Abstract
SHZ-2 is an indica rice cultivar that exhibits broad-spectrum resistance to rice blast; it is widely used as a resistance donor in breeding programs. To dissect the QTL responsible for broad-spectrum blast resistance, we crossed SHZ-2 to TXZ-13, a blast susceptible indica variety, to produce 244 BC4F3 lines. These lines were evaluated for blast resistance in greenhouse and field conditions. Chromosomal introgressions from SHZ-2 into the TXZ-13 genome were identified using a single feature polymorphism microarray, SSR markers and gene-specific primers. Segregation analysis of the BC4F3 population indicated that three regions on chromosomes 2, 6, and 9, designated as qBR2.1, qBR6.1, and qBR9.1, respectively, was associated with blast resistance and contributed 16.2, 14.9, and 22.3%, respectively, to the phenotypic variance of diseased leaf area (DLA). We further narrowed the three QTL regions using pairs of sister lines extracted from heterogeneous inbred families (HIF). Pairwise comparison of these lines enabled the determination of the relative contributions of individual QTL. The qBR9.1 conferred strong resistance, whereas qBR2.1 or qBR6.1 individually did not reduce disease under field conditions. However, when qBR2.1 and qBR6.1 were combined, they reduced disease by 19.5%, suggesting that small effect QTLs contribute to reduction of epidemics. The qBR6.1 and qBR9.1 regions contain nucleotide-binding sites and leucine rich repeats (NBS-LRR) sequences, whereas the qBR2.1 did not. In the qBR6.1 region, the patterns of expression of adjacent NBS-LRR genes were consistent in backcross generations and correlated with blast resistance, supporting the hypothesis that multiple resistance genes within a QTL region can contribute to non-race-specific quantitative resistance.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Many genes containing nucleotide-binding sites (NBS) and leucine rich repeats (LRR) function to confer race-specific resistance against plant pathogens. This discovery has accelerated the manipulation of major resistance (R) genes for disease control in multiple crops. Because of the race-specificity, however, major R gene conferred resistance is often vulnerable to adaptation by variable pathogen populations. In rice, there are approximately 600 NBS-LRR sequences (Bai et al. 2002). A large proportion of these sequences exist in chromosomal clusters. Some NBS-LRR sequences confer race-specific resistance against pathogens of bacterial blight and blast (Song and Goodman 2001; Bryan et al. 2000; Ashikawa et al. 2008; Shang et al. 2009; Liu et al. 2010), but the functions of most of the sequences are not known. Furthermore, it is not clear whether some NBS-LRR sequences might contribute to quantitative, non-race specificity resistance. An interesting question is whether NBS-LRR sequences have residual effects for disease resistance after they are “defeated” by genetic shifts in the pathogens. Ballini et al. (2008) and Vergne et al. (2008) did a comprehensive analysis of the alignment of resistance genes and defense genes and published QTL in the literature. Their results show strong evidence that regions of blast resistance QTL are enriched with major resistance genes; however, due to the low resolution of QTL mapping it prevents further statistical analysis. Thus, there remains a need to experimentally determine the phenotypic effects of the NBS-LRR genes. Such information would greatly help in the design of resistance gene combinations to provide broad-spectrum resistance.
Although the genetic basis of broad-spectrum resistance is not well understood, based on the performance records, some varieties are known to exhibit stable resistance over time (Liu et al. 2009). Wang et al. (1994) first demonstrated that the durable blast resistance observed in the traditional rice variety Moroberekan is controlled by a combination of qualitative and quantitative genes. Similarly, SHZ-2, a modern variety grown in South China, exhibits broad-spectrum resistance to multiple races of the blast pathogen because it carries a combination of QTL and major resistance genes (Zhu et al. 1996; 2003; Liu et al. 2004). However, because it is difficult to measure the level of quantitative resistance when genes conferring complete resistance are present, quantitative resistance can be only estimated based on the presence of markers linked to QTLs in the absence of the epistatic effect of major resistance genes.
Using candidate defense genes as markers, we previously showed a strong association between defense genes and resistance phenotypes in segregation analysis of RIL and advanced backcross lines (Liu et al. 2004; Manosalva et al. 2009). However, since RIL and advanced backcross lines (BC3 generation) are far from isogenic, the specific effects of individual genes or chromosomal regions are difficult to ascertain. Traditionally, dissecting the phenotypic effects of a QTL would require 7–8 backcross generations to create near-isogenic lines with and without the target region. To accelerate the development of near-isogenic lines, an approach of selecting for sister lines derived from a heterogeneous inbred family (HIF) has been proposed (Tuinstra et al. 1997).
Here, we report the use of the genome-wide genotyping and phenotypic analysis of pairs of lines derived from HIF to identify chromosomal regions carrying strong and partial resistance. Expression analysis of candidate genes within QTL regions suggested that NBS-LRR sequences are potential candidates responsible for the phenotypic effects in the selected lines. Our results support the hypothesis that individual NBS-LRR genes can play a role in quantitative resistance, and that this resistance may not be race-specific. The combination of race-specific and non-race specific effects of NBS-LRR genes may account for the broad-spectrum resistance observed in SHZ-2 that is widely used in rice breeding programs.
Materials and methods
Plant materials
Three parental lines, SHZ-2, TXZ-13, and BC-10, and their derived progeny formed the core genetic materials for dissection of blast resistance under field and greenhouse conditions. SHZ-2 is an indica cultivar with resistance to multiple races of the blast pathogen (Zhu et al. 1996; 2003; Liu et al. 2004; Manosalva et al. 2009), whereas TXZ-13 is a high-yielding indica cultivar but susceptible to blast. BC-10 is a blast resistant progeny selected from the third backcross generation (BC3F2) derived from backcrossing SHZ-2 to TXZ-13 (Liu et al. 2004).
To dissect the QTL responsible for broad-spectrum blast resistance, BC-10 was backcrossed to TXZ-13 to produce BC4F2 progeny (because BC-10 is a BC3 progeny). Because seeds of the BC4F2 lines were not enough to evaluate disease phenotypes in replicated tests under multiple conditions, we advanced the population to BC4F3 generation by single seed descent. In total, 244 BC4F3 lines were developed and they were evaluated in the greenhouse and in the blast nursery at the IRRI experimental farm in Los Baños, the Philippines.
Evaluation of blast resistance phenotypes
To establish that BC-10 has a level of blast resistance comparable to that observed in SHZ-2, we evaluated disease resistance of BC-10, SHZ-2, and TXZ-13 in multiple sites known to have diverse pathogen populations. Field test experiments (natural infection) were conducted at three locations, Yangjiang, Conghua, and Longchuan, in Guangdong Province in two crop seasons (March–July and August–November) each year from 2004 to 2008 (total ten seasons). Blast disease was measured for 15 plants randomly selected from each of the three replicated rows (approximately 60 plants) per entry using the 0–9 scale of Standard Evaluation System for Rice (International Network for Genetic Evaluation of Rice 1996). The same three parental varieties were also evaluated under natural infection in the IRRI blast nursery in the Philippines.
The disease reaction of BC4F3 lines was evaluated by both single-isolate inoculation in the greenhouse and by exposure to natural pathogen population in the field. For greenhouse inoculation, we used a virulent isolate PO6-6 which exhibits virulence to most of the rice varieties in the Philippines. A randomized complete block design with three replicates was used, with 15 plants/line/replicate being inoculated and scored as previously described (Wang et al. 1994; Liu et al. 2004). Field tests against natural inoculum were conducted in the blast nursery in 2005 and 2006. A randomized complete block design was used in field studies, with three replicates of 50 plants/line planted in 16 m × 1.2 m2 nursery plots. Three rows of diverse, susceptible varieties were planted around the plots to maintain the diversity of the pathogen population and to enhance natural infection. Diseased leaf area (DLA) was assessed at 14 days after sowing in the blast nursery using the method described by Notteghem et al. (1981).
Target preparation and hybridization in microarray-based genotyping
To identify the chromosomal introgressions of BC-10 from SHZ-2, a genome-wide genotyping microarray was used (Edwards et al. 2008). This array consists of 880 oligos that detect single feature polymorphisms (insertions and deletions) in unique sequences evenly spaced along the chromosomes (medium spacing about 250 kb). DNA was extracted from the leaves of BC-10 and TXZ-13 by CTAB method (Murray and Thompson 1980) and digested with the restriction enzyme DpnII. The digested genomic DNAs were labeled separately with Cy3 and Cy5 fluorescent dyes according to the protocol from BioPrime Array CGH Genomic labeling system, (cat# 18095-012, Invitrogen, Carlsbad, CA). The labeled targets were purified using NucleoSpin Extract II kit (Qiagen PCR purification kit, cat# 28104, Valencia, CA) according to the manufacturer’s instruction. The purified targets were denatured at 99°C for 5 min in 2XMES buffer containing Herring sperm. The denatured targets were hybridized overnight in a water bath at 50°C. Hybridized slides were washed successively for 5 min in each of three solutions, comprising 6 × SSPE plus 0.005% SDS at 50°C, 0.06 × SSPE at room temperature (RT), and 0.02 × SSC at RT. The hybridized slides were then dried by spinning at 2,000 rpm at 25°C for 2 min. Dried hybridized slides were scanned using a Perkin Elmer Microarray Scanner (ScanArray Gx, Shelton, USA).
Molecular marker analysis
To verify the results of microarray analysis, the introgression regions of SHZ-2 were monitored in all BC4F3 lines using SSR markers (from GRAMENE, http://www.gramene.org) which were located in or near to the introgression regions according to the graphical genotypes of BC-10 determined by the microarray assay. In the case where defense genes in the introgression regions were used as markers, polymorphism was assayed by TILLING (Raghavan et al. 2007) in all BC4F3 lines. Candidate defense genes, including germin-like proteins, chitinase, 14-3-3 protein, and oxalate oxidases, were identified from previous studies (Liu et al. 2004; Manosalva et al. 2009; Carrillo et al. 2009). Primer pairs were designed using Oligo Primer Analysis Software Version 5.0 based on the gene models from TIGR 5.0. Primers were synthesized by the SBS Company (Beijing, China) (Supplementary Table S1).
Development of heterogeneous inbred families
Pairs of sister lines extracted from heterogeneous inbred families (HIF) were used for determining the effect of individual QTL on blast resistance. To develop the HIF, we identified BC4F3 lines that showed segregation of resistance within a family. Single plants from these families were genotyped with markers defining the QTL regions. Progeny derived from a plant that is heterozygous at the target region yielded two lines homozygous at the locus; a homozygous line was then selfed to produce fixed lines for evaluation. We concentrated on extracting lines that were homozygous at the three QTL on chromosomes 2, 6, and 9. Lines with different allelic combinations were evaluated in three independent experiments at the IRRI blast nursery.
Genome mapping and QTLs analysis
The graphical genotyping of BC-10 was generated using the software of Graphical Genotyping (GGT) version 2.0 (http://www.dpw.wau.nl/pv/pub/ggt/www.plantbreeding.nl). Single marker analysis (Qgene software, in 3.0 versions) was used to identify markers significantly associated with blast quantitative resistance. A LOD value greater than 3.0 was interpreted as a QTL. This conventional cutoff served the purpose of identifying the location rather than the strength of a QTL. The actual phenotypic effects of individual QTL were quantified using pairs of sister lines derived from HIF.
Fungal inoculation and samples collection for RNA extraction
SHZ-2, TXZ-13, and BC-10 seedlings were inoculated at 21 days after sowing by spraying with spore suspension of 7 × 105 spores/ml of Magnaporthe oryzae isolate PO6-6 in the greenhouse. Mock inoculation was done by spraying sterile distilled water. Inoculated plants were maintained in an incubation chamber (24–26°C and 100% relative humidity) in the dark for 24 h, and then transferred to a mist room at 25°C and 100% relative humidity in the greenhouse. Each treatment was repeated at least twice. The seedlings were harvested at 0, 12, 24, 48, and 72 h after inoculation, quickly frozen in liquid nitrogen and stored at −80°C until use.
RNA Isolation and reverse transcriptase-PCR
Total RNA was extracted from green leaf tissue using the TRIzol method (Invitrogen, San Diego) according to the instruction provided by the manufacturer. RNA samples were treated with DNase I (Amplification Grade) to remove any contaminating genomic DNA. First-strand cDNA was synthesized using the SuperScript II Reverse Transcriptase kit (Invitrogen, San Diego) according to the manufacture’s instructions. The resulting first-strand cDNA was used as a template, and amplification was done using the following cycles (30 s at 94°C, 30 s at 58°C or 60.5°C, and 2 min at 72°C) followed by 10 min at 72°C. Primer sequences were designed using Oligo Primer Analysis Software Version 5.0 based on the gene models from TIGR (5.0). Primers were synthesized by the SBS Company (Beijing, China) (Supplementary Table S2).
Results
Since the quantitative dissection of the resistance in BC-10 was based upon the premise that this variety has broad-spectrum resistance, performance of BC-10 was monitored in locations, where blast is endemic and severe. In our previous study, SHZ-2 displayed complete resistance to 344 blast isolates from eight provinces in China and 49 isolates, except for isolate CA89, representing 14 lineages in Philippines (Liu et al. 2004; Chen et al. 1995). For ten subsequent cropping seasons from 2004 to 2008, BC-10 displayed consistent blast resistance in three locations in Guangdong Province (Table 1). Similar results were obtained at the IRRI blast nursery (Supplementary Table S3). The three sites in Guangdong Province were selected as “hot spot” screening sites for the rice breeding program in South China. Previous studies on pathogen populations at these sites showed a diversity of pathogen lineages (8 in Yangjiang, 6 in Conghua and 6 in Longchuan) (Zhu et al. 1996; 2003; Yang et al. 2008). The level of susceptibility shown in BC-10 was slightly higher than SHZ-2, though not statistically significant, across all three sites. TXZ-13 was significantly more susceptible than SHZ-2 and BC-10 (P < 0.001) across all locations and seasons (Supplementary Table S4). The resistance observed in SHZ-2 and BC-10 observed across locations and years were not absolute immunity, indicating that these varieties are susceptible to some pathogen isolates at the test sites. Overall, as disease pressure increased (as indicated by disease severity of TXZ-13), more diseases were observed on BC-10 and SHZ-2. For example, in Longchuan, the level of susceptibility of BC-10 was positively correlated (r = 0.72*) with that of TXZ-13, consistent with the expected behavior of a variety possessing quantitative resistance.
Chromosomal introgressions in BC-10 from SHZ-2
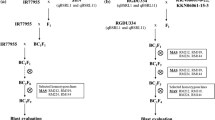
Chromosomal introgressions from SHZ-2 in BC-10 were first defined using a genome-wide genotyping microarray (Edwards et al. 2008). This array consists of 880 oligos that detect single feature polymorphisms (insertions and deletions) in sequences evenly spaced along the chromosomes (median spacing about 250 kb). Hybridization was done by pairing Cy3/Cy5 labeled DNA of BC-10 with that of the recurrent parent TXZ-13. Based on the data from four replicated hybridization experiments, we detected 39 regions of SHZ-2 introgression segments in BC-10. The regions of SHZ-2 introgression shown by microarray data were checked by SSR markers located within or near the introgression regions. Of 39 SSR markers tested, 19 were consistent with the microarray data; the other SSR markers were monomorphic and hence not informative (data not shown). Combining the microarray and SSR data, we generated the graphical genotypes of BC-10 using the software of Graphical Genotyping (GGT) version 2.0 (http://www.dpw.wau.nl/pv/pub/ggt/www.plantbreeding.nl) (Fig. 1).
Graphical genotypes of BC-10 derived from SHZ-2 × TXZ-13 showing three QTLs to blast resistance, qBR2.1, qBR6.1, and qBR9.1. The introgressions of SHZ-2 in BC-10 were detected using SFP microarrary chips (Edwards et al. 2008) and confirmed by SSR markers and gene-specific markers. The gray bar shows that the genotype of BC-10 is the same as the recurrent parent TXZ-13. The black bars are the introgressions of SHZ-2 in BC-10. On the left are single feature polymorphism (SFP) marker names and their physical positions on the each chromosome as defined by the SFP microarray (Edwards et al. 2008)
Identifying QTL regions in BC4F3 population
To determine the relationship between introgression regions and blast resistance, a BC4F3 population was produced by backcrossing BC-10 to TXZ-13. Blast resistance of the BC4F3 population was evaluated by natural infection in the blast nursery and by artificial inoculation with M. oryzae isolate PO6-6 in the greenhouse at IRRI. The distribution of DLA among the BC4F3 lines in both the blast nursery and greenhouse was skewed toward resistance (Fig. 2). Pairwise analysis of DLA on the BC4F3 lines from different experiments showed significant correlation in the levels of resistance between tests (r = 0.70** for the greenhouse versus the blast nursery, Philippines). To accelerate the identification of markers linked to quantitative resistance, 20 BC4F3 lines from each of the “tails” of the distribution curve, representing high and low DLA, and five partial resistance lines (DLA about 16–32%) were selected for association analysis. We then monitored the introgression regions of SHZ-2 alleles in 244 BC4F3 lines using SSR markers and SNPs in defense genes assayed by TILLING method (Raghavan et al. 2007). We were able to define the introgression regions of SHZ-2 in BC-10 using SFP chips because the markers used on the SFP chips were anchored to the rice physical map. In addition, we selected about 300 SSR markers (unpublished data) within or around the introgression regions to validate the SFP results. Thus, by combining the genotyping data from using microarray, SSR markers, and SNPs of specific alleles, we determined the intervals on chromosome 2, 6, and 9, which were associated with blast resistance. Following the recommended nomenclature of rice QTL (McCouch 2008), the quantitative Blast Resistance loci are tentatively named as qBR2.1, qBR6.1, and qBR9.1 (Fig. 1). The logarithm of the odds (LOD) and the contribution of the three QTLs (LOD value > 3.0) were determined by single marker analysis using Q-gene software (version 3.05) (Table 2). The qBR2.1 on chromosome 2 was identified by SSR marker RM6069, with a LOD score of 9.0. It accounted for 16.2% of the DLA variance. The qBR6.1 on chromosome 6, defined by SSR markers RM204 and RM584, spanned approximately 250 kb; it accounted for 14.9% of the DLA variance. The qBR9.1 on chromosome 9, defined by RM24022, RM7364 and RM5777, spanned 560 kb, and accounted for 22.3% of the DLA variance.
Estimating QTL effects using sister lines from heterogeneous inbred families
Based on the QTL analysis, we extracted HIF to define the phenotypic effect of the three regions on chromosomes 2, 6, and 9. From 244 BC4F3 lines tested in the blast nursery, we selected 19 families that showed segregation in DLA to determine the heterozygosity at each of the regions defined by QTL mapping. Of these families, nine contained plants that showed heterozygosity in the targeted regions on chromosomes 2, 6, and 9. These plants were selfed to produce pairs of sister lines that were homozygous at the three QTL regions (Table 3). These lines were first tested in the greenhouse by artificial inoculation using M. oryzae isolate PO6-6, and then were evaluated in the blast nursery by natural infection in two independent replicated experiments. These pairs of lines were referred to as pre-near-isogenic line (pre-NILs) because, although they differ from each other with respect to the presence of the target QTL, they are not sufficiently similar to the recurrent parent TXZ-13 to be called near-isogenic.
Pairs of lines with fixed homozygous QTL regions are summarized in Table 3. The pre-NILs derived from each HIF had significantly different DLA; pre-NILs with the same combination of QTL had similar leaf lesions and similar development of DLA (Figs. 3, 4). Lines carrying either qBR2.1 or qBR6.1, such as 18-14, 18-17 or 22-10, 22-16, and 21-7, were susceptible to M. oryzae. Lines carrying qBR2.1 and qBR6.1 (e.g., 22-1, 22-8, 18-1, 18-18, and 21-19) showed partial resistance. Lines carrying qBR9.1 (e.g., 21-8) showed strong resistance to M. oryzae isolates. Lines with qBR2.1, qBR6.1, and qBR9.1 (e.g., 22-4) showed the highest level of resistance. Taken together, the effect of qBR9.1 was estimated to contribute a 31.4% disease reduction (Table 4). Although the individual effects of qBR2.1 and qBR6.1 were relatively small, combining the two QTL could reduce disease by 19.5% (Table 4). While it is obvious that qBR9.1 is epistatic to qBR2.1 and qBR6.1, we cannot infer the epistatic relationship between qBR2.1 and qBR6.1 because significant phenotypic effect (P < 0.001) was evident under field conditions only when these two QTL were together (Supplementary Table S5).
Disease progress curves as measured by development of the diseased leaf area (DLA) of different sister lines derived from a heterogeneous inbred family (HIF-22) under natural infection in the Blast Nursery at IRRI Experimental Farm (see Table 2)
Candidate genes within QTL regions and their expression patterns
Of the three QTL identified on chromosome 2, 6, and 9, the regions defined by qBR6.1 and qBR9.1 are sufficiently narrow to allow for a search for potential candidate genes related to expression of resistance. We examined the gene models (TIGR version 5.0) in the regions delimited by qBR2.1, qBR6.1, and qBR9.1 based on the Nipponbare sequence from Gramene (http://www.gramene.org) (see Supplementary Table S6, Table S7 and Table S8 for all listing of genes defined by the three QTLs). In the qBR6.1 region, there were 36 gene models, of which four were annotated as predicted defense response-related genes (Supplementary Table S7). Two adjacent genes were predicted disease resistance genes that contained a NBS-LRR domain, including resistance protein LR10 (LOC_Os06g06850) and disease resistance RPP13-like protein 3 (LOC_Os06g06860). Within the qBR9.1 region, there were 49 gene models, of which only 13 genes had predicted functions (Supplementary Table S8). Three disease resistance genes with NBS-LRR domain were found in this region including a leucine rich repeat family protein (LOC_Os09g15850), and two NBS-LRR disease resistance proteins (LOC_Os09g15840 and LOC_Os09g16000).
To clarify the potential roles of the resistance gene-like candidates within the qRB6.1 region, we examined their expression patterns in relation to neighboring genes in SHZ-2, BC-10, and TXZ-13 (Table 5). Within this region (qRB6.1), we tested the expression pattern of nine genes including LOC_Os06g06780, LOC_Os06g06790, LOC_Os06g06810, LOC_Os06g06850, LOC_Os06g06860, LOC_Os06g06970, LOC_Os06g07030, LOC_Os06g07070, and LOC_Os06g07100 (Supplementary Table S7). The five genes adjacent to the resistance gene candidates showed similar expression patterns over all time points for SHZ-2, BC-10 and TXZ-13 (data not shown). In contrast, three resistance gene candidates showed differential response between resistant (SHZ-2, BC-10) and susceptible (TXZ-13) varieties (Table 5, LOC_Os06g06810 is included as a reference). Interestingly, the expression patterns of these four genes were nearly identical in SHZ-2 and BC-10, but distinct in TXZ-13, suggesting that the regulatory control of these genes was transmitted in an intact state from SHZ-2 to BC-10 after three backcross generations. Under qBR6.1, the disease resistance RPP13-like protein 3 (LOC_Os06g06860) was induced by M. oryzae in SHZ-2 and BC-10, but not in TXZ-13. Expression of the RPP13-like sequence was highly induced by M. oryzae at 24 h after inoculation (HAI). The expression of resistance protein LR10 (LOC_Os06g06850) in SHZ-2 and BC-10 was low at zero time point, but induced by pathogen and mock inoculations at 12 and 24 HAI. Induction was sustained by pathogen infection until 48 h. Little or no expression was observed in TXZ-13.
Under the qBR9.1 on chromosome 9, the NBS-LRR disease resistance gene (LOC_Os09g16000) was constitutively expressed in TXZ-13 at all time points. For SHZ-2 and BC-10, this candidate resistance gene expressed at a low level at 0 HAI (Fig. 5, lane 1), but was highly induced either by spraying with water or with M. oryzae PO6-6 at 12 and 24 HAI (Fig. 5, lane 2 and lane 3), but returned to the basal level at 48 HAI (Fig. 5, lane 4, lane 5). With mock inoculation, the gene was expressed at a low level at 0 HAI (Fig. 5b, lane 1), induced at 12 and 24 HAI (Fig. 5b, lane 2, lane 3), and remained induced at 48 HAI (Fig. 5b, lane 4, lane 5) with water. Two other NBS-LRR protein genes in the qRB9.1 region were LOC_Os09g15840 and LOC_Os09g15850 that corresponded to the Pi5-1 and Pi5-2, respectively, identified by Lee et al. (2009). We attempted to detect the Pi5-1 candidate (LOC_Os09g15840) by RT-PCR, but were not successful in all three genotypes, possibly due to the sequence variation between the indica and japonica varieties. The Pi5-2 candidate (LOC_Os09g15850) showed no or very low expression in SHZ-2 and BC-10 but was constitutively expressed in TXZ-13 (Table 5). Since both Pi5-1 and Pi5-2 are required for Pi5 function (Lee et al. 2009), we infer that Pi5 does not play an important role in conferring resistance in SHZ-2 or BC-10.
The expression pattern of the LOC_Os09g16000 (putative NBS-LRR disease resistance protein) gene using RT-PCR. a Expression of the LOC_Os09g16000 gene upon inoculation with PO6-6. Total RNA was extracted from leaf tissue of SHZ-2 and TXZ-13 at 0, 12, 24, 48, and 72 h after inoculation (HAI), respectively. b Expression of the LOC_Os09g16000 gene upon inoculation with sterilized water. Total RNA was extracted from leaf tissue of SHZ-2 at 0, 12, 24, 48, and 72 HAI. c Expression of the LOC_Os09g16000 gene under control conditions. Total RNA was extracted from leaf tissue of SHZ-2 at 0, 12, 24, 48, and 72 h from plants which were not inoculated. Amplification of the LOC_Os09g16000 gene and GADPH was at 35 cycles (annealing temperature: 60.5°C) and 25 cycles (annealing temperature: 58.0°C), respectively
Discussion
SHZ-2, an indica cultivar grown in China, exhibits resistance to multiple races of the rice blast pathogen, M. oryzae (Zhu et al. 1996; 2003; Liu et al. 2004; Table 1). This broad-spectrum resistance has been attributed to a combination of genes conferring complete and partial resistance (Liu et al. 2004; Liu et al. 2009). Using a combination of RFLP and SSR markers, four QTL regions were identified that together accounted for 60.3% of the phenotypic variation (Liu et al. 2004). However, the stability of disease resistance seen in SHZ-2 and in its derivatives suggested that more resistance genes or QTL are yet to be discovered in this variety. Attempts to identify additional QTL have been hindered by a low degree of polymorphism between the parental lines. This is particularly problematic in introgressing QTL into popular varieties when both the donor and recipient varieties are indica type. To help address this problem, a SFP chip, capable of detecting 880 features, was used to enable the detection of genomic variation. Our data suggested that a genome-wide scan of SFP provided a better resolution of differences between the donor and recipient genotypes. We were able to track the QTL and at the same time locate additional QTL regions. Through a combination of SFP and SSR mapping, we found three QTL regions on chromosome 2, 6, and 9 using a BC4F3 population. Based on the map positions, these three QTL regions are distinct from those identified by Liu et al. (2004), where three major resistance genes and 4 QTL regions were found on chromosomes 2, 7, 8, 10, and 12. The QTL on the chromosome 2 in this study is not in the same region containing the candidate gene 14-3-3 protein as reported by Liu et al. (2004). The QTL on chromosome 6 was not detected in Liu et al. (2004). We compared the location of qRB6.1 with the meta-QTL compiled by Ballini et al. (2008, 2009), and found that meta QTL q6G4 (1.22 Mb) and q6G9 (0.33 Mb) are located beside qRB6.1 (3.16-3.4 Mb). The qBR9.1 on chromosome 9 was also located near known resistance genes Pi15 and Pi5/Pi3/Pii (Lin et al. 2004; Jeon et al. 2003; Yi et al. 2004; Lee et al. 2009). The expression pattern of Pi5-2 suggested that it is unlikely that qBR9.1 was solely contributed by Pi5. However, because the meta QTLs are defined using published data, it is difficult to pinpoint their locations precisely. Unless allelic tests are done, we cannot be certain that the QTLs identified in this study are different from those identified by others. This highlights the need to assign phenotypic value to chromosomal regions, where multiple QTLs have been mapped by different studies (Ballini et al. 2008; Vergne et al. 2008).
We show that pairs of pre-NILs from different HIF were highly suitable for estimating the phenotypic contribution of each new QTL region to resistance. Pre-NIL lines with different combinations of the three QTL were identified (Table 3), and these enabled estimates of the phenotypic effects of individual QTL and their interactions. In terms of phenotypic variance, the three QTL on chromosome 2, 6, and 9, appeared to function additively to confer resistance to blast. qBR2.1 and qBR6.1 accounted for 16.2 and 14.9% of the DLA variance, respectively, whereas qBR9.1 accounted for 22.3%. Together the three QTLs accounted for 53.3% of the DLA variance in the original mapping population. However, in terms of disease reduction, neither qBR2.1 nor qBR6.1 individually gave protection against blast at the seedling stage (Table 4). Effects on disease reduction were only measurable when the two QTL were combined. The cumulative effects of these QTLs, which were manifested as reduced disease over time, were assessed by following disease progression over 2 weeks on sister lines planted in the field (Fig. 4). This could be attributed to reduced initial inoculum, leading to reduced disease progression. Addition of small QTL effects could be amplified under field conditions because inoculum would be reduced resulting in fewer new leaf tissue infections in each cycle. We only assessed disease at the seedling stage, and, therefore, cannot predict if the same level of disease reduction occurred at the panicle stage. These same lines are being evaluated in blast hot spots in Guangdong, China to determine the correlation between leaf and neck blast severity (Zhu XY, unpublished data).
The narrow chromosomal regions defined by qBR6.1 and qBR9.1 allowed us to explore candidate genes underlying the QTL. Within these two regions, we found five resistance gene-like sequences with NBS-LRR domains. These resistance gene-like sequences are expected to code for proteins that interact with avirulence effectors of specific pathogen strains. In general, each R-gene and avirulence effector gene interaction is expected to give a strong response, thus accounting for the qualitative resistance provided by a major R gene. It is also common to find clusters of NBS-LRR genes in the genome (Meyers et al. 1999; Bai et al. 2002; Richly et al. 2002; Liu et al. 2010). A question of practical interest is whether non-race-specific quantitative resistance is a manifestation of the collective specific resistance provided by the clusters of NBS-LRR genes. In our experiments, the lines were evaluated under field conditions in the blast nursery, where a diverse pathogen population was maintained through the planting of susceptible varieties throughout the year (Chen et al. 1995; C. Vera Cruz, IRRI, personal communication). It is conceivable that the interactions between multiple resistance genes and diverse pathogen strains could trigger a gradation of defense responses resulting in a phenotypic expression of quantitative resistance. The genetic materials produced by this study can be used to address this question.
As a first step to identify the candidate genes within the QTLs, we examined the expression patterns of the resistance genes and defense response genes by RT-PCR (Table 5). The expression analysis supports the genotyping data showing that BC-10 and SHZ-2 have the same allele in the QTL regions, and that these are different from TXZ-13. The expression patterns of SHZ-2 and BC-10 were nearly identical for the differentially expressed genes. In the qBR6.1 region, the patterns of expression of two adjacent genes are similar in SHZ-2 and BC-10, suggesting that coordinated expression patterns are transmitted from the resistant parent SHZ-2 to the backcross progeny BC-10. Since disease resistance phenotype was evaluated against diverse pathogen strains under field conditions, this result supports the hypothesis that resistance genes within a QTL region can contribute to non-race-specific quantitative resistance. On the other hand, the expression patterns of individual genes within qBR9.1 do not suggest a simple relationship between resistance gene expression and resistance (Table 5, Fig. 5). The allele of LOC_Os09g16000 gene in qBR9.1 was strongly and constitutively expressed in TXZ-13 but only weakly expressed in SHZ-2 and BC-10. However, strong induction of expression was seen in SHZ-2 and BC-10 by both pathogen inoculation and mock inoculation. It appears the allele of SHZ-2 is relatively more responsive to environmental perturbations. Interestingly, a similar pattern of expression was observed in Pib, a blast-resistance gene that was up-regulated by environmental conditions, such as humidity and darkness and by chemical signals such as probenazole (Wang et al. 1999). Although the expression of Pib was low in uninoculated leaves, it increased in expression at 12 and 24 HAI after inoculation with both compatible and incompatible races of M. oryzae (Wang et al. 1999, 2001). It was hypothesized that it may be advantageous for the rice plant to respond first to environmental changes that are conducive to pathogen infection; the activation of defense genes and specific resistance genes may thereby prime the plant to respond to pathogen attack (Wang et al. 1999, 2001). Since we did not include multiple environmental factors (e.g., humidity, light, or temperature) in our expression analysis, we cannot speculate whether the SHZ-2 allele is also induced by environmental factors. More detailed work is needed to determine whether the inductive property of the SHZ-2 allele is important for the resistance phenotype.
There is increasing evidence that coordinated expression of multiple functionally related genes within a small chromosomal region could be an important mechanism for achieving quantitative resistance. Ashikawa et al. (2008) showed that two adjacent NBS-LRR genes (Pikm1-TS and Pikm2-TS) within a 131-kb region were required for the expression of specific resistance previously considered to be controlled by a single locus Pikm in variety Tsuyuake. Similarly, Lee et al. (2009) found that the complete function of Pi5 was conferred by two adjacent genes Pi5-1 and Pi5-2 within a 130 kb region. In a case of basal defense genes, Manosalva et al. (2009) demonstrated that members of a multi-gene family encoding germin-like proteins can play a role in quantitative resistance to blast and sheath blight. In such a complex locus, the phenotypic contributions of individual members could be small, however, collectively, they produce measurable effects. The correlation between resistance phenotype and expression patterns of candidate resistance genes in qBR6.1 is consistent with the hypothesis that NBS-LRR genes are contributors to quantitative resistance; however, direct evidence is needed to show that the expression of patterns are causally related to phenotypic resistance. As demonstrated by Manosalva et al. (2009), systematic silencing of functionally related genes can be a sound approach to determine whether the NBS-LRR genes collectively contribute to quantitative resistance.
Our study showed that the extraction of pre-NIL from HIF, when done in breeding populations with good adaptability, has the advantage of shortening the time in developing near-isogenic lines that are sufficiently fixed for quantifying QTL contribution. Because whole genome genotyping is becoming routine, the use of HIF provides a rapid approach to isolate pre-NILs for phenotypic comparison (Tuinstra et al. 1997; Perchepied et al. 2006). Being able to quantify the phenotypic contributions of QTL is critical for adoption of marker-assisted selection for quantitative resistance in breeding. While the use of marker-assisted selection for major resistance genes is common place for rice (Fjellstrom et al. 2004; Jia et al. 2002; Yi et al. 2004), use of markers for the selection of quantitative disease resistance has not been widely adopted. One reason is that breeders are not convinced of the breeding value of QTL for disease resistance, and hence have little incentive to invest in marker-assisted selection for QTL. We demonstrated that the accumulation of QTL with moderate effects can result in significant disease reduction, thus supporting the notion that small QTL can contribute to stability of the variety against blast. Because resistance of the pre-NILs was assayed under natural conditions with diverse pathogen strains, these QTLs could be suitable for deployment in different environments. However, it is important to emphasize that durability is a retroactive assessment of a variety based on its performance over time and space (Johnson 1981). SHZ-2 was identified as a “durable” resistance variety based on its historical performance. Although our analysis shows that multiple QTL could contribute to the stable resistance observed in SHZ-2 and its derivatives, we have yet to examine how effective individual QTLs are under commercial production. In order to test the effectiveness of these QTL, the HIF lines or NIL derived from them will be tested in multiple locations.
References
Ashikawa I, Hayashi N, Yamane H, Kanamori H, Wu JZ, Matsumoto T, Ono K, Yano M (2008) Two adjacent NBS-LRR class genes are required to confer Pikm-specific rice blast resistance. Genetics 180:2267–2276
Bai JF, Pennill LA, Ning JC, Lee SW, Ramalingam J, Webb CA, Zhao BY, Sun Q, Nelson JC, Leach JE, Hulbert SH (2002) Diversity in nucleotide binding site—leucine-rich repeat genes in cereals. Genome Res 12:1871–1884
Ballini E, Morel JB, Droc G, Price A, Courtois B, Notteghem JL, Tharreau D (2008) A genome-wide meta-analysis of rice blast resistance genes and quantitative trait loci provides new insights into partial and complete resistance. Mol Plant Microbe Interact 21:859–868
Ballini E, Vergne E, Tharreau D, Notteghem JL, Morel JB (2009) ARCHIPELAGO: towards bridging the gap between molecular and genetic information in rice blast disease resistance. In: Wang GL, Valent B (eds) Advances in genetics, genomics and control of rice blast disease. Springer, Netherlands, pp 417–425
Bryan GT, Wu KS, Farrall L, Jia YL, Hershey HP, McAdams SA, Faulk KN, Donaldson GK, Tarchini R, Valent B (2000) A single amino acid difference distinguishes resistant and susceptible alleles of the rice blast resistance gene Pi-ta. Plant Cell 12:2033–2045
Carrillo MGC, Goodwin PH, Leach JE, Leung H, Vera Cruz CM (2009) Phylogenomic relationships of rice oxalate oxidases to the cupin superfamily and their association with disease resistance QTL. Rice 2:67–79
Chen DH, Zeigler RS, Leung H, Nelson RJ (1995) Population structure of Pyricularia grisea at two screening sites in the Philippines. Phytopathology 85:1011–1020
Edwards JD, Janda J, Sweeney MT, Gaikwad AB, Liu B, Leung H, Galbraith DW (2008) Development and evaluation of a high-through, low-cost genotyping platform based on oligonucleotide microarrays in rice. Plant Methods 4:13
Fjellstrom R, Conaway-Bormans CA, McClung AM, Marchetti MA, Shank AR, Park WD (2004) Development of DNA markers suitable for marker assisted selection of three Pi genes conferring resistance to multiple Pyricularia grisea pathotypes. Crop Sci 44:1790–1798
International Network for Genetic Evaluation of Rice (1996) Standard evaluation system for rice, 4th edn. International Rice Research Institute, Los Banos, Laguna, Philippines, p 52
Jeon JS, Chen D, Yi GH, Wang GL, Ronald PC (2003) Genetic and physical mapping of Pi5 (t), a locus associated with broad-spectrum resistance to rice blast. Mol Genet Genomics 269:280–289
Jia YL, Wang ZH, Singha P (2002) Development of dominant rice blast Pi-ta resistance gene markers. Crop Sci 42:2145–2149
Johnson R (1981) Durable resistance: definition of, genetic control, and attainment in plant breeding. Phytopathology 71:567–568
Lee SK, Song MY, Seo YS, Kim HK, Ko S, Cao PJ, Suh JP, Yi G, Roh JH, Lee S, An G, Hahn TR, Wang GL, Ronald P, Jeon JS (2009) Rice Pi5-mediated resistance to Magnaporthe oryzae requires the presence of two coiled-coil-nucleotide-binding-leucine-rich repeat genes. Genetics 181:1627–1638
Lin F, Liu Y, Wang L, Liu X, Pan QH (2004) A high-resolution map of the rice blast resistance gene Pi15 constructed by sequence-ready markers. Plant Breed 126:287–290
Liu B, Zhang SH, Zhu XY, Yang QY, ShZh Wu, Mei MT, Mauleon R, Leach JE, Mew T, Leung H (2004) Candidate defense genes as predictors of quantitative blast resistance in rice. Mol Plant Microbe Interact 17:146–152
Liu B, Zhu XY, Zhang SH, Wu JL, Han SS, Cho YC, Roh JH, Leach JE, Liu Y, Madamba S, Bordeos A, Baraoidan M, Oña I, Vera Cruz CM, Leung H (2009) What it takes to achieve durable resistance to rice blast? In: Wang GL, Valent B (eds) Advances in genetics, genomics and control of rice blast disease. Springer, Netherlands, pp 385–402
Liu J, Wang X, Mitchell T, Hu Y, Liu X, Dai LY, Wang GL (2010) Recent progress and understanding of the molecular mechanisms of the rice-Magnaporth oryzae interaction. Mol Plant Pathol 11:419–427
Manosalva PM, Davidson RM, Liu B, Zhu XY, Hulbert SH, Leung H, Leach JE (2009) A germin-like protein gene family functions as a complex quantitative trait locus conferring broad-spectrum disease resistance in rice. Plant Physiol 149:286–296
McCouch SR, CGSNL (2008) Gene nomenclature system for rice. Rice 1:72-84
Meyers BC, Dickerman AW, Michelmore RW, Sivaramakrishnan S, Sobral BW, Young ND (1999) Plant disease resistance genes encode members of an ancient and diverse protein family within the nucleotide-binding superfamily. Plant J 20:317–332
Murray MG, Thompson WF (1980) Rapid isolation of high molecular weight plant DNA. Nucl Acids Res 8:4321–4325
Notteghem JL, Chatel M, Dechanet RD (1981) Analyze of two characteristics of rice resistance to Pyricularia oryzae. In: Comptes-rendus du symposium sur la resistance du riz a la pyriculariose. IRAT-GERDAT, Montpellier, France, pp 301–318
Perchepied L, Kroj T, Tronchet M, Loudet O, Roby D (2006) Natural variation in partial resistance to pseudomonas syringae is controlled by two major QTLs in Arabidopsis thaliana. PLoS One 1(123):1–10
Raghavan C, Naredo MEB, Wang HH, Atienza G, Liu B, Qiu FL, McNally KL, Leung H (2007) Rapid method for detecting SNPs on agarose gels and its application in candidate gene mapping. Mol Breed 19:87–101
Richly E, Kurth J, Leister D (2002) Mode of amplification and reorganization of resistance genes during recent Arabidopsis thaliana evolution. Mol Biol Evol 19:76–84
Shang JJ, Tao Y, Chen XW, Zou Y, Lei CL, Wang J, Li XB, Zhao XF, Zhang MJ, Lu ZK, Xu JC, Cheng ZK, Wan JM, Zhu LH (2009) Identification of a new rice blast resistance gene, Pid3, by genome-wide comparison of paired NBS-LRR genes and their pseudogene alleles between the two sequenced rice genomes. Genetics 182:1303–1311
Song FM, Goodman RM (2001) Molecular biology of disease resistance in rice. Physiol Mol Plant Pathol 59:1–11
Tuinstra MR, Ejeta G, Goldsbrough PB (1997) Heterogeneous inbred family (HIF) analysis: a method for developing near-isogenic lines that differ at quantitative trait loci. Theor Appl Genet 95:1005–1011
Vergne E, Ballini E, Droc G, Tharreau D, Nottéghem JL, Morel JB (2008) Archipelago: a dedicated resource for exploiting past, present, and future genomic data on disease resistance regulation in rice. Mol Plant Microbe Interact 21:869–878
Wang GL, Mackill DJ, Bonman M, McCouch SR, Champoux MC, Nelson RJ (1994) RFLP mapping of genes conferring complete and partial resistance to blast in a durably resistant rice cultivar. Genetics 136:1421–1434
Wang ZX, Yano M, Yamanouchi U, Iwamoto M, Monna L, Hayasaka H, Katayose Y, Sasaki T (1999) The Pib gene for rice blast resistance belongs to the nucleotide binding and leucine-rich repeat class of plant disease resistance genes. Plant J 19:55–64
Wang ZX, Yamanouchi U, Katayose Y, Sasaki T, Yano M (2001) Expression of the Pib gene rice-blast-resistance gene family is up-regulated by environmental conditions favouring infection and chemical signals that trigger secondary plant defense. Plant Mol Biol 47:653–661
Yang JY, Chen S, Zeng LX, Li YL, Chuan-ying Li, Zhu XY (2008) Race specificity of major rice blast resistance genes to Magnaporthe grisea isolates collected from indica rice in Guangdong, China. Rice Sci 15:311–318
Yi G, Lee SK, Hong YK, Cho YC, Nam MH, Kim SC, Han SS, Wang GL, Hahn TR, Ronald PC (2004) Use of Pi5(t) markers in marker-assisted selection to screen for cultivars with resistance to Magnaporthe grisea. Theor Appl Genet 109:978–985
Zhu XY, Yang QY, Huo CB, Wu SZ (1996) Studies on the qualitative and quantitative resistance of rice cultivars to blast disease. Chin J Rice Sci 10:181–184
Zhu XY, Yang QY, Liu B, Zhang SH, Wu SZ (2003) The characteristics of resistance to rice blast of SHZ-2 and its derivative varieties. Guangdong Agric Sci 2:37–40
Acknowledgments
This work was supported in part by grants from the Generation Challenge Program (HL), USDA-NRI (DWG, JEL, HL), USAID Linkage Program grants, the MOST Key International Collaboration Project (2006DFB33320, BL, HL), NSFC-IRRI Project (30821140350, BL, HL), and the Guangdong International Collaboration Project (2007A050100037, BL).
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by M. Wissuwa.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Liu, Y., Zhu, X.Y., Zhang, S. et al. Dissecting quantitative resistance against blast disease using heterogeneous inbred family lines in rice. Theor Appl Genet 122, 341–353 (2011). https://doi.org/10.1007/s00122-010-1450-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00122-010-1450-2