Abstract
Characterization of genetic diversity is of great value to assist breeders in parental line selection and breeding system design. We screened 770 maize inbred lines with 1,034 single nucleotide polymorphism (SNP) markers and identified 449 high-quality markers with no germplasm-specific biasing effects. Pairwise comparisons across three distinct sets of germplasm, CIMMYT (394), China (282), and Brazil (94), showed that the elite lines from these diverse breeding pools have been developed with only limited utilization of genetic diversity existing in the center of origin. Temperate and tropical/subtropical germplasm clearly clustered into two separate groups. The temperate germplasm could be further divided into six groups consistent with known heterotic patterns. The greatest genetic divergence was observed between temperate and tropical/subtropical lines, followed by the divergence between yellow and white kernel lines, whereas the least divergence was observed between dent and flint lines. Long-term selection for hybrid performance has contributed to significant allele differentiation between heterotic groups at 20% of the SNP loci. There appeared to be substantial levels of genetic variation between different breeding pools as revealed by missing and unique alleles. Two SNPs developed from the same candidate gene were associated with the divergence between two opposite Chinese heterotic groups. Associated allele frequency change at two SNPs and their allele missing in Brazilian germplasm indicated a linkage disequilibrium block of 142 kb. These results confirm the power of SNP markers for diversity analysis and provide a feasible approach to unique allele discovery and use in maize breeding programs.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Maize (Zea mays L.) is one of the most important food crops globally and a model system for the study of genetics, evolution, and domestication. Detailed knowledge of the relationship between maize breeding lines is important not only for parental selection but also for genetic analysis and breeding system design. Diversity analysis of germplasm collections can be carried out using data at morphological, geographical, molecular (DNA, sequence, gene) and functional levels (Buckler et al. 2006). This type of analysis has five important outputs for crop genetics researchers and breeders (Xu 2003; Xu et al. 2003): differentiating cultivars and classifying inbred lines into heterotic groups; identifying gaps and redundancy in germplasm collections; monitoring genetic shifts that occur during germplasm conservation, regeneration, domestication, and breeding; identifying novel and superior alleles for improvement of agronomic traits; and constructing a representative subset or core collection. With the large-scale availability of gene-based single nucleotide polymorphism (SNP) makers in maize, allele mining, and diversity analysis based on functional sequence variation are now highly feasible and powerful approaches.
Maize is a widely grown C4 crop with a high rate of photosynthetic activity leading to high grain and biomass yield potential. It is predominantly a cross-pollinating species, a feature that has contributed to its broad morphological variability and geographical adaptability. Agro-geographically, maize is classified into three types, temperate, tropical, and subtropical. Maize germplasm can also be classified based on endosperm and kernel constitution (waxy, sweet, and pop corn), kernel color and texture (flint, dent, floury), environment in which it is grown, maturity class, and use. The economically most important types of maize are grown for feed, food, fodder, and fuel. In the tropics, grain is still primarily grown for human consumption.
A collection of maize germplasm, which is to be used as a source of genetic diversity, must be well characterized with well-defined distinct groups for efficient management and effective exploitation in breeding programs. Crosses between genetically divergent lines generally produce better-performed hybrids than crosses between closely related parents (Tracy and Chandler 2006). Genetic diversity analysis of maize germplasm can be used to classify accessions into heterotic groups, so that maize lines within each group have a high level of genetic similarity and a high level of dissimilarity compared to other groups. Commercial maize hybrids are typically created from crosses between inbred lines from opposite and complementary heterotic groups in order to maximize hybrid performance. The International Maize and Wheat Improvement Center (CIMMYT) has generated tropical maize germplasm of dent and flint types with distinct sets of genes. Dent and flint core accessions have been tentatively named as heterotic groups A and B, respectively. New accessions are testcrossed with inbred testers selected from A and B groups to determine their specific heterotic patterns. The hybrid maize breeding program at the Brazilian Agricultural Research Corporation (Embrapa), Brazil was established in the late 1970s using kernel type as a major criterion for classifying lines into distinct heterotic groups. Thus, flint and dent inbreds have been managed as two separate heterotic groups. Twenty-eight open pollinated varieties (OPVs), representing part of the tropical maize diversity used in Brazil, have been separated into dent and flint types after being classified into four heterotic groups based on yield-specific combining ability (Parentoni et al. 2001). In temperate germplasm, such as the Chinese maize lines tested in this study, heterotic groups have been established over a long period and proven useful in hybrid breeding. Chinese maize lines are generally classified into four to six major heterotic groups according to the pedigree information and combining ability. Using 111 SSR markers, 84 parental lines of 71 hybrids widely grown across the China Corn Belt were classified into 5 major heterotic groups: Lancaster, Reid, Sipingtou (SPT), Zi330, and Tem-tropic I (Teng et al. 2004). Recently, while analyzing 187 commonly used Chinese maize inbred lines, six subgroups were identified, BSSS (Iowa Stiff Stalk Synthetic population), PA (group A germplasm derived from modern US hybrids), PB (group B germplasm derived from modern US hybrids), Lancaster (Lancaster Surecrop), LRC (derivatives from Lvda red coda Chinese landrace), and SPT (derivatives from Tangshan Sipingtou Chinese landrace) (Xie et al. 2008). However, another recent analysis resulted in their 288 inbred lines being subdivided into four subgroups according to major empirical germplasm origin in China; i.e., Lancaster, Reid, SPT (Sipingtou) and P (introduced from Pioneer hybrids), which assigned LRC to SPT group and combined BSSS and PA into Reid group (Wang et al. 2008). Thus, the creation of heterotic groups in some types of germplasm is still somewhat subjective and individual breeding programs need to select the approach, which best serves their respective breeding strategy and goals.
Information from testcrosses, pedigrees, and morphological traits of inbred lines from within a breeding program is all useful for defining heterotic groups. However, heterotic relationships of germplasm from maize breeding programs across the world cannot be easily classified due to its diverse origins (different breeding practices and breeding goals), and incomplete or insufficient pedigree information (as many breeding programs prefer to keep such information confidential). Thus, in such cases, molecular marker analyses provide an important alternative approach for estimating genetic relationships. Restriction fragment length polymorphism (RFLP) and simple sequence repeat (SSR) or microsatellite markers have been used to examine genetic diversity in maize landraces, modern OPVs, and inbred lines adapted to nontemperate growing areas to find unique sources of allelic diversity (Warburton et al. 2008), characterize the genetic structure and diversity of 260 important tropical and temperate maize inbred lines using SSR markers (Liu et al. 2003), construct “phylogenetic” trees and define potential heterotic groups within medium to late maturing Iranian maize inbred lines (Choukan et al. 2006) and within Chinese germplasm (Teng et al. 2004; Wang et al. 2008; Xie et al. 2008), assign tropical Asian maize inbred lines to potential heterotic groups (Yuan et al. 2001), classify 116 European and North American maize inbreds into the dent and flint groups (Smith et al. 1997), and cluster temperate, tropical, and subtropical lines into major breeding groups (Liu et al. 2003). Furthermore, these molecular markers have been extensively used for analysis of genotypic frequencies for identification of deviations at individual loci (Reif et al. 2004) and for characterization of molecular variation within or between populations (Dubreuil and Charcosset 1998; Warburton et al. 2002, 2008; Reif et al. 2004, 2006; Vigouroux et al. 2008) and for analysis of correlation between genetic distance and hybrid performance, and specific combining ability (Melchinger 1999; Betrán et al. 2003).
Several studies have demonstrated a decline in genetic diversity across the elite temperate maize breeding pool over the past century (Duvick et al. 2004). However, this can be easily reversed by use of landraces and wild-related species that contain untapped sources of biotic and abiotic stress resistance that should provide useful new alleles for future maize improvement (Hoisington et al. 1999). After analyzing more than 100 maize inbred lines and teosinte accessions with 462 SSRs, Vigouroux et al. (2005) concluded that many alleles in the progenitor species of maize (teosinte) are not present in modern maize. Moreover, a significant difference in the overall level of genetic variation in the maize cultivars of industrialized countries as compared to landraces and wild relatives has been found. However, genetic diversity of germplasm developed in developing country breeding programs from both temperate and tropical areas has not been well studied, particularly regarding characterization of alleles and their frequencies among germplasm collections including elite lines from different parts of the world.
A very large number of SNP markers are now available in maize, many of which have been developed from the DNA sequence of known genes. For this reason, SNP markers are now the assay of choice for a variety of tasks in maize improvement including genetic diversity analysis, linkage map construction, marker–trait association or quantitative trait locus (QTL) mapping, and marker-assisted selection (MAS). SNPs have two major advantages over other molecular markers: they are the most abundant form of genetic variation within genomes (Zhu et al. 2003), and a wide array of technologies have now been developed for high throughput genotyping (Fan et al. 2006). In addition, SNP markers can be used for revealing genetic diversity at the sequence level within functionally relevant genes. Characterization of alleles and their frequencies at SNP loci provide an opportunity for discovery of functional diversity, particularly when candidate gene-based markers are used.
There are two SNP genotyping platforms that are of particular importance for maize breeders: the GoldenGate assay for medium-density genotyping that contains 96–1,536 SNPs per array, and the Infinium assay for high-density genotyping that contains up to 500 K SNPs per array (Fan et al. 2006; http://www.illumina.com). A custom oligo pool assay (OPA) containing 1,536 SNPs has been developed for the GoldenGate assay. This OPA has already been used to genotype the nested association mapping (NAM) population consisting of 5,000 RILs derived from 25 families, and 1,106 of these SNPs were successfully mapped to an integrated linkage map (http://www.panzea.org). More recently, a refined OPA has been developed using these 1,106 mapped SNPs plus 430 new SNPs selected from the Panzea database to replace those SNPs from the original OPA that could not be mapped or did not provide good quality data (Yan et al. 2009). A third OPA has been developed through a collaboration between CIMMYT and Molecular and Functional Diversity Team of the USA-NSF Maize Genome Project, which contains SNP markers developed from candidate genes associated with mechanisms and pathways known to influence drought tolerance.
In maize, the frequency of SNPs and distribution of DNA polymorphisms were examined at 18 maize genes in 36 maize inbreds (Ching et al. 2002). It was found that the frequency of nucleotide changes was high, on average one polymorphism per 31 bp in noncoding regions and one polymorphism per 124 bp in coding regions. By comparing SNP diversity between maize and teosinte in 774 genes (Wright et al. 2005), it was concluded that maize accessions had a far lower genetic diversity consistent with products of artificial selection and crop improvement. In linkage mapping, 133 SNPs were mapped onto the intermated B73 × Mo17 high-resolution mapping population (Vroh et al. 2006). Based on an empirical comparison of SSRs and SNPs in assessment of maize diversity and relatedness (Hamblin et al. 2007), it was found that SSRs performed better at clustering germplasm into populations than did a set of 847 SNPs or 554 SNP haplotypes, and SSRs provided more resolution in measuring genetic distance based on allele-sharing. It was suggested that large numbers of SNP would be required to replace highly polymorphic SSRs in studies of diversity and relatedness.
In this study, 770 maize inbred lines representing both temperate and tropical/subtropical maize germplasm were obtained from breeding programs in Brazil and China, as well as CIMMYT breeding programs in Mexico, Zimbabwe and Kenya, and genotyped using a 1,536-SNP chip. The main objectives of this research were to screen core sets of SNP markers showing a high level of polymorphism and providing high-quality data for large-scale characterization of maize breeding lines, to analyze the genetic diversity presented in a broad and diverse set of maize inbred lines to determine the level of genetic diversity that has been tapped by modern plant breeding, to explore the genetic distinction between germplasm from breeding programs in diverse global locations as revealed by allelic frequency differences, and to investigate the relationships between these germplasm subsets and its association with pedigrees and breeding history.
Materials and methods
Plant materials
A total of 770 maize inbred lines representing temperate, tropical, and subtropical maize germplasm from breeding programs and germplasm collections in diverse global locations were chosen for characterization. Lines were chosen due to their biotic stress resistance or abiotic stress tolerance including 394 lines from CIMMYT maize breeding programs in Mexico, Zimbabwe, and Kenya, 282 lines from China Agricultural University, Sichuan Agricultural University and the Chinese Academy of Agricultural Sciences, and 94 from Embrapa, Brazil (Table 1). All the CIMMYT maize lines tested were tropical/subtropical types, the majority of which (309) were white kernel types, and many were developed from the same populations. In contrast, the majority of Chinese maize lines tested was temperate types with just 14 tropical lines that were derived directly and solely from tropical germplasm. The Chinese germplasm tested also included parental genotypes of widely grown commercial hybrids, as well as 34 high oil content line selections from populations developed by Song and Chen (2004) at China Agricultural University, 28 Chinese landraces, and four high pro-vitamin A lines. All the Brazilian lines tested were yellow kernel types, comprising 89 tropical types, 4 subtropical types, and 1 temperate haploid inducer line (W23). The names of all tested lines together with kernel color and texture characters, environment adaptation, and heterotic group assignment (where available) are listed in Table S1 and summarized in Table 1. For data analysis, the 770 lines used in this study were arranged in contrasting groups as follows: temperate lines (269) versus tropical/subtropical lines (501), yellow (377) versus white kernel (341) (Subset 1 in Table S1), and dent (172) versus flint texture (255) (Subset 2 in Table S1). For the convenience of description, however, we call all tropical and subtropical maize collectively as tropical/subtropical throughout the rest of this paper as in many cases they cannot be well distinguished.
SNP genotyping
All maize lines tested in this study, except those from Embrapa and China Agricultural University, were planted during the 2007 wet season at CIMMYT’s Maize Experimental Station in Tlaltizapan (State of Morelos, Mexico). After removal of off-types and mixtures, leaf samples were harvested from 15 plants from each line and bulked for DNA extraction. For all the samples, DNA extraction was based on a CTAB procedure (CIMMYT Applied Molecular Genetics Laboratory 2003).
Designability rank score was given to each SNP by Illumina, with the score ranging from 0 to 1.0, where a rank score of <0.4 has a low success rate, 0.4 to <0.6 has a moderate success rate, and >0.6 has a high success rate for the conversion of a SNP into a successful GoldenGate assay. A total of 384 SNPs with a designability rank score of 0.4 or higher were selected to be included in the OPA, except when multiple SNPs were located on the same STS, in which case only the SNPs with the highest designability rank score was selected (the others were not used).
The OPA for SNP genotyping used in this study was developed under the framework of the Molecular and Functional Diversity Team of the USA-NSF Maize Genome Project (Yan et al. 2009). A total of 1,106 SNP markers developed previously by the project have been integrated into an existing linkage map (http://www.panzea.org). In this study, these 1,106 mapped-SNPs were combined with a further 430 SNPs selected from the panzea database to develop a new OPA chip containing 1,536 SNPs on the basis of having a designability score higher than 0.60, which was given to each SNP by Illumina with the score ranging from 0 to 1.0. A score of >0.6 has a high success rate for the conversion of a SNP into a successful GoldenGate assay. The original sequences used to develop these SNP markers were used for a BlastN (Altschul et al. 1990) search against the B73 genome sequence (reported at http://www.maizesequence.org, December 2008) enabling in silico map positions to be generated for 1,326 of the SNP markers. The number of SNP markers per chromosome ranged from 81 (chromosome 10) to 231 (chromosome 1) with just 20 SNP markers remaining unmapped (see http://www.panzea.org for details).
SNP genotyping was carried out using an Illumina BeadStation 500 G (Illumina, Inc., San Diego, CA, USA) at the Cornell University Life Sciences Core Laboratories Center and the protocols described by Fan et al. (2006). A total of 5 μL of 50 ng/μL genomic DNA was used to make single-use DNA as required by the genotyping system which was arranged in Sentrix Array Matrices (SAMs) each with 96 samples comprising one negative control (water) sample and one repeatability control (duplicate) sample in addition to 94 genotype samples. The GoldenGate assay using a SNP chip containing the 1,536 markers described above was then hybridized to the SAMs for genotype analysis of the 770 maize lines. Allele calling for each locus was carried out using Illumina BeadStudio genotyping software with a no-call threshold of 0.25, which is the lower bound for a reliable genotype call. Data not automatically dealt with by the software were manually scored.
Data analysis
The polymorphism information content (PIC) value, described by Botstein et al. (1980), was used to refer to the relative value of each marker with respect to the amount of polymorphism exhibited. PIC value was estimated by
where P ij and P ik are the frequencies of the jth and kth alleles for marker i, respectively, and the summation extends over n alleles. Heterozygosity and gene diversity were calculated to quantify the genetic variation in the maize lines sampled. The heterozygosity value reflects the proportion of heterozygous loci detected in a single maize line while the gene diversity is defined as the probability that two alleles randomly chosen from the test sample are different. The common biased estimator of the gene diversity for marker i can be obtained using the above equation by dropping the last item.
Allele frequency was calculated for each locus across each of the three geographically distinct sets of maize germplasm within each of the defined subsets: tropical/subtropical versus temperate, white versus yellow kernel, and dent versus flint texture. Statistical significance of differences in allele frequency was based on the P value from Fisher’s exact test (Fisher 1922). The genetic distance between genotypes was computed using the Rogers (1972) genetic distances (RD) and cluster analysis was then carried out using the neighbor-joining tree (NJ) method (Saitou and Nei 1987). Groups and subgroups were identified from the resultant phylogenetic tree. All of the above calculations were performed with PowerMarker software (Liu and Muse 2005).
The population structure of the 770 genotypes was investigated using the STRUCTURE software (Pritchard et al. 2000). The dataset was tested for a number of subpopulations ranging from k = 1–12. Three runs for each k value were performed using the admixture model and correlated allele frequencies (Falush et al. 2003). The burn-in length and iterations were all set to 500,000. In the model-based method, membership coefficients (Q values) for each inbred line were estimated to have its memberships in multiple subgroups. Inbred lines with membership probabilities ≥0.60 were assigned to the corresponding subgroup and lines with membership probabilities <0.60 were assigned to a mixed subgroup. In addition, principal component analysis (PCA) was conducted using TASSEL 2.1 software (Yu et al. 2006; Bradbury et al. 2007) to visualize the genetic relationships between the maize lines tested and to compare different subsets of germplasm with contrasting origins, kernel colors, kernel textures, and environmental adaptation. SNP markers showing significant different segregation patterns in different subsets of germplasm and between different established heterotic groups were identified. Shared allele frequency (the proportion of the loci with identical alleles between two inbred lines), was calculated in order to carry out pairwise comparisons of maize lines that were known to be closely related to each other based on pedigree information.
Allele frequencies within different subsets of germplasm and within groups identified by STRUCTURE, PCA and clustering analyses were used to identify SNP markers detecting unique, missing, and under-representative alleles solely in specific subsets of germplasm. SNP markers detecting significantly different allelic frequencies can be used to distinguish closely related inbred lines and heterotic groups. Unique alleles are defined as those that can be only found in one germplasm subset but not in the others. Missing alleles are defined as those completely lacking in a specific subset of germplasm while existing in all others at a regular frequency. Under-representative alleles are defined as those with very low frequencies in one collection (with only one or two lines containing the allele) but existing normally in others.
Results
Characterization of SNP and germplasm diversity
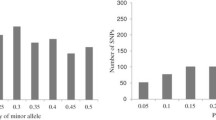
All SNP data (770 lines by 1,536 SNPs) were scored using Illumina BeadStudio genotyping software. An unambiguous output from this software is shown in Fig. S1 where there are three clearly separated clusters representing AA, BB, and AB genotypes. The designability score ranged from 0.1 to 1.0 with the majority (62%) of SNP markers with designability scores of more than 0.6. This very high level of designability score may be due to all the tested material being inbred lines. SNPs with designability scores lower than 0.3 are considered of poor quality so that they cannot be called correctly by automatic scoring or manually re-clustering. In addition, SNP markers that were monomorphic or had missing data points more than 20% in the diversity panel of inbred lines were removed from further analyses. As a result, a total of 1,034 SNPs (67%) were called successfully with high quality. Base changes involved A/C (181), A/G (661), A/T (73), and C/G (119), and A/G transition accounted for 63.9% of the informative SNPs. A list of these informative SNP loci including chromosome, base change, minor allele frequency (MAF), heterozygosity, gene diversity, and PIC is provided in Table S2. Of the 1,034 markers, only 8.8% (91/1,034) showed MAF less than 0.05. Approximately 54.3% (561/1,034) of SNPs had an MAF more than 0.2, which were selected as markers with normal allele frequencies. In addition, 193 (18.7%) showed almost equal allele frequencies (with MAF close to 0.5) for two alternative alleles (Fig. 1).
In the 770 inbred lines, 2,068 alleles were detected at the 1,034 marker loci, each with two alleles as expected. The average PIC was 0.259, ranging from 0.003 to 0.375 (Table S2) with a peak distribution between 0.350 and 0.375 (Fig. 1). Estimated gene diversity varied from 0.003 to 0.500 with an average of 0.321. On average, the inbred lines exhibited heterozygosity at 4.3% of the genetic loci.
Further data analyses have been taken to test if the levels of genetic diversity identified among different germplasm collections matter with the SNP chip that was developed for maximizing the genetic polymorphism between the temperate parent B73 and other parents in the panel for development of the NAM population. Three subsets of SNPs were selected from the chip for genetic diversity analysis. The first subset was selected for high quality with 449 SNPs selected based on three factors: high designability scores (>0.6), normal MAF (>0.2), and high PIC values (>0.25) (Table S2, Subset A). Two subsets (Table S2, Subset A and B) were obtained by excluding the markers with allele frequency difference between temperate and tropical/subtropical germplasm larger than 10 and 5%, respectively. In addition, to understand the influence of possible biased sampling method in collecting germplasm on genetic diversity, a subset of the Chinese (mainly temperate) germplasm collection was formed by excluding 28 landraces and 34 high-oil lines that might have made an additional contribution to the genetic diversity in elite Chinese maize germplasm. All these subsets selected for SNPs and germplasm were compared with the entire counterparts for PIC and gene diversity (Table 2).
Using all 1,034 informative SNPs and 770 inbred lines, Chinese germplasm were found to show the highest average PIC value and gene diversity, followed by CIMMYT germplasm, whereas the Brazilian germplasm showed the lowest (Table 2). Analyses with the three subsets of SNP markers provide consistent results that the differences in detected genetic diversity between germplasm collections were significantly reduced and as a result, the estimates for PIC and gene diversity from Chinese germplasm are only slightly higher than others. On the other hand, using 449 high-quality markers significantly increased the estimates of PIC and gene diversity for all germplasm collections, which are about 30% higher than those obtained from other marker subsets. Excluding landraces and high-oil inbreds from Chinese germplasm slightly decreased the estimates for PIC and gene diversity, but did not change the order of genetic diversity in different germplasm collections, indicating that the Chinese germplam collection did not contain highly diverse landraces or populations that should have significantly contributed to the level of genetic diversity detected in this study. In all cases, the Brazilian maize lines tested in this study appear to have relatively narrow genetic base as revealed by their estimates for both PIC and gene diversity, which probably is related to the small number of inbreds sampled that do not represent the whole inbred line collection from Embrapa, but were mainly selected for biotic and abiotic stresses. As the 449 high-quality markers provided the highest estimates for PIC and gene diversity and also the results comparative to those obtained from other selected marker subsets, these markers should be recommended for future use or optimization of SNP chips by combining with high-quality markers identified from other chips.
Population structure
An admixture model-based clustering method in the STRUCTURE was implemented to infer population structure for all 770 tested lines and it was run for the number of fixed subgroups k from 1 to 12. The results indicate that the model overestimated the number of groups, and thus no suitable k could be chosen from the Ln probability and used directly for group determination. However, when k = 2, there was a big increase in the probability compared to k = 1 so that two major groups can be claimed (Fig. 2). When the results were compared with the known pedigrees of the inbred lines for each run of different ks, the model-based groups were largely consistent with known pedigrees when k = 8, so that the 770 tested lines can be mainly divided into eight groups (Fig. 2). Five of these groups (Group 1, 2, 3, 5, and 6) include all Chinese maize inbred lines and correspond to the five major germplasm origins in China, i.e., PA, BSSS, PB, Lancaster and SPT. The other three groups (4, 7, and 8) including maize lines predominantly from CIMMYT and Brazil majorly representing tropical/subtropical germplasm, which are named as Tropics 1, 2, and 3, respectively (Fig. 2).
Population structure in the 770 maize lines shown as membership coefficients (Q values). Each horizontal bar represents one maize line, which is partitioned into up to k colored segments. Color codes are as follows: BSSS, red; Lancaster, green; SPT, blue; Tropic 1, yellow; PB, purple red; PA, light blue; Tropic 2, orange; Tropic 3, brown
Group 1 belongs to BSSS heterotic group, consisting of 19 Chinese inbred lines that have B73 in their pedigrees. Group 2, Lancaster, comprised 30 high-oil inbred lines, 52 Chinese normal inbred lines and 1 CIMMYT inbred line. The majority of these Chinese normal inbred lines are closely related to Mo17 and Zi300 pedigrees. Group 3, SPT, included 34 Chinese inbred lines, which were mainly derived from Chinese local germplasm such as Huangzao 4. Group 5, PB or Tem-tropic I, comprised 34 Chinese inbred lines, most of which were related to US hybrid P78599. Group 6 was PA heterotic group including 38 Chinese inbred lines derived from Ye478 and Shen5003. Group 4 (named Tropics 1) contained 26 large grain borer (LGB) resistant inbred lines bred by CIMMYT, Kenya. Group 7, also called as Tropics 2, consisted of 21 tropical inbred lines bred by CIMMYT, Zimbabwe. Group 8, Tropics 3, was the largest group, including 434 inbred lines, of which 330 were collected from CIMMYT, 92 from Brazil, and 12 from China. Out of the 12 Chinese inbred lines, four were derived from CIMMYT germplasm, such as S37 and SW1611, which are related to Suwan2 pedigree, and four other lines adapted to tropical environment. In addition to the inbred lines that were clearly assigned with probability >0.60 to a single group (population), 81 inbred lines (10.5% of the total) could not be clearly assigned to any of these groups. These lines, which is called mixture group, include 63 from China, 16 from CIMMYT and two from Brazil. Most of them are either derived from crosses between tropical and temperate germplasm or derived from landraces. Out of two lines from Brazilian collection, the w23 was derived from a public temperate haploid inducer line W23 (Wisconsin 23) and LPF9-01-2-1-1-1 is a subtropical line and adapted to the south of Brazil. Inbred lines with proportional memberships in the model-based groups are provided in Table S3.
Clustering analysis
In order to gain further insight into the genetic diversity among different groups of the global maize collection, a neighbor-joining tree based on Roger’s genetic distance was constructed. The 770 tested lines were grouped into two major groups (Fig. S2 for details; Fig. 3 for the framework), which is consistent with structure analysis at k = 2. In one major group, 270 Chinese lines, representing temperate germplasm, were grouped along with 15 CIMMYT and 3 Brazilian lines. Four of the CIMMYT lines in the group have some lines from old gene pools such as EV7992, G16, and G27 in their pedigrees. One Brazilian line, additional to the two lines to the mixture group, was adapted to the subtropical region. The 270 Chinese lines in this major group can be subdivided to five heterotic groups such as Lancaster, SPT, PA, PB, and BSSS, which was consistent with the result of structure analysis, plus an additional group, called LRC, which included inbred lines with Dan340 and Zi330 in their pedigrees and were assigned to Lancaster group by structure analysis. The clustering result is consistent with heterotic groups established based on the pedigree information and combining ability. Some inbred lines that were assigned to the mixed group by structure analysis were also allotted to one of these six groups (Fig. 3). The second major group, which represents tropical/subtropical germplasm, comprised 482 inbred lines largely from CIMMYT and Brazil, and 7 of the 12 from China in this group with tropical backgrounds. Unlike the first major group, these 482 lines cannot be further distinguished well into subgroups that can be explained based on their environmental adaptation, kernel color, kernel texture, or heterotic response. However, out of the 89 Brazilian yellow tropical inbred lines, 55 were grouped separately from other tropical materials, and could be clustered into four groups well assigned based on pedigree, kernel type and yield combining ability (Fig. 3). Additionally, the lines closely related in pedigree usually did cluster together.
Neighbor-joining (NJ) tree for the 770 maize inbred lines based on Roger’s genetic distance. A full NJ tree, sectional tree representing Chinese germplasm, Brazilian germplasm, and PA and PB heterotic group are shown from left to right. A version of this tree with the names of the individual inbred lines can be found in Fig. S2
Principal component analysis
Principal component analysis results showed good agreement with both structure and cluster analyses. PCA on the entire set of 770 inbred lines (Fig. 4a) exhibited a clear separation of temperate and tropical/subtropical germplasms. Temperate maize lines from China dispersed on the plot, compared to the tropical/subtropical lines from CIMMYT and Brazil that clustered around the original point of the plot with roughly equal contribution by the first two principal components. However, PCA did not separate germplasm well by kernel color or texture (data not shown). PCA graph on CIMMYT inbred lines (Fig. 4b) indicated that the first PC well separated these lines into two groups. Group 1 included 26 LGB resistant inbred lines from Kenya, which was fully consistent with the Group 4 identified in structure analysis. These lines can be traced back to a few genebank accessions from Cuban and Guadalupe races (Cuban flint and coastal tropical flint). The other group contains the rest of inbred lines from CIMMYT. Brazilian inbred lines also were well divided into two groups by the first PC, where Group 1 was composed by flint lines essentially derived from L3, and Group 2 was mainly represented by dent lines derived from L228-3, two important inbred lines for Embrapa’s breeding program (Fig. 4c). Chinese inbred lines were differentiated into four distinct groups (Fig. 4d), including the three groups (PA, PB and BSSS) that were consistent with cluster and structure analysis, plus the fourth one that mixes two groups, Lancaster and SPT, which were distinguishable by structure and clustering analyses.
Genetic distances and allele differentiation among germplasm groups
Genetic distances
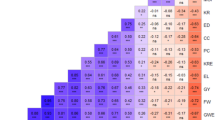
Based on the comprehensive population structure analyses above, genetic distances were calculated among seven groups (Table 3). Two of the groups represented CIMMYT and Brazilian collections; the other five are Chinese groups that were identified in structure analysis. The largest genetic distance was observed between temperate and tropical/subtropical lines (Chinese vs. CIMMYT/Brazilian lines), and the smallest between CIMMYT and Brazil groups, both of which belong to tropical/subtropical lines and the latter are largely developed using the former germplasm. Interestingly, a high level of differentiation was observed among the Chinese groups.
Allele frequencies
To reveal genetic difference among different sets of maize germplasm, comparative analysis of allele frequencies was performed for three pairwise comparisons: temperate versus tropical/subtropical lines, yellow versus white kernel (Subset 1 in Table S1), and dent versus flint texture (Subset 2 in Table S1). Of the 1,034 SNPs, a significant difference in allele frequency (P < 0.01) was observed for 779 (75%) SNPs in the temperate versus tropical/subtropical lines, 643 (62%) SNPs in yellow versus white kernel, and 352 (34%) SNPs in dent versus flint texture. Figure 5a shows a distribution of allele frequency difference observed in the three pairwise comparisons, ranging from 2.5 to 60%. In temperate versus tropical/subtropical comparison, 535 (51.7%) SNPs fell into the class with allele frequency difference larger than 10% (P < 0.01). The most remarkable difference was 60% at SNP PZA00416.7 for allele C (30% for temperate vs. 90% for tropical/subtropical lines), with 19 SNP markers with allele frequency difference larger than 40%. In yellow versus white kernel comparison, 369 (35.7%) SNPs had significant difference (>10%), with the largest difference of 36% at PHM1899.157 for allele G (35% for yellow vs. 71% for white lines) and 10 SNPs with allele frequency difference larger than 30%. In dent versus flint texture comparison, 223 (21.6%) SNPs belonged to the class with a difference larger than 10%, with the largest difference of 23% at SNP PZA01316.1 for allele A (60% for dent vs. 83% for flint texture) and 14 SNPs with allele frequency difference larger than 20%. These results indicate distinct genetic differences among the three pairwise germplasm comparisons and the strongest differentiation was observed between temperate and tropical/subtropical lines, the second between yellow and white kernel, and the least between dent and flint texture (see Fig. 5b for X–Y plots for allele frequencies in the pairwise comparisons). For the SNPs with extremely different allelic frequencies between two germplasm collections or groups, one of their alleles must be less frequent in one collection while highly frequent in the other.
Differentiation of allele frequencies between maize germplasm. a Difference in allele frequency between maize germplasm collections: temperate versus tropical/subtropical lines, white versus yellow kernels, and dent versus flint textures. b X–Y plots for allele frequencies in pairwise comparisons of germplasm
In order to distinguish different germplasm collections, ten SNP markers with most significant allelic difference were selected from each of six pairwise germplasm comparisons. Three of the pairwise comparisons are for germplasm from different collections (Table 4). The largest average allelic frequency difference for the top ten differences was between China and Brazil (0.55), followed by China versus CIMMYT (0.50). The smallest allelic frequency difference was between CIMMYT and Brazil (0.44). The three other pairwise comparisons are for germplasm of different types (Table 5). As expected from genetic diversity analysis, the largest allelic frequency difference was between temperate versus tropical/subtropical lines with an average of 0.51 for the top ten differences, followed by white versus yellow kernels (0.33). The smallest allelic frequency difference was between dent and flint textures (0.21).
To understand whether SNP alleles have been differentiated between contrasting heterotic groups as the result of selection for hybrid performance, two distinct groups determined by structure and PCA analyses, PA, and Lancaster, were taken as an example. These two groups, which are consistent with their heterotic groups, had a large genetic distance (0.2889, Table 3) and were well separated in PCA analyses (Fig. 4d). By comparing allele frequencies between these two groups and with the entire set of Chinese germplasm, 202 (19.5%) SNP markers were found to show allele frequency differences larger than 50%. Thirty-eight of these SNP markers showed allele frequency differences larger than 70% (Table 6). These differences are even much larger than those between temperate and tropical/subtropical lines where the largest allele frequency difference detected was 60%. For each of these SNP markers, one heterotic group had a much higher allelic frequency while the other had a much lower allelic frequency than the entire Chinese collection, suggesting a significant allele differentiation between two opposite heterotic groups. Change of allele frequencies in opposite directions at so many loci between heterotic groups indicates that long-term selection for hybrid performance may have resulted in allele differentiation that relates to the improved hybrid performance.
There are several pairwise markers that were tightly linked and showed the same tendency in the change of allele frequencies between the two heterotic groups. For example, two markers on chromosome 3 (67 bp apart), PHM3352.19 and PHM3352.21, where were developed from a unigene named PCO123854 (Zea mays PCO123854 mRNA sequence), both showed high allele A frequencies in PA group (0.82 and 0.79) but low frequencies in Lancaster group (0.06 and 0.05). Two markers on chromosome 4, PZA03254.1 and PZA03587.1, tightly linked (142 kb apart), showed similar allele frequency differences (0.75 and 0.76) between the two heterotic groups.
To compare pairwise lines that are close to each other in pedigree, the top ten pairs of inbred lines, each clustered closely in the dendrogram, were selected for further analysis. The share allele frequency (SAF), the proportion of alleles shared by two compared lines across all genotyped marker loci, were calculated (Table 7). For each comparison, a higher degree of genetic similarity (SAF) indicates a smaller genetic difference. SAFs of 80% or higher were detected across all comparisons with an average of 87.6%. The most similar pairwise maize lines, Chang7-2 versus CN_BI_10_03 [(Chang7-2 × Huangzao4) × Chang7-2BC4F2-24-33 L × 24-33], shared alleles at 98.5% of marker loci. These two maize lines had close relatedness as Chang7-2 was the recurrent parent for the backcrossed introgression line, CN_BI_10_03.
Missing and unique alleles in different germplasm collections
There is only one missing allele identified in tropical/subtropical germplasm (allele A at PHM1960.37, chromosome 3) but highly frequent (16.4%) in the temperate germplasm. The same allele was also missing in CIMMYT germplasm while it was frequently present in Chinese germplasm. On the contrary, no allele was missing in the Chinese collection but frequently existing in the CIMMYT or Brazilian collection. Compared with CIMMYT germplasm, Brazilian germplasm lacked the allele C at PZA00235.9 (chromosome 1), which was present in 12.7% of the CIMMYT germplasm. Table 8 lists the markers and their chromosomes for the missing alleles identified for Brazilian germplasm. There are 23 and 8 of such alleles that were highly frequent in Chinese (10%) and CIMMYT (5%) germplasm but completely lacking in Brazilian germplasm. Interestingly, the two closely linked markers on chromosome 4 (142 kb apart), PZA03254.1 and PZA03587.1, both of which detected significant allelic frequency difference between heterotic groups PA and Lancaster, each had one allele lacking in the Brazilian germplasm but presenting in 20.9% of Chinese germplasm. This indicates that there is a linkage disequilibrium (LD) block, which brackets these two markers 142 kb apart.
By comparing allele frequencies of a specific germplasm collection with those in the entire germplasm set (770 lines), 11 unique alleles were identified that only existed in that specific germplasm collection but not in others. Two and nine of such unique alleles were found to only exist in CIMMYT and Chinese germplasm, respectively (Table 9). Most of the unique alleles identified in Chinese germplasm came from landraces, high-oil maize lines, introduced maize lines including B73 and Mo17, and parental lines for three recombinant inbred line (RIL) populations. Thirteen landraces hosted five of the nine unique alleles. B73 and its two RIL populations hosted four alleles. Ye478 and its backcross introgression lines hosted the unique allele A at PHM1960.37. The markers that had significant allelic frequency difference among germplasm collections and missing or unique alleles identified above can be combined and used for characterization of maize germplasm.
Discussion
Utilization of the random-gene SNP chip developed for the nested association mapping
In the present study, we have shown that a 1,536-random SNP GoldenGate assay can be used successfully for maize genotyping. A total of 1,034 SNPs involving 4 base changes were obtained with high polymorphism and good quality as revealed by 770 diverse maize lines. These markers were well distributed on the ten maize chromosomes, and average distance between two SNPs reached a level of 1 cM in an integrated NAM map (McMullen et al. 2009). These SNPs were originally developed for detection of as much as possible the genetic polymorphisms in 27 diverse inbred maize lines that were used as parental genotypes to develop the NAM population. The common parent, B73, is a temperate maize inbred line while the 26 other parents were selected to represent genetic diversity in both temperate and tropical/subtropical maize germplasm (Yu et al. 2008). Markers developed to be polymorphic in one set of germplasm are likely to provide a biased estimate of diversity in another set of germplasm, which is called ‘ascertainment bias.’ The number of SNP alleles unique to the ascertained groups would be the indication of the degree of bias. Our results indicate that the bias, if exists, could be tolerable, as unique alleles could be identified from CIMMYT germplasm and most unique alleles in Chinese germplasm were from landraces, high-oil maize lines and introduced lines.
To test if the chip developed for the NAM population has any preference in revealing genetic diversity, we developed the subsets of SNPs after removal of those showing high allelic frequency difference between temperate and tropical/subtropical germplasm collections. The level of genetic diversity in temperate germplasm has been significantly reduced compared to only a small reduction for tropical/subtropical germplasm, indicating that the preference of this SNP chip to temperate germplasm can be corrected to some extent by removing the preference markers. With the possibility of bias, however, only way to fully remove the bias is to use unbiased methodology, such as de novo sequencing of loci from all germplasm groups.
Selection for better polymorphism between temperate and temperate or tropical/subtropical germplasm provides some advantages for this random gene-based chip in genetics and breeding applications. Because of the preference, this SNP chip should be more powerful for genetic diversity analysis of temperate germplasm, and it will distinguish temperate germplasm from each other better than tropical/subtropical germplasm. Furthermore, the chip can be used for, with limited or no negative influence at all on, the rest of analyses that have been done in this study including grouping via STRUCTURE, clustering, and PCA analyses. For allelic frequency analyses, however, this chip actually provides more opportunities of finding missing and unique alleles from comparison of germplasm collections.
Information on the allelic frequencies of the SNP markers contained in this SNP chip in a large number of maize germplasm should help determining the usefulness of this OPA for analysis of a broader range of maize germplasm. The 770 inbred lines used in this study is the largest set of inbred lines that have been tested so far to cover diverse temperate and tropical/subtropical maize germplasm including different kernel color and textures. The results provide a robust test of the suitability of this SNP chip for genetics and breeding applications. Based on three criteria, designability scores above 0.6, MAF exceeding 0.2, and PIC values larger than 0.25, 449 high-quality SNP markers were selected (Table S2 Subset A). This subset of markers has been proven the best in terms of the estimation of PIC and gene diversity and the detection of genetic diversity with least preference to temperate germplasm, compared to other subsets selected based on the allelic frequency differences alone. We recommend this subset of SNPs for use in genetics and breeding as well as for the development of new SNP chips with optimized allele frequency and simplification of the cumbersome manual scoring process by automated scoring.
SNP versus SSR markers for genetic diversity and genetic analyses
Single nucleotide polymorphism markers occur at much higher density in the genome (Zhu et al. 2003). In maize, there is a SNP every 60–104 bp (Ching et al. 2002; Tenaillon et al. 2001). SNPs are amenable to high-throughput methods such as genotyping arrays, and have lower genotyping error rates (Fan et al. 2006; Gupta et al. 2008). On the other hand, SNPs are distributed in the whole genome (either intergenic or intragenic). Compared to SSR, SNP markers could be more functional-related.
Simple sequence repeats being multi-allelic markers can provide many times the numbers of alleles per locus because of high mutational rate and a virtually unlimited number of new alleles that can be created by the slippage process. However, one important factor that has been neglected is that it is very difficult to distinguish real SSR alleles from those created by error scoring, artifact bands, and false alleles because of the difficulty in comparing cross-lab or experiment scores, which could have contributed greatly to the number of ‘alleles’ detected at a SSR locus, particularly when the number of alleles is large and the repeat length difference between two alleles is small. On the other hand, SSRs are prone to homoplasy/heteroplasy (Bruford and Wyne 1993), i.e., same underlying DNA sequence alleles can be associated with different SSR lengths or vice versa, which is fine for estimating ancestral relationships, but usually overestimates sequence diversity due to faster evolution of SSRs relative to SNPs.
According to Laval et al. (2002) (k − 1) times more biallelic markers are needed to achieve the same genetic distance accuracy as a set of SSRs with k alleles. If this assumption is right, the 1,034 SNP markers used in this study could be translated into 54–115 SSRs each with 10–20 alleles. It has been showed that in genetic diversity analysis, ten or more SNPs had information content equal to just one SSR, in terms of the number of alleles detected (Yan et al. 2009). A comparative analysis indicated that 89 SSRs performed better at clustering 259 inbred lines into populations than did a set of 847 SNPs or 554 SNP haplotypes, and SSRs provided high resolution in measuring genetic distance based on allele-sharing (Hamblin et al. 2007). Two recent reports showed that measures of distance based on SSR and SNP markers were well-correlated only for the small sets of individuals that were closely related (Hamblin et al. 2007; Jones et al. 2007). It has been anticipated that if enough SNPs are used the difference would be overcome (Hamblin et al. 2007). The results from this study have confirmed the heterotic groups that have been revealed by SSR markers and pedigrees, indicating that the SNP markers provide a powerful discrimination on diverse germplasm. SNPs also can be used to distinguish closely related germplasm as indicated by the ten pairs of inbred lines (Table 7) that shared alleles at most tested loci but still showed different alleles at others.
Use of molecular markers in genetic research has different requirements and both rare and unique alleles are important. In linkage-based genetic mapping, markers with both frequent and rare alleles can be used because the parental lines have been selected for polymorphism at each locus. For LD-based mapping, however, markers with rare alleles (e.g. allele frequency <0.1) or two alleles with significant allelic frequencies (0.90 vs. 0.10) will not provide enough power for association detection as the probability for individuals with one of the alleles will be very low. For the SNPs contained in the current chip, they were designed based on the probability of detecting high polymorphism among 27 lines. As a result, very few rare alleles had been considered and thus two SNP alleles can be detected with high frequencies. Therefore, this chip should be more suitable for LD-based association mapping.
We would like to argue that comparison of SSR and SNP markers should not be based on allele numbers alone. The number of SNP alleles (less alleles per locus but more loci), compared to the same number of SSR alleles (more alleles per locus but less loci), apparently provides a better genome coverage as much more SNP loci are involved. Not all comparisons available for these two types of markers have taken into consideration of the genome coverage. In genetic diversity criteria, more weight should have been given to the number of loci than the number of alleles. If so, 100 SSRs should have less power than 1,000 SNPs in the detection of genome-wide genetic diversity if each SSR has ten times of alleles in SNPs. An extreme example would be one SSR marker with 40 alleles versus 20 SNP markers each with two alleles. The former only detects allelic variation at one locus while the latter can have two loci on each maize chromosome detecting 20 loci and their interlocus variation. On the other hand, when more SNP markers are used (than the number used in this study), the reliability of SNP analysis would increase. For example, Yu et al. (2009) revealed that kinship estimation was more sensitive to the number of markers used than population structure estimation in terms of model fitting. In our study, 1,034 informative SNP markers provide heterotic groups completely consistent with those determined based on pedigree and combining ability and the results from SSR-based grouping, indicating the efficiency and power of SNP markers in genetic diversity analysis.
Impacts of selection and breeding history on genetic diversity and germplasm grouping
Genetic diversity in maize as in other crops has been reduced during domestication (Doebley et al. 1984; Hilton and Gaut 1998; Vigouroux et al. 2005). Maize contains 75% of the variation found in its progenitor (Z. mays ssp. parviglumis) (Eyre-Walker et al. 1998). For SSR, maize has 88% of the gene diversity and 76% of the number of alleles found in teosinte (Vigouroux et al. 2005). A genetic bottleneck during the domestication of maize as well as ongoing selection by breeders is expected to result in diversity reduction across the whole genome as well as selected loci (Rafalski and Ananiev 2009). At the maize Y1 gene, selection for the yellow endosperm phenotype preferred in some markers resulted in >10-fold reduction of diversity (Palaisa et al. 2003). The effect of this selective sweep extends to several hundred kilobytes (Palaisa et al. 2004). Reduction of diversity and other effects indicative of selection has also been observed in the starch pathway (Whitt et al. 2002) and in the anthocyanin regulatory locus c1 (Hanson et al. 1996). By survey of over 1,000 genes, eight of them were identified showing evidence of selection with strongly reduced diversity (Yamasaki et al. 2005). However, a recent study showed that variation might not be declining in global maize germplasm (Warburton et al. 2008). Globally, maize germplasm are being mixed up more and as a result, population differentiation is declining but overall diversity is being maintained because of the mixing. With high throughput sequencing or chip technology, scanning of all maize genes will allow identification of a more complete set of genes under natural and artificial selection, with which selective diversity enhancement by introgressions from other germplasm collections, landraces or even teosinte, would become possible.
Pedigree and molecular marker information showed that changes in genetic diversity had occurred during each decade for a set of maize hybrids that were widely grown in the central US Corn Belt. For Pioneer brand hybrids widely grown from 1930 to 1999 contributions from some founders has risen, then fallen; some were low and disappeared, while others reappeared; and contributions from others persisted at low levels (Smith et al. 2004). By the 1980s, contributions from 26% of both founders and landraces had become extinct. When older (1930s and 1940s) hybrids were compared with recent (1990s and 2000s) hybrids, SSR alleles showed large qualitative and quantitative differences (Feng et al. 2006). For example, 23% of SSR alleles were found only in the older hybrids, while 30% of SSR alleles were found only in the recent hybrids. Duvick et al. (2004) showed that the allele numbers reached the highest during 1960s and has since declined in recent era hybrids. On the other hand, efforts directed at increasing heterotic yield have increased genetic distance between two heterotic groups (Cooper et al. 2004; Tracy and Chandler 2006). Allele frequencies at many loci have changed in opposite directions in different heterotic groups such as Stiff Stalk Synthetic (SSS) and Non-Stiff Stalk (NSS). SSR data also supplied evidence that divergent artificial selection on flowering time may have played a role in generating population structure (Remington et al. 2001). Our results indicate that two major Chinese heterotic groups, PA and Lancaster, showed allele frequency difference of over 70% at 38 SNP loci. This effect deepens the non-random population structure of maize germplasm collection (Rafalski and Ananiev 2009). In China, the Reid germplasm were introduced from the US during 1950–1970s and Lancaster germplasm, such as Mo17, were introduced from the US in 1970s. Those US Corn Belt germplasm have played an important role in Chinese hybrid maize breeding with many inbred lines developed. In addition, Pioneer hybrids were introduced into China in the late 1980s, with a new group of inbreds bred that was defined as “P” or Tem-tropic I. Introduction of these germplasm and development of new lines from them have significantly enhanced Chinese maize germplasm bases and also increased the differentiation from opposite heterotic groups.
The majority of Brazilian tropical inbred lines tested in this study were clustered into four heterotic groups, including two dent and two flint lines, as predicted based on yield combining ability (Parentoni et al. 2001). The major flint group, which is coincident with the heterotic group 4, comprises 18 lines, 17 of which are related to the flint heterotic pattern and were derived from L3. The second flint cluster, which is comparable to the Brazilian heterotic group 2, includes eight flint lines, one dent line and one line with unknown origin, most of which were derived from Swan-1 germplasm. The third group was represented by the dent heterotic group related to the L228-3. For the 21 inbred lines in this group, only one was in disagreement with its origin. This group was mainly derived from Tuxpeño germplasm, which was described as Brazilian heterotic group 1 (Parentoni et al. 2001). The second dent cluster comprises seven dent lines and one line with unknown origin from Brazil, one from CIMMYT and two from China. The clustering improved the genetic diversity resolution compared with PCA. However, the heterotic group defined by PCA was the major source for hybrid development from Embrapa.
Some CIMMYT lines developed recently can be grouped into two major heterotic groups, A and B, by breeders using well-known heterotic testers. However, CIMMYT maize lines tested in this study could not be divided into groups with significant genetic difference, although 394 maize lines from CIMMYT global maize breeding programs were tested. This confirms the previous reports that no clear clustering or heterotic patterns could be identified in either lowland tropical maize (Xia et al. 2004) or subtropical, tropical midaltitude and highland maize populations (Xia et al. 2005). Other studies of tropical maize, which were considered extremely diverse, have found that SSR variation does not provide evidence of population structure other than among individuals closely related by pedigree (Warburton et al. 2002, 2005). It has been suggested that relatedness among highly diverged maize lines is difficult to measure accurately regardless of the marker system (Hamblin et al. 2007). In this study, however, most CIMMYT inbred lines tested are elite germplasm and they are not so diverse compared to Chinese temperate germplasm that has been clearly grouped consistently with their heterotic patterns. Therefore, that there was no evidence for distinct population structure within CIMMYT inbred lines may not matter with the level of genetic diversity but with breeding history. These may be explained by the mixed origin of the populations used to extract those lines and the breeding program that initially emphasized developing germplasm with a wide genetic base (for creation of OPVs) and only has begun to develop heterotic groups since early 1990s. On the other hand, the CIMMYT heterotic groups are not created the same way as the temperate heterotic groups. Temperate lines are bred to be heterotic among groups while CIMMYT lines are tested after breeding and are usually assigned to a heterotic pattern with specific other inbreds. CIMMYT inbred lines have been developed frequently by incorporating genetic diversity mainly existing in Latin American (Mexican, Central American, and Caribbean, and southern cone countries) and Asia, US and European germplasm as well. Although CIMMYT maize lines cannot be grouped as different heterotic groups, they can be used as one of the parents to be crossed with other maize lines developed outside CIMMYT to produce hybrids with good yield potential. CIMMYT lines can be also used as a gene pool for mixed novel and unique alleles. This has been supported by this study that the genetic distance between CIMMYT lines and Chinese BSSS group examined was the largest among all comparisons. However, in the long run, CIMMYT breeding programs should pay more attention to heterotic pattern establishment so that high-performance hybrids can be produced using two CIMMYT lines as parents. For this purpose, a well-designed study should be conducted for a systematic survey for heterotic patterns and classify them based on both molecular markers and their combining ability and pedigree. Differentiation among CIMMYT maize lines should be increased so that heterotic patterns can become distinct, as more effort in breeding is put into selection for better hybrid performance. This prediction has been supported by Pioneer’s long-term selection for heterotic patterns in its breeding program which resulted in significant increase of genetic diversity between heterotic groups (Cooper et al. 2004).
SNP-marker assisted germplasm evaluation
As indicated by Mikel and Dudley (2006) and concurred by Smith (2007) that regular assays of genetic diversity of maize hybrids are necessary and that the diversity of maize germplasm needs to be broadened, marker-assisted germplasm evaluation will play an important role in worldwide maize genetics studies and plant breeding. The required resolution in germplasm evaluation depends on research objectives. Genetic studies are more interesting in understanding of population structure, while breeding studies have their focus on heterotic grouping. In genetic analysis, the major objective of classification, for example, is to precisely evaluate the genetic relationship among tested individuals and then use this information in other genetic analyses, and thus a refined or high-resolution classification is required. In plant breeding, however, the major objective is to evaluate combining ability and construct heterotic groups and thus, only a rough-scale of classification is required.
The relationship of genetic diversity with the location of maize origin can be predicted but has not been fully proved in this study as we found that Chinese temperate maize germplasm have a (or even slight higher) level of genetic diversity compared to the tropical/subtropical lines developed by CIMMYT, although CIMMYT maize breeding programs have been based on the germplasm sources collected from its host country, Mexico, which is the center of origin for maize. This indicates that CIMMYT maize breeding might have used only a part of genetic diversity existing in Mexican maize germplasm while much of genetic diversity hidden in Mexican landraces has not been tapped into for breeding modern maize lines. This also indicates that elite maize inbreds developed by using genetic diversity available from the center of origin does not necessarily have to be more diverse compared to those developed by the breeding programs located in non-center countries. In this study, the greatest extent of genetic divergence was identified between temperate and tropical/subtropical germplasm, compared to those between yellow and white kernels and between dent and flint textures. Over 75% of the SNP markers showed significant allele frequency difference between temperate and tropical/subtropical germplasm, and eight and five alleles were found to exclusively exist in temperate and tropical/subtropical germplasm, respectively, if all rare alleles are considered. This indicates that there is a great potential for breeding programs with focus on one type of germplasm by utilizing genetic variation in another.
Germplasm that host rare and unique alleles provide an opportunity of using these alleles in genetics studies and plant breeding. Search for such alleles have been reported in other crops including rice (Xu et al. 2004). However, the availability of similar information in maize is limited. In this report, we identified many alleles that have significantly different allelic frequencies among different maize germplasm, and many alleles that are highly frequent in one specific germplasm collection but are lacking or rare in others. These alleles can be first used to distinguish different germplasm collections. Second, markers hosting these alleles can be exploited for candidate gene discovery and function characterization using the sequence information and the associated phenotypic difference. Third, germplasm containing these alleles can be used for plant breeding if association between these alleles and phenotypic traits can be validated.
As SNPs can be developed from both intergenic and intragenic, allele mining can be combined with functional diversity analysis. Gene-based markers can be directly associated with the genetic difference identified in segregating populations or a panel of diverse lines used in LD-based association mapping. We found that two SNPs developed from the same candidate gene associated with divergence of two opposite heterotic groups, PA and Lancaster. Two other SNPs within a LD block of 142 kb were found to be associated with allele frequency change and allele missing in Brazilian germplasm. Methods to ascertain allele function include marker-assisted backcrossing, genetic transformation, transient expression assays, and association analysis using an independent set of germplasm from that used to identify the allele. As more genetic variation is functionally validated, the next big step in this area is the development of bioinformatics tools to compare sequence variation with protein and functional domain variation or with public databases including associated phenotype data, in order to predict which sub-selections of SNP haplotype variants have the maximum likelihood of providing beneficial phenotypic variation in the target trait (Xu et al. 2009). It is likely that SNPs in promoter and non-coding regions will also be important for predictive phenotype analysis. The same methodology used in association mapping may also be used for allele mining of the diverse core subsets of maize created from breeder’s lines, genebank accessions, and wild relatives.
References
Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215:403–410
Betrán FJ, Ribaut JM, Beck D, Gonzalez de León D (2003) Genetic diversity, specific combining ability, and heterosis in tropical maize under stress and nonstress environments. Crop Sci 43:797–806
Botstein DR, White RL, Skolnick M, Davis RW (1980) Construction of a genetic linkage map in man using restriction fragment length polymorphisms. Am J Hum Genet 32:314–331
Bradbury PJ, Zhang Z, Kroon DE, Casstevens TM, Ramdoss Y, Buckler ES (2007) TASSEL: software for association mapping of complex traits in diverse samples. Bioinformatics 23:2633–2635
Bruford MW, Wyne RK (1993) Microsatellites and their application to population genetic studies. Curr Opin Genet Dev 3:939–943
Buckler ES, Gaut BS, McMullen MD (2006) Molecular and functional diversity of maize. Curr Opin Plant Biol 9:172–176
Ching A, Caldwell KS, Maurine Dolan JM, Smith OS, Tingey S, Morgante M, Rafalski AJ (2002) SNP frequency, haplotype structure and linkage disequilibrium in elite maize inbred lines. BMC Genet 3:19
Choukan R, Hossainzadeh A, Ghannadha MR, Warburton ML, Talei AR, Mohammadi SA (2006) Use of SSR data to determine relationships and potential heterotic groupings within medium to late maturing Iranian maize inbred lines. Field Crops Res 95:212–222
CIMMYT Applied Molecular Genetics Laboratory (2003) Laboratory Protocols, 3rd edn. Mexico, D.F, pp 7–11
Cooper M, Smith OS, Graham G, Arthur L, Feng L, Podlich DW (2004) Genomics, genetics, and plant breeding: a private sector perspective. Crop Sci 44:1907–1913
Doebley JF, Goodman MM, Stuber CW (1984) Isoenzymatic variation in Zea (Gramineae). Syst Bot 9:203–218
Dubreuil P, Charcosset A (1998) Genetic diversity within and among populations: a comparison between isozyme and nuclear RFLP loci. Theor Appl Genet 96:577–587
Duvick DN, Smith JSC, Cooper M (2004) Changes in performance, parentage, and genetic diversity of successful corn hybrids, 1930–2000. In: Smith CW, Betran J, Runge ECA (eds) Corn: origin, history, technology, and production. Wiley, New York, pp 65–97
Eyre-Walker A, Gaut RL, Hilton H, Feldman DL, Gaut BS (1998) Investigation of the bottleneck leading to the domestication of maize. Proc Natl Acad Sci USA 95:4441–4446
Falush D, Stephens M, Pritchard JK (2003) Inference of population structure using multilocus genotype data: linked loci and correlated allele frequencies. Genetics 164:1567–1587
Fan JB, Gunderson KL, Bibikova M, Yeakley JM, Chen J, Wickham Garcia E, Lebruska LL, Laurent M, Shen R, Barker D (2006) Illumina universal bead arrays. Methods Enzymol 410:57–73
Feng L, Sebastian S, Smith S, Cooper M (2006) Temporal trends in SSR allele frequencies associated with long-term selection for yield of maize. Maydica 51:293–300
Fisher RA (1922) On the interpretation of χ2 from contingency tables, and the calculation of P. J R Stat Soc 85:87–94
Gupta PK, Rustgi S, Mir RR (2008) Array-based high-throughput DNA markers for crop improvement. Heredity 101:5–18
Hamblin MT, Warburton ML, Buckler ES (2007) Empirical comparison of simple sequence repeats and single nucleotide polymorphisms in assessment of maize diversity and relatedness. PLoS ONE 12:e1367
Hanson MA, Gaut BS, Stec AO, Fuerstenberg SI, Goodman MM, Coe EH, Doebley JF (1996) Evolution of anthoryanin biosynthesis in maize kernels: the role of regulatory and enzymatic loci. Genetics 143:1395–1407
Hilton H, Gaut BS (1998) Speciation and domestication in maize and its wild relatives: evidence from the globulin-1 gene. Genetics 150:863–872
Hoisington D, Khairallah M, Reeves T, Ribaut JM, Skovmand B, Taba S, Warburton M (1999) Plant genetic resources: what can they contribute toward increased crop productivity? Proc Natl Acad Sci USA 96:5937–5943
Jones ES, Sullivan H, Bhattramakki D, Smith JSC (2007) A comparison of simple sequence repeat and single nucleotide polymorphism marker technologies for genotypic analysis of maize (Zea mays L.). Theor Appl Genet 115:361–371
Laval G, San Cristobal M, Chevalet C (2002) Measuring genetic distances between breeds: use of some distances in various short term evolution models. Genet Sel Evol 34:481–507
Liu K, Muse SV (2005) PowerMarker: an integrated analysis environment for genetic marker analysis. Bioinformatics 21:2128–2129
Liu K, Goodman M, Muse S, Smith JS, Buckler E, Doebley J (2003) Genetic structure and diversity among maize inbred lines as inferred from DNA microsatellites. Genetics 165:2117–2128
McMullen MM, Kresovich S, Villeda HS, Bradbury P, Li H, Sun Q, Flint-Garcia S, Thornsberry J, Acharya C, Bottoms C, Brown P, Browne C, Eller M, Guill K, Harjes C, Kroon D, Lepak N, Mitchell SE, Peterson B, Pressoir G, Romero S, Rosas MO, Salvo S, Yates H, Hanson M, Jones E, Smith S, Glaubitz JC, Goodman M, Ware D, Holland JB, Buckler ES (2009) Genetic properties of the maize nested association mapping population. Science 325:737–740
Melchinger AE (1999) Genetic diversity and heterosis. Chapter 10. In: Coors JG, Pandey S (eds) The genetics and exploitation of heterosis in crops. CSSA, Madison
Mikel MA, Dudley JW (2006) Evolution of North American dent corn from public to proprietary germplasm. Crop Sci 46:1193–1205
Palaisa K, Morgante M, Williams M, Rafalski A (2003) Contrasting effects of selection on sequence diversity and linkage disequilibrium at two phytoene synthase loci. Plant Cell 15:1795–1806
Palaisa K, Morgante M, Tingey S, Rafalski A (2004) Long-range patterns of diversity and linkage disequilibrium surrounding the maize Y1 gene are indicative of an asymmetric selective sweep. Proc Natl Acad Sci USA 101:9885–9890
Parentoni SN, Magalhães JV, Pacheco CAP, Santos MX, Abadie T, Gama EEG, Guimarães PEO, Meirelles WF, Lopes MA, Vasconcelos MJV, Paiva E (2001) Heterotic groups based on yield-specific combining ability data and phylogenetic relationship determined by RAPD markers for 28 tropical maize open pollinated varieties. Euphytica 121:197–208
Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155:945–959
Rafalski A, Ananiev E (2009) Genetic diversity, linkage disequilibrium and association mapping. In: Bennetzen JL, Hake SC (eds) Handbook of maize: genetics and genomics. Springer, New York, pp 201–219
Reif JC, Xia XC, Melchinger AE, Warburton ML, Hoisington DA, Beck D, Bohn M, Frisch M (2004) Genetic diversity determined within and among CIMMYT maize populations of tropical, subtropical, and temperate germplasm by SSR markers. Crop Sci 44:326–334
Reif JC, Warburton ML, Taba S, Hoisington D, Crossa J, Franco J, Xia X, Muminović J, Bohn M, Frisch M, Melchinger AE (2006) Grouping of accessions of Mexican races of maize revisited with SSR markers. Theor Appl Genet 113:177–185
Remington DL, Thornsberry JM, Matsuoka Y, Wilson LM, Whitt SR, Doebley J, Kresovich S, Goodman MM, Buckler ES (2001) Structure of linkage disequilibrium and phenotypic associations in the maize genome. Proc Natl Acad Sci USA 98:11479–11484
Rogers JS (1972) Measures of genetic similarity and genetic distance. In: Studies in genetics VII. University of Texas Publication 7213, Austin
Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425
Smith S (2007) Pedigree background changes in US hybrid maize between 1980 and 2004. Crop Sci 47:1914–1926
Smith JSC, Chin ECL, Shu H, Smith OS, Wall SJ, Senior ML, Mitchell SE, Kresovich S, Ziegle J (1997) An evaluation of the utility of SSR loci as molecular markers in maize (Zea mays L.): comparisons with data from RFLPs and pedigree. Theor Appl Genet 95:163–173
Smith JSC, Duvick DJ, Smith OS, Cooper M, Feng L (2004) Changes in pedigree backgrounds of Pioneer brand maize hybrids widely grown from 1930 to 1999. Crop Sci 44:1935–1946
Song TM, Chen SJ (2004) Long term selection for oil concentration in five maize populations. Maydica 49:9–14
Tenaillon MI, Sawkins MC, Long AD, Gaut RL, Doebley JF et al (2001) Patterns of DNA sequence polymorphism along chromosome 1 of maize (Zea mays ssp. mays L.). Proc Natl Acad Sci USA 98:9161–9166
Teng WT, Cao QS, Chen YH, Liu XH, Men SD, Jing XQ, Li JS (2004) Analysis of maize heterotic groups and patterns during past decade in China. Agric Sci Chin 3:481–489 (in Chinese)
Tracy WF, Chandler MA (2006) The historical and biological basis of the concept of heterotic patterns in Corn Belt Dent maize. In: Lamkey KR, Lee M (eds) Plant breeding: The Arnel R. Hallauer international symposium. Blackwell, Ames, pp 219–233
Vigouroux Y, Mitchell S, Matsuoka Y, Hamblin M, Kresovich S, Smith JSC, Jaqueth J, Smith OS, Doebley J (2005) An analysis of genetic diversity across the maize genome using microsatellites. Genetics 169:1617–1630
Vigouroux Y, Glaubitz JC, Matsuoka Y, Goodman MM, Sánchez G, Doebley J (2008) Population structure and genetic diversity of new world maize races assessed by DNA microsatellites. Am J Bot 95:1240–1253
Vroh BI, McMullen MD, Sanchez-Villeda H, Schroeder S, Gardiner J, Polacco M, Soderlund C, Wing R, Fang Z, Coe EH Jr (2006) Single nucleotide polymorphisms and insertion–deletions for genetic markers and anchoring the maize fingerprint contig physical map. Crop Sci 46:12–21
Wang R, Yu Y, Zhao J, Shi Y, Song Y, Wang T, Li Y (2008) Population structure and linkage disequilibrium of a mini core set of maize inbred lines in China. Theor Appl Genet 117:1141–1153
Warburton ML, Xia XC, Crossa J, Franco J, Melchinger AE, Frisch M, Bohn M, Hoisington DA (2002) Genetic characterization of CIMMYT maize inbred lines and open pollinated populations using large scale fingerprinting methods. Crop Sci 42:1832–1840
Warburton ML, Ribaut JM, Franco J, Crossa J, Dubreuil P, Betrán FJ (2005) Genetic characterization of 218 elite CIMMYT inbred maize lines using RFLP markers. Euphytica 142:97–106
Warburton ML, Reif JC, Frisch M, Bohn M, Bedoya C, Xia XC, Crossa J, Franco J, Hoisington D, Pixley K, Taba S, Melchinger AE (2008) Genetic diversity in CIMMYT nontemperate maize germplasm: landraces, open pollinated varieties, and inbred lines. Crop Sci 48:617–624
Whitt SR, Wilson LM, Tenaillon MI, Gaut BS, Buckler ES (2002) Genetic diversity and selection in the maize starch pathway. Proc Natl Acad Sci USA 99:12959–12962
Wright SI, Vroh BI, Schroeder SG, Yamasaki M, Doebley JF, McMullen MD, Gaut BS (2005) The effects of artificial selection on the maize genome. Science 308:1310–1314
Xia XC, Reif JC, Hoisington DA, Melchinger AE, Frisch M, Warburton ML (2004) Genetic diversity among CIMMYT maize inbred lines investigated with SSR markers: I. Lowland tropical maize. Crop Sci 44:2230–2237
Xia XC, Reif JC, Melchinger AE, Frisch M, Hoisington DA, Beck D, Pixley K, Warburton ML (2005) Genetic diversity among CIMMYT maize inbred lines investigated with SSR markers: II. Subtropical, tropical midaltitude, and highland maize inbred lines and their relationships with elite US and European maize. Crop Sci 45:2573–2582
Xie CX, Warburton M, Li MS, Li XH, Xiao MJ, Hao ZF, Zhao Q, Zhang SH (2008) An analysis of population structure and linkage disequilibrium using multilocus data in 187 maize inbred lines. Mol Breed 21:407–418
Xu Y (2003) Developing marker-assisted selection strategies for breeding hybrid rice. Plant Breed Rev 23:73–174
Xu Y, Ishii T, McCouch S (2003) Marker-assisted evaluation of germplasm resources for plant breeding. In: Mew TW, Brar DS, Peng S, Hardy B (eds) Rice science: innovations and impact for livelihood. Proceedings of the 24th international rice research conference, 16–19 September 2002, Beijing, China. International Rice Research Institute, Chinese Academy of Engineering, and Chinese Academy of Agricultural Sciences, Manila, Philippines, pp 213–229
Xu Y, Beachell H, McCouch SR (2004) A marker-based approach to broadening the genetic base of rice (Oryza sativa L.) in the US. Crop Sci 44:1947–1959
Xu Y, Skinner DJ, Wu H, Palacios-Rojas N, Araus JL, Yan J, Gao S, Warburton ML, Crouch JH (2009) Advances in maize genomics and their value for enhancing genetic gains from breeding. Int J Plant Genomics 2009:957602. doi:10.1155/2009/957602
Yamasaki M, Tenaillon MI, Vroh BI, Schroeder SG, Sanchez-Villeda H, Doebley JF, Gaut BS, McMullen MD (2005) A large-scale screen for artificial selection in maize identifies candidate agronomic loci for domestication and crop improvement. Plant Cell 17:2859–2872
Yan J, Yang X, Shah T, Sánchez-Villeda H, Li J, Warburton M, Zhou Y, Crouch JH, Xu Y (2009) High-throughput SNP genotyping with the GoldenGate assay in maize. Mol Breed (in press)
Yu J, Pressoir G, Briggs WH, Vroh Bi I, Yamasaki M, Doebley JF, McMullen MD, Gaut BS, Nielsen DM, Holland JB, Kresovich S, Buckler ES (2006) A unified mixed-model method for association mapping that accounts for multiple levels of relatedness. Nat Genet 38:203–208
Yu J, Holland JB, McMullen MD, Buckler ES (2008) Genetic design and statistical power of nested association mapping in maize. Genetics 178:539–551
Yu J, Zhang Z, Zhu C, Tabanao DA, Pressoir G, Tuinstra MR, Kresovich S, Todhunter RJ, Buckler ES (2009) Simulation appraisal of the adequacy of number of background markers for relationship estimation in association mapping. Plant Genome 2:63–77
Yuan LX, Fu JH, Zhang SH, Liu XZ, Peng ZB, Li XH, Warburton M, Khairallah M (2001) Heterotic grouping of maize inbred lines using RFLP and SSR markers. Acta Agron Sin 27:149–156
Zhu YL, Song QJ, Hyten DL, Van Tassell CP, Matukumalli LK, Grimm DR, Hyatt SM, Fickus EW, Young ND, Cregan PB (2003) Single-nucleotide polymorphisms in soybean. Genetics 163:1123–1134
Acknowledgments
We thank Drs. Marilyn Warburton and Rodomiro Ortiz for their critical review and suggestions which much improved the manuscript, Dr. Kevin Pixley for constructive discussion, and Eva Huerta Miranda, Carlos Martinez Flores, Martha Hernandez Rodríguez, Alberto Vergara Alva, Maria Asunción Moreno Ortega, and Jose Simon Pastrana Marias for lab and field assistance. The maize breeding program team from Embrapa (Drs. Paulo Evaristo O. Guimarães, Cleso A.P. Pacheco, and Lauro J.M. Guimarães) provided breeding materials and related information for the 94 Brazilian maize germplasm. This work at CIMMYT is funded by the Rockefeller Foundation, Bill and Melinda Gates Foundation, and European Community, and through other attributed or unrestricted funds provided by the members of the Consultative Group on International Agricultural Research (CGIAR) and national governments of USA, Japan, and UK. The Brazilian research was supported by Embrapa, FAPEMIG, The Generation Challenge Program and The McKnight Foundation—CCRP. Yanli Lu was supported by China Scholarship Council for her research at CIMMYT as a joint PhD student.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by T. Luebberstedt.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Table S1
List of 770 maize inbred lines used in the study. Information provided in this supplemental table include ID, brief name, sample name, pedigree, origin, adaptation, kernel color, kernel texture, and the subsets that were used to compare allele frequencies in yellow versus white (Subset 1), dent versus flint (Subset 2) comparisons. (XLS 180 kb)
Table S2
Summary statistics for the 1034 informative SNP markers identified from the 1536 SNP chip. Information provided in this supplemental table include SNP order, SNP index, SNP name, SNP, chromosome, physical position, contig, minor allele frequency, heterozygosity, gene diversity, and polymorphic information content (PIC). Subset A represents 449 high-quality SNPs; Subsets B and C represent 499 and 279 SNP markers after excluding markers that showed allelic frequency difference larger than 10% and 5% between temperate and tropical/subtropical germplasm collections, respectively. (XLS 267 kb)
Table S3
Inbred lines with their proportional memberships in the model-based subgroups determined by STRUCTURE. Eight groups/clusters were identified. Information about membership of inbred lines corresponding to each cluster is provided. (XLS 178 kb)
Fig. S1
A successful score with obvious three clusters as shown by a single nucleotide polymorphism (SNP) marker, PZA00566.5. AA and BB indicate the homozygous clusters, AB indicates heterozygotes. A plot located outside of cluster was regarded as missing. (TIFF 1790 kb)
Fig. S2
Cluster dendrogram constructed for 770 maize inbred lines genotyped with 1034 SNP markers. Two major groups are identified as “China” and “CIMMYT and Brazil”. Six groups are identified within “China” by different colors as SPT, LRC, PA, PB, Lancaster, and BSSS. (TIFF 5065 kb)
Rights and permissions
About this article
Cite this article
Lu, Y., Yan, J., Guimarães, C.T. et al. Molecular characterization of global maize breeding germplasm based on genome-wide single nucleotide polymorphisms. Theor Appl Genet 120, 93–115 (2009). https://doi.org/10.1007/s00122-009-1162-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00122-009-1162-7