arcsine value, resulting in 5 classes. Class is distinguished by resistance rate (Class 1 = 81–100% resistance, Class 2 = 61–80% resistance, Class 3 = 41–60% resistance, Class 4 = 21–40% resistance, Class 5 = 1–20% resistance)
Trichome density on the main stem and calyx
Trichome density in the F1 population was decreased by roughly half as compared with CM334. Frequency distribution of the trichome density on the main stem was considerably skewed to glabrous in 100 F2 individuals of a mapping population. However, the frequency was normally distributed in 81 F2 individuals when glabrous ones were excluded. To more precisely evaluate trichome density, stem, leaf, and calyx of segregating F2 individuals were carefully observed. The calyx was selected for clearly differentiating of trichome density in different plants by direct counting. Since the whole region of the calyx had quite a number of trichomes in many F2 individuals, one typical sepal from a calyx that had little variation in area among F2 individuals was finally chosen as a representative organ for trichome density in the aerial parts of the whole plant. The average number of trichomes on the calyx of CM334 was 28 when 100 flowers were examined, while no trichomes were observed in the number of calyces from Chilsungcho. The F1 hybrids had average 15 trichomes on the calyx (data not shown). The frequency distribution of trichome density on the calyx was higher than that of trichome-bearing (T) to glabrous (G) on the main stem in segregating crosses of CM334, Chilsungcho, and their progenies skewed toward glabrous in both F2 and F3 families. However, the distribution was normal when glabrous plants were excluded from F2 and F3 population tested (Fig. 2a–d). In addition, the absence of trichomes on a part of the calyx was occasionally observed at very low frequency in F2 and F3 population bearing trichomes on the main stem (data not shown).
QTL mapping of trichome density
Compared to IM, CIM is known to be more powerful due to a reduction in the bias of a QTL around markers tested and no treatment of the effects of other QTL as residual variance (Mei et al. 2004). Accordingly, CIM was employed to estimate and interpret QTL for trichome density on the main stem and calyx. For trichomes on the main stem, the empirical threshold values for F2 and F3 phenotype data were 14.62 and 2.51, respectively. A major QTL (Ptel1) was detected on stem trichome density in the F2 and F3 populations. The QTL was on LGs corresponding to pepper chromosome 10 and accounted for 81% of the phenotypic variation for the trait. The mode of gene action of Ptel1 was expected to be partially dominant. For trichomes in the calyx, the threshold values were 2.41 and 2.43 for the F2 and F3 populations, respectively. A total of 11 QTL were found on ten different LGs, among which LG15 included the two QTL each with phenotypic variations ranging from 5 to 81%. Ptel1 was implicated in trichome density on both the main stem and calyx of the F2 and F3 populations. Ptel2 on LG2 was specific to the trichomes on the calyx. The other QTL were specific to each experimental generation. Markers having the highest LOD score were located in LG24. m96 and EAMC192 markers located near Ptel4 and Ptel8 were originated from Chilsungcho. All the other QTL related with trichome, exclusive Ptel4 and Ptel8 were from CM334 (Table 2).
Identification of QTLs in PepMoV resistance
QTL for PepMoV-SNU1 resistance were preliminarily analyzed by one-way ANOVA. Two markers (TG420 and EAMC91) were significantly associated with the resistance. To verify the accurate number and position of resistance QTL in the pepper map, further analysis with CIM function was carried out in Windows QTL Cartographer 2.0 (Wang et al. 2004), resulting in two QTL (Pep1 and Pep2) with 34 and 57% of total phenotypic variance for the resistance, respectively. These QTLs were located on linkage group (LG) 24 (Table 2).
Epistatic analysis for trichome and PepMoV resistance
Using SAS Proc GLM (Hanrahan et al. 1990) and type-III sums of squares, a probability of P < 0.0001 was used as a threshold for claiming the presence of a putative QTL. Digenic interactions were analyzed using the two-way ANOVA in which the closest markers for QTL and remnant markers for the linkage map were used on factors 1 and 2, respectively. For trichomes on the main stem, HpmsE031 interacted with HpmsE123, EAMC138, and EAMG15. For trichomes on the calyx in F2 population, there were two interactions. The m104 and HpmsE031 markers interacted with m96 and HpmsE123 markers, respectively.
For PepMoV resistance significant interactions were between TG420 and two background markers, HpmsE128 and EAMG80 (Table 3).
Relationship between PepMoV resistance and stem trichome formation
LG 24 corresponding to pepper chromosome 10 was newly constructed with 17 markers which included Ptl1, Tsca, and Tco (Kim et al. 2010). The LG24 covered 125.3 cM, on which TG420B2 was co-localized with Pst112 restriction fragment length polymorphism marker that was significant for PepMoV resistance in one-way ANOVA (data not shown). Using the phenotype data of the average percentage of PepMoV infection and trichome density in the main stem, CIM revealed a geometrical correlation among major QTL for both traits. All QTL originated from CM334 were located in 21 cM interval, in which Ptel1 and Pep1 were most closely linked (Fig. 3).
Map location and LOD profiles of the QTLs for trichome density and PepMoV resistance on linkage group 24 using a recombinant population from the cross between CM334 and Chilsungcho. Genetic distance was calculated using the Kosambi function (Kosambi 1944). a Genetic map of pepper chromosome 10 constructed using 100 individuals of an F2 population from CM334 × Chilsungcho. b CIM results from the integrated map. QTL intervals of trichome density and PepMoV resistance are presented as solids bars
Discussion
Trichome density on the leaf and stem of Arabidopsis thaliana has been known to be modulated by combinations between major genes common in diverse species and many minor genes specific to accessions of Arabidopsis plants. Especially, the interaction between GL1 and TTG1 influences glabrous phenotype (Marks 1997). In pepper, the presence or absence of trichome on main stem to be apparently controlled by single gene. However, protrusions of immature trichome were occasionally found in the glabrous category when more precise scale was adapted to distinguish glabrous one in F2 individuals from CM334 and Chilsungcho cross. This means the glabrous without any protrusions may be controlled by at least two genes. In this study, there was just one QTL of Ptel1 for the trichome density on pepper main stem. The cause may come from small size mapping population limiting the potential for QTL detection. CIM analysis deduced two peaks with the distance of 18.3 cM on LG24, which may be independent QTLs for trichome density on main stem although the population size is too small to convince. Therefore, a fine mapping needs for dissecting the doubtful region in the near future. The trichome density on the calyx is more complex compared to that of stem trichome. The QTLs in F2 and F3 mapping populations were quite difference in their map location, suggesting minor QTLs except for Ptel1 and Ptel2 may be false positive QTLs or undefined factors although these QTLs have significant value for CIM analysis. Indeed, the presence or absence of trichome on calyx in F2 mapping individuals in this study occasionally did not coincide with those of trichome on the main stem. Thus, trichome density on the pepper main stem and calyx might be controlled by a QTL specifically to each organ in the presence of Ptel1.
PepMoV resistance has been known to be qualitatively controlled by Pvr4 in CM334 (Caranta et al. 1999; Grube et al. 2000). However, in this study, the PepMoV resistance was quantitatively inherited in the cross combinations between CM334 and its paternal parents (Table 1). The segregating ratio of PepMoV resistance in F2 families in Table 1 was more accordance with 13:3 than 3:1 under two genes model being one dominant and one recessive. Furthermore, the PepMoV resistance in the F3 families from F2 mapping population could not be clearly dissected to two types of 75% resistance and 25% susceptible. The infection frequency was normally distributed with a little skewness toward resistance in CM334 and Chilsungcho F3 population (Fig. 2e). CIM analysis for PepMoV resistance showed that two QTLs were located on chromosome 10, among which one QTL, Pep1 may correspond to Pvr4 which is reported early in CM334. This result indicated that PepMoV resistance was inherited with at least two genes including Pvr4 in the CM334 and Chilsungcho mapping population.
Analysis for Pearson correlation with the phenotype data for PepMoV resistance displayed close relation with trichome bearing on the main stem (r = 0.83 in F2 and r = 0.68 in F3) (data not shown). These results indicate that trichome-bearing pepper plants have a high rate of PepMoV resistance. Some examples showing correlation between trichome and disease resistance can be found in pepper and rice. The presence of trichomes on stems and leaves related with resistance to P. capsici in pepper (Egea-Gilabert et al. 2008) and significant correlations were reported between tillering ability and rice yellow mottle virus resistance on chromosome 12 in rice (Albar et al. 1998). Consequently, this study suggests that trichomes could be used for enhanced selection marker for PepMoV resistance in pepper breeding program. Upcoming positional cloning and complementation of Ptel1 and Pvr4 using genome sequence of this region will provide an insight into the molecular association of trichome-bearing and Potyvirus resistance in pepper plants.
References
Ager FJ, Ynsa MD, Domnguez-Sol JR, Lopez-Mart MC, Gotor C, Romero LC (2003) Nuclear micro-probe analysis of Arabidopsis thaliana leaves. Nucl Instrum Methods Phys Res 210:401–406
Albar L, Lorieux M, Ahmadi N, Rimbault I, Pinel A, Sy AA, Fargette D, Ghesquiere A (1998) Genetic basis and mapping of the resistance to rice yellow mottle virus. I. QTLs identification and relationship between resistance and plant morphology. Theor Appl Genet 97:1145–1154
Caranta C, Thabuis A, Palloix A (1999) Development of a CAPS marker for the Pvr4 locus: a tool for pyramiding potyvirus resistance genes in pepper. Genome 42:1111–1116
Caranta C, Pflieger S, Lefebvre V, Daubeze AM, Thabuis A, Palloix A (2002) QTLs involved in the restriction of cucumber mosaic virus (CMV) long-distance movement in pepper. Theor Appl Genet 104:586–591
Dalin P, Bjorkman C (2003) Adult beetle grazing induces willow trichome defence against subsequent larval feeding. Oecologia 134:112–118
De Givry S, Bouchez M, Chabrier P, Milan D, Schiex T (2005) CARHTA GENE: multipopulation integrated genetic and radiation hybrid mapping. Bioinformatics 21:1703–1704
Doganlar S, Frary A, Daunay MC, Lester RN, Tanksley SD (2002) Conservation of gene function in the Solanaceae as revealed by comparative mapping of domestication traits in eggplant. Genetics 161:1713–1726
Dogimont C, Palloix A, Daubze AM, Marchoux G, Selassie KG, Pochard E (1996) Genetic analysis of broad spectrum resistance to potyviruses using doubled haploid lines of pepper (Capsicum annuum L.). Euphytica 88:231–239
Egea-Gilabert C, Bilotti G, Requena ME, Ezziyyani M, Vivo-Molina JM, Candela ME (2008) Pepper morphological traits related with resistance to Phytophthora capsici. Biol Plant 52:105–109
Elle E, van Dam NM, Hare JD (1999) Cost of glandular trichomes, a “resistance” character in Datura wrightii Regel (Solanaceae). Evolution 53:22–35
Espigares T, Peco B (1995) Mediterranean annual pasture dynamics: impact of autumn drought. J Ecol 83:135–142
Grube RC, Blauth JR, Arnedo Andres MS, Caranta C, Jahn MK (2000) Identification and comparative mapping of a dominant potyvirus resistance gene cluster in Capsicum. Theor Appl Genet 101:852–859
Han JH, Choi HS, Kim DH, Lee HR, Kim BD (2006) Biological, physical and cytological properties of pepper mottle virus-SNU1 and its RT-PCR detection. Plant Pathol J 22:155–160
Han JH, Shin JS, Kim YH, Kim BD (2007) Improvement of antigen blotting in a tissue blot immunobinding assay for the detection of two chili pepper viruses. J Microbe Biotechnol 17:1885–1889
Hanrahan LP, Mirkin I, Olson J, Anderson HA, Fiore BJ (1990) SMRFIT: a statistical analysis system (SAS) program for standardized mortality ratio analyses and Poisson regression model fits in community disease cluster investigations. Am J Epidemiol 132:116–122
Johnson HB (1975) Plant pubescence: an ecological perspective. Bot Rev 41:233–258
Kennedy GG (2003) Tomato, Pests, Parasitoids, and Predators: tritrophic interactions involving the genus Lycopersicon. Annu Rev Entomol 48:51–72
Kim HJ, Han JH, Kwon JK, Park M, Kim BD, Choi D (2010) Fine mapping of pepper trichome locus 1 controlling trichome formation in Capsicum annuum L. CM334. Theor Appl Genet 120:1099–1106
Kosambi DD (1944) The estimation of map distance from recombination values. Ann Eugen 12:172–175
Lee HR, Bae IH, Park SW, Kim HJ, Min WK, Han JH, Kim KT, Kim BD (2009) Construction of an integrated pepper map using RFLP, SSR, CAPS, AFLP, WRKY, rRAMP, and BAC end sequences. Mol Cells 27:21–37
Levin DA (1973) The role of trichomes in plant defense. Quart Rev Biol 48:3–15
Marks MD (1997) Molecular genetic analysis of trichome development in Arabidopsis. Annu Rev Plant Biol 48:137–163
Mei M, Syed NH, Gao W, Thaxton PM, Smith CW, Stelly DM, Chen ZJ (2004) Genetic mapping and QTL analysis of fiber-related traits in cotton (Gossypium). Theor Appl Genet 108:280–291
Min WK, Han JH, Kang WH, Lee HR, Kim BD (2008) Reverse random amplified microsatellite polymorphism reveals enhanced polymorphisms in the 3 end of simple sequence repeats in the pepper genome. Mol Cells 26:250–257
Murphy JF, Blauth JR, Livingstone KD, Lackney VK, Jahn MK (1998) Genetic mapping of the pvr1 locus in Capsicum spp. and evidence that distinct potyvirus resistance loci control responses that differ at the whole plant and cellular levels. Mol Plant Microbe Interact 11:943–951
Skaltsa H, Verykokidou E, Harvala C, Karabourniotis G, Manetasi Y (1994) UV-B protective potential and flavonoid content of leaf hairs of Quercus ilex. Phytochemistry 37:987–990
Tanksley SD, Ganal MW, Prince JP, De-Vicente MC, Bonierbale MW, Broun P, Fulton TM, Giovannoni JJ, Grandillo S, Martin GB (1992) High density molecular linkage maps of the tomato and potato genomes. Genetics 132:1141–1160
Voorrips RE (2002) MapChart: software for the graphical presentation of linkage maps and QTLs. J Hered 93:77–78
Wang S, Basten CJ, Zeng Z (2004) Window QTL Cartographer. V. 2.0 Program in Statistical Genetics. North Carolina State University, North Carolina
Wilkins TA, Rajasekaran K, Anderson DM (2000) Cotton biotechnology. Crit Rev Plant Sci 19:511–550
Zeng ZB (1994) Precision mapping of quantitative trait loci. Genetics 136:1457–1468
Acknowledgments
This work was supported by a grant (project No. 609002-5) from the Screening Center for Disease Resistant Vegetable Crops of Technology Development Program for Agriculture and Forestry, Ministry for Food, Agriculture, Forestry and Fisheries and the Crop Functional Genomics Center, 21st Frontier Research Program of MEST of the Korean government.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by I. Paran.
Rights and permissions
About this article
Cite this article
Kim, H.J., Han, JH., Kim, S. et al. Trichome density of main stem is tightly linked to PepMoV resistance in chili pepper (Capsicum annuum L.). Theor Appl Genet 122, 1051–1058 (2011). https://doi.org/10.1007/s00122-010-1510-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00122-010-1510-7