Abstract
Genetic improvement in yield and fiber quality is needed for worldwide cotton production. Identification of molecular markers associated with fiber-related traits can facilitate selection for these traits in breeding. This study was designed to identify associations between SSR markers and fiber traits using an exotic germplasm population, species polycross (SP), derived from multiple crosses among Gossypium tetraploid species. The SP population underwent 11 generations of mixed random mating and selfing followed by 12 generations of selfing. A total of 260 lines were evaluated for fiber-related traits under three environments in 2005 and 2006. Large genotypic variance components in traits were identified relative to components of genotype × environment. Eighty-six primer pairs amplified a total of 314 polymorphic fragments among 260 lines. A total of 202 fragments with above 6% allele frequency were analyzed for associations. Fifty-nine markers were found to have a significant (P < 0.05, 0.01, or 0.001) association with six fiber traits. There were six groups identified within the population using structure analysis. Allele frequency divergence among six groups ranged from 0.11 to 0.27. Of the 59 marker–trait associations, 39 remained significant after correction for population structure and kinship using a mixed linear model. The effect of population sub-structure on associations was most significant in boll weight among the traits analyzed. The sub-structure among the SP lines may be caused by natural selection, the breeding method applied during development of inbred lines, and unknown factors. The identified marker–trait associations can be useful in breeding and help determine genetic mechanisms underlying interrelationships among fiber traits.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Increasing competition in global textile market has resulted in the foreign market becoming the primary customer of USA cotton. Eighty percent of cotton produced in the USA was exported in 2006 (MacDonald and Meyer 2007). International customers demand higher fiber strength than the domestic market previously required. Since yield is always a top priority in breeding to keep profits for cotton growers, it is a great challenge for cotton breeders to continuously improve fiber quality while maintaining its productivity. Progress to break the negative association between lint yield and fiber quality using traditional breeding methods has been limited (Smith and Coyle 1997). Identification of molecular markers associated with fiber-related traits can facilitate selection and improve our understanding of genetic mechanisms underlying interactions among fiber-related traits.
Association mapping is a new approach for identification of QTLs associated with the quantitative traits. This approach is alternative to the traditional linkage-based QTL mapping. These two approaches are different in their requirement for linkage information among markers before the association analysis is done. The first step in the traditional QTL approach is to identify polymorphic markers and make a linkage map in a specific genetic population segregating between contrasting phenotypes. Next, the regions associated with the quantitative traits are identified in the linkage groups based on marker–phenotype association. The first step in association mapping is to identify those markers with unequal distribution of alleles among the individuals ranked for a trait such as agronomic performance. Under this circumstance, the terminology ‘association’ can be defined as the cases when polymorphic fragments are identified with significant higher frequencies in one type of phenotype than the contrasting phenotype (Schafer and Hawkins 1998). Significant trait-associated markers can be further located on a chromosome if mapped markers are used for the analysis.
The advantages of association analysis over traditionally linkage-based QTL mapping have been reviewed in several reports (Pritchard and Rosenberg 1999; Pritchard et al. 2000; Flint-Garcia et al. 2003; Skøt et al. 2005). First, the cost of evaluation for traits such as agronomic performance in replicated experiments is usually large. Unlike linkage-based QTL mapping, association studies can be conducted either in a collection of breeding lines, cultivars, natural populations, or a breeder’s population derived from multiple crosses. Phenotypic data are often available from evaluation trials of these breeding lines or cultivars. More importantly, since association is analyzed in genetic populations with diverse genetic backgrounds, a large number of alleles can be analyzed that provides better opportunities to identify associations between markers and traits. QTLs identified in this way are more likely to reveal the genetic diversity that exists in plant germplasm. The disadvantages of this approach are also obvious: (1) associations identified in comparisonwise tests could be Type I errors; (2) associations could be caused by population structure; and (3) there would be a lack of linkage information among the markers identified for significant associations. In non-random mating populations, artifacts occurred when an association was identified for the markers that were not linked to the QTLs (Pritchard and Rosenberg 1999; Flint-Garcia et al. 2003). In these studies, this type of artifact has been attributed to the presence of population stratification caused by gene drift, founder effects, or selection. False association may be detected between markers with different allele frequencies and different levels of a phenotype among sub-populations although there is no underlying physical linkage. This problem could be partially solved by use of random mating population for association analysis. Increasing recombination rate could reduce the linkage disequilibrium between QTLs and the unlinked or loosely linked markers to some extent. However, extensive random mating would also hinder association analysis since the identification of association depends on the detection of linkage disequilibrium between markers alleles and QTLs for the traits, and therefore, the size of linkage blocks (Xie et al. 2008).
Association studies were first applied in human populations to identify loci controlling diseases susceptibility (Risch and Merikangas 1996; Schafer and Hawkins 1998). In plants, this approach has been used to identify DNA polymorphisms associated with phenotypes including flowering time in maize (Zea mays L.) (Thornsburry et al. 2001) and perennial ryegrass (Lolium perenne L.) (Skøt et al. 2005), cold tolerance in perennial ryegrass (Sackville Hamilton et al. 2002; Skøt et al. 2002), salt tolerance in wild barley (Hordeum spontaneum) (Ivandic et al. 2003), resistance to late blight and maturity in potato (Solanum tuberosum ssp tuberosum) (Gebhardt et al. 2004), and resistance to pachymetra root rot, leaf scald Fiji leaf gall, and smut in sugarcane (Saccharum officinarum) (Wei et al. 2006). In cotton, the first attempt of association mapping was reported by Kantartzi and Stewart (2008). In that study, 30 SSR marker–trait associations were identified in 56 Gossypium arboretum germplasm accessions introduced from different regions worldwide.
In a current study, 260 germplasm lines derived from multiple crosses among tetraploid species in Gossypium (Zeng et al. 2007) were used for analyzing associations between SSR markers and fiber-related traits. This population underwent 11 generations of mixed random mating and selfing followed by 12 generations of selfing. The objectives are to evaluate the genotypic variation in yield components and fiber quality, screen for polymorphic SSR markers, identify population sub-structures, and analyze the associations between SSR markers and fiber-related traits by accounting for population structure identified in the germplasm and pairwise kinship among individual lines. The identified molecular markers associated with fiber traits will be useful in breeding for genetic improvement of lint yield and fiber quality and analysis of interrelationships among fiber traits, and therefore, help break or reduce negative associations between lint yield and fiber quality.
Materials and methods
The population and planting
An exotic population, designated as species polycross (SP), was initiated by P.A. Miller in 1967. The origin and development of this population were described in detail in a previous study (Zeng et al. 2007). Briefly, the SP population was developed by crossing 12 cultivars and strains in G. hirsutum with the other four tetraploid species, G. barbadense L., G. tomentosum Nutt., G. mustelinum Watt., and G. darwinii Watt.. The exact crossing patterns were unknown. The F 2 and subsequent progenies were maintained in an isolated field plot surrounded by woods in Raleigh, NC. Bee activity was known to be high in this area. The SP population was maintained by natural pollination from 1968 to 1978. Natural crosses were estimated to exceed 50% during this period of time. A sub-population of 2,000 plants was grown in Stoneville, MS from 1979 to 2004 under an environment with predominant self-pollination. The sub-population was maintained and advanced by harvesting one boll from each plant and bulking of bolls for planting of the next generation. The plants of this sub-population were grown almost every 2 years during this period. By 2004, this sub-population underwent 11 years of random mating followed by 12 years of predominant selfing. In 2004, 260 plants were randomly sampled from this sub-population and 15–20 bolls were collected from each plant. The seeds of each plant were grown as one line in a row in 2005 for evaluation trials.
Two hundred and sixty lines were evaluated at two field locations during summer 2005 and one field location during summer 2006. The experimental design was a complete randomized block with two locations and two replicates at each location in 2005, and one location with one replication in 2006. For purpose of statistical analysis, the factors of location and year were combined as a factor of environment. In this way, environments were defined as Environment 1 = 2005, Field Location 1; Environment 2 = 2005, Field Location 2; Environment 3 = 2006, Field Location 1. The lines were grown in single row plots, 4.6 m long and 1.0 m row space. In 2005, plants were planted on April 19 at Field Location 1 and May 5 at Field Location 2. In 2006, plants were planted on April 18 at Field Location 1. Thirty bolls from each plot were sampled by hand and ginned through a laboratory saw gin. Seed weight and lint weight from the sample of each plot were measured and recorded separately for individual plots. Yield components including lint percent and boll weight were calculated from the samples. Lint samples were submitted to StarLab, Knoxville, TN for analysis of fiber quality. Fiber strength was measured by a stelometer as the force per tex required breaking a bundle of fibers. Elongation was the percentage of elongation at the point of break in strength determination. Fiber span lengths were measured as the average length of the longest 2.5% of the fibers scanned.
SSR markers
Leaves were collected from field plots in summer 2005. Young leaves were collected from five plants of each line, bulked, and freeze dried. DNA was extracted from 40 to 50 mg of grounded freeze dried leaf tissue following the protocol reported by Paterson (Paterson et al. 1993). The sequences of SSR primers were downloaded from CMD (Cotton Marker Database, www.cottonmarker.org/cgi-bin/panel.cgi) based on the polymorphic information from a standard screening panel including upland cotton cultivars and other tetraploid species (Blenda et al. 2006).
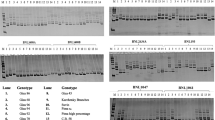
The primers were labeled with NED (7′8′-benzo-5′fluoro-2′,4,7-trichloro-5-carboxyfluorescein), HEX (4,7,2′,4′,5′,7′-hexacloro-6-carboxyfluorescein), or 6FAM (6-carboxyfluorescein) by Applied Biosystems. PCR amplification was performed in 10 μl volumes containing 5 mM Tris–HCL, 25 mM KCL, 0.75 mM MgCl2, 0.1 mM for each dNTP, 0.1 μM each of forward and reverse primers, 0.15 units of JumpStart Taq DNA polymerase (Sigma), and 10 ng of template DNA. Thermal cycler PTC-100 (MJ Research, Inc.), was used for PCR amplification in the following cycles: 5 min at 94°C followed by 15 s at 94°C for initial denaturation, 30 s at 65°C, and 1 min at 72°C for 11 cycles with a decrease of 1° for annealing temperature in each cycle, 40 cycles for 15 s at 95°C, 30 s at 55°C, and 1 min at 72°C, and 30 min at 72°C for final extension. The amplified DNA fragments with fluorescence labels were separated as peaks by capillary electrophoresis using ABI 3730XL DNA Sequencer 96-capillary automated sequencer (Applied Biosystems). Allele calling was performed using Genemapper Software v. 4.0 (Applied Biosystems). The fragment with the highest fluorescence intensity was called as an allele when SSR products stuttered. The sizes of the alleles were automatically determined as base pairs to two decimal places by the software. The alleles were scored by their sizes. The germplasm lines were genotyped as allele sizes at all SSR. Polymorphic Information Content (PIC) of each SSR primer pair and stepwise mutation index were calculated using software PowerMarker 3.25 (Liu and Muse 2005).
Statistical, population sub-structure, and kinship analysis
The GLM procedure of the Statistical Analysis System (version 6; SAS Institute; Cary, NC) was used for the analysis of variance (ANOVA) on all data coupled with the RANDOM statement to test the factors of genotype and genotype × environment. Estimation of variance components of yield components and fiber properties was calculated from mean squares across environments (Bernardo 2002). Broad-sense heritability (h 2) was calculated from variance components (Bernardo 2002) using the equation described by Fehr (1987). Confidence intervals (90%) of heritability were calculated according to Knapp et al. (1985).
Population sub-structures among 260 SP lines were analyzed using the software STRUCTURE 2.2 (Pritchard et al. 2007). In grouping, the length of running time was 100,000 and replication after burning was 50,000. Admixture model was chosen as an ancestry model. Allele Frequencies Correlated was chosen as allele frequency model. For structure analysis, the individuals were coded using a two-row format: x i,1 j , x i,2 j , in which i represents individual at locus j as described by Pritchard et al. (2007). The individuals with probability >0.50 were assigned to the respective groups and the individuals with probability <0.50 were artificially assigned to a group designated as “mixed group.” The population was analyzed for genetic diversity and genetic differentiation in the population using the software AFLP-SURV 1.0 (Vekemans 2002). In doing so, the data of SSR were converted to dominant type, i.e., 1 and 0 for presence and absence of the bands to conduct analysis using software AFLP-SURV. Genetic diversity, Ht, and Wright’s F st values (Wright 1951) were estimated from marker loci in the population by the approach described by Lynch and Milligan (1994). Wright’s F st index was defined as the proportion of genetic variation between groups to the genetic variation of the overall population, i.e., genetic variation between groups plus genetic variation within group. Under extreme circumstance, F st = 0 when all groups are same for their allele frequency.
Means of fiber-related traits were compared by marker classes, i.e., presence and absence of the marker, using t tests to initially determine the association between markers and traits. The tests were conducted by PROC MULTTEST procedures (SAS Institute) for permutation adjustment. The procedures were repeated 1,000 times by shuffling the data (permutation = 1000). The purpose of permutation was to set up threshold values for significant (P < 0.05) rejection of the null hypothesis that there is no association between markers and traits.
The P values in permutation tests were converted to Bayes factor as strength of evidence against the null hypothesis or strength to support alternative hypothesis. Bayes factor is an evidence-based framework for statistical interference. Unlike P value which is a probability under null hypothesis, Bayesian frame depends on the probability of the observed data along with interference and decisions (Goodman 1999). By definition, Bayes factor is a likelihood ratio: probability (given the null hypothesis)/probability (given the alternative hypothesis). Based on an explanation by Ball (2005), size of P-value required for strength of evidence can be determined by reference to Bayes factor for specific sample size and experiment setup. That means “the larger the sample size, the smaller the P-values need to be to correspond to a given strength of evidence.” For convenience, Bayes factor was structured the same way as P values, i.e., the smaller the Bayes factor, the less the support for the null hypothesis. Minimum Bayes factor is the smallest amount of evidence to support null hypothesis or strongest evidence against the null hypothesis (Goodman 1999). The Minimum Bayes factor was calculated by the equation described by Goodman (1999):
where z is the number of standard errors from the null effect. In t tests, z was substituted by t values. If converted Minimum Bayes factor is larger than 0.05, the strength of evidence against null hypothesis will be moderate or weak. Otherwise, say smaller than 0.01, the strength of evidence against null hypothesis will be strong or very strong.
Significant marker–trait associations identified in t tests were further tested using two different models, a general linear model (GLM) and a mixed linear model (MLM) in TASSEL 2.0.1 (Bradbury et al. 2007). In GLM, population sub-structures in SP germplasm were incorporated into the model as covariates. In MLM model, association was estimated by simultaneous accounting multiple levels of population structure (Q matrix) and relative kinship among the individuals (K matrix) as described by Yu et al. (2006). A data file of Q matrix was created with K = 6 as determined by Structure 2.2. The K matrix was created by the calculation of pairwise kinship coefficients using the procedures of Loiselle et al. (1995) and Ritland (1996) using TASSEL (personal communication, Zhiwu Zhang, Maize Genetics Laboratory, Cornell Univ.).
Results
Two hundred and sixty lines were evaluated under three environments. Large genotypic effects were identified for most traits (Table 1). Genotypic variance exceeded error variance for most traits except boll weight and elongation. Genotypic × environmental variance was small relative to genotypic variance for all traits except boll weight. Broad-sense heritability was high for most of the traits except boll weight (Table 1).
Eighty-six primer pairs amplified a total of 314 polymorphic fragments among 260 lines. One hundred and twelve of these fragments had allele frequency lower than 6%. In order to avoid the possible overestimation of association on markers with low frequency, these fragments were not included in the association analysis. Means of traits by marker classes were only analyzed for the remaining 202 fragments.
The number of substructures was determined by running the software Structure with different K (the number of groups) values ranging from 1 to 10. The results showed that the likelihood, i.e., p(X|K) as defined in the program, increased with the increase of K and peaked at K = 6. Therefore, the number of six, i.e., six groups, was chosen as the number of groups. Bar plot of the subpopulation structure in the 260 lines (Fig. 1) showed that a large number of lines were strongly assigned, i.e., the lines with at least 50% of single ancestral genetic background, to each of the six groups. The proportions of 260 lines assigned to different groups were asymmetric. Allele frequency divergence among groups measured as net nucleotide distance (Pritchard et al. 2007) (Table 2) showed that allele frequency distance ranged from 0.11 (between Group 1 and Group 5) to 0.27 (between Group 5 and Group 6). These results provide evidence for the existence of population substructure among the 260 lines.
Bar plot of sub-population structures in 260 SP lines analyzed using Structure 2.2 by point estimate (k = 6). Groups are represented in different colors as shown in figure legends. Each column represents one SP line and partitioned into segments representing admixture of ancestral composition. The length of segments represents the percentage of a single ancestral background in that line. The columns (260 in total) were assigned to six groups
Two hundred and nine lines were assigned to the six groups with probability 50% or higher. These six groups, designated as Group1, Group2, Group3, Group 4, Group 5, and Group 6, consisted of 55, 27, 25, 47, 36, and 19 lines, respectively. The remaining 51 lines failed to group with a probability higher than 50%. These 51 lines with mixed ancestral genetic backgrounds were artificially assigned to the “mixed group.” In order to further analyze population sub-structure among 260 SP lines, pairwise distances among these groups were calculated by Wright’s F st index (Table 3). The distances between groups ranged from 0.04 (between Group 1 and the “Mixed Group”) and 0.36 (between Group 5 and Group 6). Generally, smaller genetic distances were observed between the “Mixed Group” and the other groups than those between groups with relatively simple genetic backgrounds. In overall population, i.e., 260 SP lines, H t values and Wright’s F st index were determined to be highly significant (P < 0.0001), 0.36 and 0.21, respectively. These results suggest that SP germplasm is significantly structured with significant genetic distance among population substructures. The means of fiber-related traits except strength were significantly different (P < 0.01) among the groups (Table 4).
The means of yield components and fiber properties of 260 lines classed by markers were compared between “1” and “0” alleles, i.e., presence and absence of the marker, using experimentwise t tests. A total of 23 SSR markers were identified significantly (P = 0.05, 0.01, or 0.001) associated with lint percent and boll weight (Table 5). Another 36 SSR markers were identified significantly (P = 0.05, 0.01, or 0.001) associated with micronaire, elongation, strength, and 2.5% span length (Table 6). Only one marker–trait association, i.e., BNL3569175-micronaire, was identified with Minimum Bayes factor larger than 0.05 (Table 6). Minimum Bayes factors were less than 0.01 for 67% of the remaining marker–trait associations. These results indicate strong evidence against the null hypothesis that there is no significant association between markers and traits.
After calculation for population structure and kinship using MLM, 12 of the 23 marker–trait associations for yield components survived stringent correction and remained significant (P < 0.05) (Table 5). Twenty-seven of the 36 associations for the four fiber traits remained significant (P < 0.05) (Table 6) after correction. Over half of the marker–trait associations in boll size became non-significant after correction, while all associations in elongation remained significant. When correction was calculated using a GLM model only accounting for population structure, significance level of associations was generally higher than that using MLM model account for both Q and K matrices (Tables 5 and 6). The most significant effects of kinship on association were observed in boll weight and micronaire by comparisons of the corrections between GLM and MLM. Four marker–trait associations in boll weight and three marker–trait associations in micronaire became non-significant after kinship was accounted for corrections in MLM. The most significant effect of population sub-structure on associations was observed in boll weight. Five of the 14 marker–trait associations in boll weight became non-significant after correction by accounting only population structure in GLM.
Discussion
The SP population was derived from multiple crosses among five tetraploid species in Gossypium followed by 11 generations of partial random mating and 12 generations of predominant selfing. The genotypic variation and uniqueness of this population due to its diverse genetic background have been described in a previous report (Zeng et al. 2007). In that study, highly significant genotypic effects were identified for yield, yield components, and fiber properties. Some lines in SP population had desirable combination between yield and fiber quality with good yield comparable to high yielding cultivars Deltapine 555BR and Stoneville 4892BR and good fiber quality comparable to Phytogene72 and FiberMax960B2R. In consistent with previous report, results in current study indicate that large genotypic effects for yield components and fiber properties exist in the SP population. Large genotypic effects relative to genotype (G) × environment (E) and high broad-sense heritability indicate the potential to identify molecular markers associated with fiber-related traits in SP population. The results also indicate a relatively small G × E for fiber traits. Small interactions of G × E for fiber traits indicate that three environments are sufficient for association studies of fiber-related traits because of large genotypic effects.
Association mapping has not been extensively applied in molecular breeding currently in cotton. The main concerns over the use of this approach include the false rejection of the null hypothesis that there is no association, and false associations related to population structure. In the current study, it was clearly shown that Type I errors were well controlled by setting up stringent threshold values for significance, i.e., P < 0.05, using permutation testing during experimentwise t tests. A P-value of 0.05 in the experimentwise t tests was approximately equivalent to a P-value of 0.005 in the comparisonwise t tests in this experiment. Furthermore, the P-values were converted to Minimum Bayes factors over half of which were less than 0.01 with only one larger than 0.05. Since Bayes factor can validate P values for a given sample size and experiment design (Ball 2005), the small minimum Bayes factors in the current study indicate either moderate or strong strength of evidence for H1 with large sample size of 260 lines in the experiments. As a result, 59 marker-trait associations were apparent for yield components and fiber properties.
These associations were further analyzed for possible false association due to population substructures. The structure analysis identified six groups with significant genetic differentiation among them. There were about 20% of the lines failed to group with high probability. These lines were thought to have mixed ancestral genetic backgrounds. The large number of lines with mixed ancestral genetic backgrounds is consistent with partial random mating of 11 generations before the development of inbred lines. Significant differences of trait means among the groups indicate the influence of population substructure on associations between markers and traits. The lines with a marker could be associated with high fiber quality and the lines without this marker could be associated with low fiber quality in one of groups (positive association). The reverse could be observed in other groups (negative association). When allele frequency of a marker is same among these groups, association with traits should not be identified for this marker. When allele frequency is different among groups, false associations could be significant because of these substructures. After correction accounting for population structure and kinship using MLM, 39 of the 59 marker–trait associations remained significant (P < 0.05). Therefore, these markers–trait associations are not false ones caused by population substructures or relatedness among individual lines. Some markers remained significant in association with boll weight and micronaire after correction by accounting for population structure in GLM model. However, these marker–trait associations became non-significant after the factor of kinship was accounted. These results indicate a moderate level of kinship effect on association analysis in this germplasm population. Low to moderate level of structure effects on association were observed among different traits. Population substructure affects marker–trait association most significantly in boll weight, while it affects associations least significantly in micronaire and elongation among the traits analyzed. Therefore, corrections accounting for both population substructure and kinship are necessary for the determination of associations in molecular markers with fiber-related traits in this germplasm population.
The exact cause of genetic differentiation within SP population is unknown since there was no selection applied to the population during the development. Genetic drift, mutation, and natural selection are major factors causing changes in gene frequency among individuals in a population. Although 12 generations of selfing during germplasm development was long enough to allow mutation to occur, a relatively large population size, i.e., 2,000 plants, was maintained throughout the period of selfing. This population size should maintain gene frequency in the population to some extent against genetic drift. The moderate allele frequency divergence, 0.11–0.27 (net nucleotide distance) (Table 2), among the six groups observed in 260 lines may be caused by natural selection due to fitness of exotic genes in the local environments. Although partial random mating was initiated from F 2 generation, some exotic genes could have been lost from F 1 to F 2 generation. In later generations, especially during the 12 selfing generations for the development of inbred lines, the alleles which conferred increased fitness to the local environments would have been naturally selected. In addition, breeding method applied during the development of inbred lines may have played some effects for the genetic differentiation. For example, single bolls were collected from individual plants and bulked in each generation during the development of inbred lines. The plants with greater seeds per boll or bigger bolls would have higher chance to be represented in the subsequent generation. After 12 generations of selfing, many plants advanced in the germplasm population possessed high seeds per boll or big bolls and became genetically different from the rest of plants. These factors with other unknown ones could have contributed to the genetic differentiation among plants in this germplasm population.
Tremendous efforts have been reported to identify and map QTLs controlling fiber traits in the last 10 years (Jiang et al. 1998; Shappley et al. 1998; Ulloa and Meredith 2000; Kohel et al. 2001; Zhang et al. 2003; Mei et al. 2004; Lacape et al. 2005; Shen et al. 2005; Zhang et al 2005; Abdurakhmonov et al. 2007). A large number of QTLs for fiber-related traits have been identified. For example, 80 QTLs for 10 fiber-related traits were mapped to 22 chromosomes using multiple backcross populations derived from a cross between G. hirsutum and G. barbadense (Lacape et al. 2005). Most of these QTLs have been localized to chromosomes by linkage groups using chromosome substitution lines which lack single chromosomes or single chromosome arms. It would be unpractical to compare the QTLs identified at different laboratories due to different marker systems, markers, or genetic populations screened. There is also a lack of repeatability for these QTLs among genetic populations due to complexity of the fiber traits and significant effects of G × E within different genetic populations, locations, and generations (Paterson et al. 2003; Lacape et al. 2005). Nevertheless, some “QTL-rich” chromosomes have been repeatedly identified for the same QTLs among different laboratories. These chromosomes include Chromosomes 12, 23, 18, and 26 for lint percent (Zhang et al. 2005; Abdurakhmonov et al. 2007) and Chromosomes 4, 18, and 26 for fiber length (Kohel et al. 2001; Mei et al. 2004; Lacape et al. 2005; Zhang et al. 2005). Chromosome locations of some fiber trait-associated markers identified in the current study are in agreement with the “QTL-rich” chromosomes in literature. These markers include BNL1317191 (Chromosome 9/23) and JR295108 (Chromosome 12/26) associated with lint percent and BNL569144 (Chromosome 18Lo), BNL2495195 (Chromosome 26Lo), and CIR170162 (Chromosome 26) with 2.5% span length. In addition, strength-associated BNL2986158 (Chromosome 16), elongation-associated BNL2960148 (Chromosome 10), and length-associated BNL2495195 (Chromosome 26) are consistent with a QTL map, a comprehensive study of QTLs associated with fiber quality, recently reported by Lacape et al. (2005). Finally, micronaire-associated BNL3408130 (Chromosome 3Sh or 17Lo) was also reported in another association study by Kantartzi and Stewart (2008) in Gossypium arboreum accessions. The remaining fiber trait-associated markers were identified on chromosome locations where the related QTLs have not been reported previously.
The results in this study imply potential application of these trait-associated markers in cotton breeding. Since marker–trait associations may be identified in breeder’s populations, breeders can use these molecular markers directly for genetic improvement of lint yield and fiber quality in their breeding programs. Results also imply that molecular markers associated with fiber-related traits can be used to determine genetic mechanisms underlying interrelationships among the traits. Usually, analysis of interrelationships among fiber-related traits is not simple due to confounding effects among the phenotypes analyzed. If molecular markers can be identified associated with these traits through association mapping, these markers may help dissect the traits and unravel the interrelationships since molecular markers such as SSR can be relatively easily located on chromosomes. Analysis of the correlated traits based on chromosome locations of the associated markers would be less confounding than analysis solely based on phenotypes.
In summary, SP germplasm is a desirable genetic resource to screen for marker–trait associations. There were six groups identified in 260 lines by Structure 2.2 with different allele frequency divergence among the groups. There were 59 SSR markers identified significantly associated with six fiber-related traits in experimentwise t tests. Thirty-nine of these marker–trait associations remained significant after correction by accounting for population substructure and relatedness among individual lines. Population substructure affects association analysis differently among traits with most significant effect in boll weight and least effect in micronaire and elongation in the SP germplasm population.
References
Abdurakhmonov LY, Buriev ZT, Saha SA, Pepper E, Musaev JA, Almatov A, Shermatov SE, Kushanov FN, Mavlonov GT, Reddy UK, Yu JZ, Jenkins JN, Kohel RJ, Abdukarimov A (2007) Microsatellite markers associated with lint percentage trait in cotton, Gossypium hirsutum. Euphytica 156:141–156
Ball RD (2005) Experimental designs for reliable detection of linkage disequilibrium in unstructured random population association studies. Genetics 170:859–873
Bernardo R (2002) Breeding for quantitative traits in plants. Stemma Press, Woodbury, Minnesota
Blenda A, Scheffler J, Scheffler B, Palmer M, Lacape JM, Yu JZ, Jesudurai C, Jung S, Muthukumar S, Yellambalase P, Ficklin S, Staton M, Eshelman R, Ulloa M, Saha S, Burr B, Liu S, Zhang T, Fang D, Pepper A, Kumpatla S, Jacobs J, Tomkins J, Cantrell R, Main D (2006) CMD: a cotton microsatellite database resource for Gossypium genomics. BMC Genomics 7:132
Bradbury PJ, Zhang Z, Kroon DE, Casstevens TM, Ramdoss Y, Buckler ES (2007) TASSEL: software for association mapping of complex traits in diverse samples. Bioinformatics 23:2633–2635
Fehr WR (1987) Principles of cultivar development. Vol. 1. Theory and technique. Macmillan Publishing Company, New York
Flint-Garcia SA, Thornsberry JM, Buckler ES (2003) Structure of linkage disequilibrium in plants. Ann Rev Plant Biol 54:357–574
Gebhardt C, Ballvora A, Walkemeier B, Oberhagemann P, Schüler K (2004) Assessing genetic potential in germplasm collections of crop plants by marker-trait association: a case study for potatoes with quantitative variation of resistance to late blight and maturity type. Mol Breed 13:93–102
Goodman SN (1999) Toward evidence-based medical statistics, 2: The Bayes factor. Annals Internal Medicine 130:1005–1013
Ivandic V, Thomas WTR, Nevo E, Zhang Z, Forster BP (2003) Associations of simple sequence repeats with quantitative trait variation including biotic and abiotic stress tolerance in Hordeum spontaneum. Plant Breed 122:300–304
Jiang CX, Wright RJ, El-Zik KM, Paterson A (1998) Polyploid formation created unique avenues for response to selection in Gossypium (cotton). Proc Natl Acad Sci USA 95:4419–4424
Kantartzi SK, Stewart JMcD (2008) Association analysis of fibre traits in Gossypium arboretum accessions. Plant Breed 127:173–179
Knapp SJ, Stroup WW, Ross WM (1985) Exact confidence intervals for heritability on a progeny mean basis. Crop Sci 25:192–194
Kohel RJ, Yu J, Park YH, Lazo GR (2001) Molecular mapping and characterization of traits controlling fiber quality in cotton. Euphytica 121:163–172
Lacape JM, Nguyen TB, Courtois B, Belot JL, Giband M, Gourlot JP, Gawryziak G, Rouques S, Hau B (2005) QTL analysis of cotton fiber quality using multiple Gossypium hirsutum × Gossypium barbasense backcross generations. Crop Sci 45:123–140
Liu K, Muse SV (2005) PowerMarker: Integrated analysis environment for genetic marker data. Bioinformatics 21:2128–2129
Loiselle BA, Sork VL, Nason J, Graham C (1995) Spatial genetic structure of a tropical understory shrub, Psychotria officinalis (Rubiaceae). Am J Bot 82:1420–1425
Lynch M, Milligan BG (1994) Analysis of population genetic structure with RAPD markers. Mol Ecol 3:91–99
MacDonald S, Meyer LA (2007) A new world for U.S. cotton producers. United States Department of Agriculture, Economic Research Service. Available at http://www.ers.usda.gov/AmberWaves/April07/Findings/Cotton.htm Cited 15 Sept 2008
Mei M, Syed NH, Gao W, Thaxton PM, Smith CW, Stelly DM, Chen ZJ (2004) Genetic mapping and QTL analysis of fiber-related traits in cotton) Gossypium). Theor Appl Genet 108:280–291
Paterson AH, Brubaker CL, Wendel JF (1993) A rapid method for extraction of cotton (Gossypium spp.) genome DNA suitable for RFLP or PCR analysis. Plant Mol Biol Rep 11:122–127
Paterson AH, Saranga Y, Menz M, Jiang CX (2003) QTL analysis of genotype × environment interactions affecting cotton fiber quality. Theor Appl Genet 106:384–396
Pritchard JK, Rosenberg NA (1999) Use of unlinked genetic markers to detect population stratification in association studies. Am J Hum Genet 65:220–228
Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure from multilocus genotype data. Genetics 155:945–959
Pritchard JK, Wen X, Falush D (2007) Documentation for structure software: Version 2.2. Department of Human Genetics, University of Chicago; Department of Statistics, University of Oxford. Available at http://pritch.bsd.uchicago.edu/software
Risch N, Merikangas K (1996) The future of genetic studies of complex human diseases. Science 273:1516–1517
Ritland K (1996) Estimators for pairwise relatedness and individual inbreeding coefficients. Genet Res 67:1175–1185
Sackville Hamilton NR, Skøt L, Chorlton KH, Thomas ID, Mizan S (2002) Molecular genecology of temperature response in Lolium perenne: 1. Preliminary analysis to reduce false positives. Mol Ecol 11:1855–1863
Schafer AJ, Hawkins JR (1998) DNA variation and the future of human genetics. Nat Biotech 16:33–39
Shappley ZW, Jenkins JN, Zhu J, McCarty JC Jr (1998) Quantitative trait loci associated with agronomic and fiber traits of upland cotton. J Cotton Sci 2:153–163
Shen X, Zhang T, Guo W, Zhu X, Zhang X (2005) Mapping fiber and yield QTLs with main, epistatic, and QTL × environment interaction effects in recombinant inbred lines of upland cotton. Crop Sci 46:61–66
Skøt L, Sackville Hamilton NR, Mizen S, Chorlton KH, Thomas ID (2002) Molecular genecology of temperature response in Lolium prenne: 2. Association of AFLP markers with ecogeography. Mol Ecol 11:1865–1867
Skøt L, Humphreys MO, Armstead I, Heywood S, Skøt KP, Sanderson R, Thomas ID, Chorlton KH, Sackville NR (2005) An association mapping approach to identify flowering time genes in natural populations of Lolium perenne (L.). Mol Breed 15:233–245
Smith CW, Coyle GG (1997) Association of fiber quality parameters and within-boll yield components in upland cotton. Crop Sci 37:1775–1779
Thornsburry JM, Goodman MM, Doebley J, Kresovich S, Nielsen D, Buchler ES (2001) Dwarf8 polymorphisms associate with variation in flowering time. Nat Genet 28:286–289
Ulloa M, Meredith WR (2000) Genetic linkage map and QTL analysis of agronomic and fiber quality traits in an intraspecific population. J Cotton Sci 4:161–170
Vekemans X (2002) AFLP-SURV version 1.0. Distributed by the author. Laboratoire de Génétique et Ecologie Végétale, Université Libre de Bruxelles, Belgium
Wei X, Jackson PA, McIntyre CL, Aitken KS, Croft B (2006) Association between DNA markers and resistance to disease in sugarcane and effects of population substructure. Theor Appl Genet 114:155–164
Wright S (1951) The genetical structure of populations. Ann Eugenics 15:323–354
Xie C, Warburton M, Li M, Li X, Xiao M, Hao Z, Zhao Q, Zhang S (2008) An analysis of population structure and linkage disequilibrium using multilocus data in 187 maize inbred lines. Mol Breed 21:407–418
Yu J, Pressior G, Briggs WH, Bi IV, Yamasaki M, Doebley JF, McMullen MD, Gaut BS, Nielsen DM, Holland JB, Kresovich S, Buckler ES (2006) A unified mixed-model for association mapping that accounts for multiple levels of relatedness. Nat Genet 38:203–208
Zeng L, Meredith WR, Boykin DL, Taliercio E (2007) Evaluation of an exotic germplasm population derived from multiple crosses among Gossypium tetraploid species. J Cotton Sci 11:118–127
Zhang T, Yuan Y, Yu J, Guo W, Kohel RJ (2003) Molecular tagging of a major QTL for fiber strength in upland cotton and its marker-assisted selection. Theor Appl Genet 106:262–268
Zhang ZS, Xiao YH, Luo M, Li XB, Luo XY, Hou L (2005) Construction of a genetic linkage map and QTL analysis of fiber-related traits in upland cotton (Gossypium hirsutum L.). Euphytica 144:91–99
Acknowledgments
The authors acknowledge MSA Genomics Laboratory and Sheron Simpson for running the genotyping samples. We also thank Dr. David M. Stelly (Texas A&M University, College Station, TX), Dr. Brian Scheffler, Jodi Scheffler, Dr. Jeffery Ray, Dr. Anne Gillen, and Dr. Nacer Bellaloui (USDA-ARS, Stoneville, MS) for useful discussion of the research or reviewing the manuscript, Dr. Zhiwu Zhang (Maize Genetics Laboratory, Cornell University) for consult on software TASSEL, and Ms. LaTonya Holmes for excellent technical support.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by J. Bradshaw.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Zeng, L., Meredith, W.R., Gutiérrez, O.A. et al. Identification of associations between SSR markers and fiber traits in an exotic germplasm derived from multiple crosses among Gossypium tetraploid species. Theor Appl Genet 119, 93–103 (2009). https://doi.org/10.1007/s00122-009-1020-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00122-009-1020-7