Abstract
We characterized three lesion mimic necS1 (necrotic Steptoe) mutants, induced by fast neutron (FN) treatment of barley cultivar Steptoe. The three mutants are recessive and allelic. When infected with Puccinia graminis f. sp. tritici pathotypes MCC and QCC and P. graminis f. sp. secalis isolate 92-MN-90, all three mutants exhibited enhanced resistance compared to parent cultivar Steptoe. These results suggested that the lesion mimic mutants carry broad-spectrum resistance to stem rust. In order to identify the mutated gene responsible for the phenotype, transcript-based cloning was used. Two genes, represented by three Barley1 probesets (Contig4211_at and Contig4212_s_at, representing the same gene, and Contig10850_s_at), were deleted in all three mutants. Genetic analysis suggested that the lesion mimic phenotype was due to a mutation in one or both of these genes, named NecS1. Consistent with the increased disease resistance, all three mutants constitutively accumulated elevated transcript levels of pathogenesis-related (PR) genes. Barley stripe mosaic virus (BSMV) has been developed as a virus-induced gene-silencing (VIGS) vector for monocots. We utilized BSMV-VIGS to demonstrate that silencing of the gene represented by Contig4211_at, but not Contig10850_s_at caused the necrotic lesion mimic phenotype on barley seedling leaves. Therefore, Contig4211_at is a strong candidate for the NecS1 gene, which encodes a cation/proton exchanging protein (HvCAX1).
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Plants have developed complex signaling and defense mechanisms to protect themselves from pathogens. One of the resistance reactions is the hypersensitive response (HR), which is characterized by rapid death of cells directly in contact with, or in close proximity to, the invading pathogen. Positive regulation of pathogen recognition initiates a stereotypic set of physiological responses, including influx of calcium and H+, efflux of K+ and Cl−, oxidative burst, defense gene activation, and HR (Nimchuk et al. 2003).
HR is a form of programmed cell death (PCD) that involves genetically defined signaling pathways, which function to rigorously control the process. Lesion mimic mutants are one of a group of plant mutants that develop spontaneous necrotic lesions in the absence of pathogen infection; thus, they are important for understanding disease resistance pathways and HR in plants (Lorrain et al. 2003). A large number of dominant and recessive mutants have been identified in maize (Johal et al. 1995) and Arabidopsis (Lorrain et al. 2003). Lesion mimic mutants, characterized at the gene level, include lls1 (lethal leaf spots) in maize (Gray et al. 1997), which encodes a non-heme iron-binding protein; hlm1 (HR-like lesion mimic) in Arabidopsis, which encodes the cyclic nucleotide-gated ion channel CNGC4 (Balague et al. 2003) and nec1 in barley, which also encodes the homologue of the Arabidopsis HLM1 gene (Rostoks et al. 2006). The phenotypes of two other Arabidopsis lesion-mimic mutants dnd1 and cpn1, both of which exhibit increased disease resistance to pathogens, were shown to be caused by mutations in genes that encode calcium-related proteins: CNGC2, a cation channel that can conduct calcium and copine, a calcium-dependent, phospholipid-binding protein (Clough et al. 2000; Jambunathan et al. 2001).
These studies implicated ion fluxes and calcium in cell death signaling. Calcium is an important secondary messenger involved in plant disease resistance responses. Ca2+ acts as a signaling molecule, communicating primary recognition events to multiple downstream responses such as phytoalexin production, induction of defense genes, and HR. Transient increases in cytosolic free calcium (Ca2+) concentrations are essential for the conversion of signals, such as red light, touch, cold shock, and pathogen infection into adapted biological responses (Bowler et al. 1994; Knight et al. 1991, 1996; Pooviah and Reddy 1993; Sanders et al. 1999). During these biological responses, Ca2+ may be mobilized from the plant vacuole, endoplasmic reticulum or mitochondria, the predominant storage compartments for Ca2+, to act as an intracellular signaling ion. Vacuolar Ca2+/H+ exchange has been observed in various plant species (Evans and Williams 1998; Maeshima 2001). The first plant Ca2+/H+ antiporter to be cloned, Arabidopsis CATION EXCHANGER1 (CAX1), was identified in a yeast suppression screen through its ability to suppress the Ca2+ hypersensitivity of a yeast vacuolar Ca2+-transport mutant (Hirschi et al. 1996). The Arabidopsis cax1 mutant exhibited altered plant development, perturbed hormonal responses, and ion homeostasis (Cheng et al. 2003).
Rpg1 is a stem rust resistance gene in barley (Hordeum vulgare L.) that confers resistance to many, but not all pathotypes of the wheat stem rust pathogen, Puccinia graminis f. sp. tritici (Pgt) (Brueggeman et al. 2002). However, this gene is not effective against isolates of the rye stem rust pathogen, P. graminis f. sp. secalis (Pgs), which attacks both rye and barley. The cultivar (cv.) Steptoe is susceptible to all tested pathotypes of wheat and rye stem rust. Several lesion mimic mutants in a Steptoe background have been characterized (Rostoks et al. 2003). Among these, FN044, FN211, and FN303 showed increased resistance to stem rust compared to the wild type susceptible cv. Steptoe. Therefore, we named these 3 necrotic Steptoe mutants, necS1-1 (FN044), necS1-2 (FN211), and necS1-3 (FN303), respectively. Here, we report that these necS1 mutants are resistant to Pgt pathotypes MCC and QCC and also Pgs isolate 92-MN-90, but not to stripe rust (Puccinia striiformis f. sp. hordei), suggesting broad-spectrum stem rust resistance in an Rpg1-independent manner. Transcript-based cloning identified two candidate genes for NecS1, and a barley stripe mosaic virus (BSMV) virus-induced gene silencing (VIGS) experiment suggested that Contig4211_at is a candidate for the NecS1 gene. The cloning of the candidate NecS1 gene demonstrated that it encodes a cation/proton exchanging protein (HvCAX1) that may be involved in HR signaling.
Materials and methods
Disease phenotyping
Seed of the necS1-1, necS1-2, and necS1-3 mutants, wild type cv. Steptoe, and stem rust resistant (Chevron and Q/SM20) and susceptible (Hiproly) controls were planted in plastic cones filled with a 50:50 mixture of soil and Metro Mix 200 (Vermiculite, peat moss, Perlite, and sand) and fertilized with Osmocote 14-14-14 (Scott’s Company, Marysville, OH: 1.4 g per cone) and Peters Dark Weather 15-0-15 formulation (Scott’s Company, Marysville, OH: 150 g per gallon at 1/16 dilution). Chevron is the original source of the Rpg1 gene and is resistant to many pathotypes of Pgt, but not Pgs. Q/SM20 is a line derived from the Q21861/SM8901 population. It carries the gene rpg4 which confers resistance to Pgt pathotype QCC (among other Pgt pathotypes) and Rpg5 (formerly RpgQ) which confers resistance to isolates of Pgs (Sun and Steffenson 2005). Plants were grown in a growth chamber (20–22°C with 14 h photoperiod provided by 160 W VHO fluorescent and 60 W incandescent lamps) and inoculated with rust when the first leaves were fully expanded, 7 days after planting. A concentration of 5.4 mg of urediniospores per ml of oil (Soltrol 170, Phillips Petroleum, Bartlesville, OK) was applied at a rate of approximately 0.025 mg per plant with an atomizer pressured at 25–30 KPa (Kilo Pascals). The protocols for the infection period were as described by Sun and Steffenson (2005). After the infection period, plants were returned to controlled environment growth chambers. The incubation temperature was 19–21°C for Pgt pathotype QCC and Pgs isolate 92-MN-90. For Pgt pathotype MCC, the temperature was set to 22–25°C because this environment is more conducive for the separation of genotypes with and without the Rpg1 gene.
Twelve to 14 days after inoculation, the infection types (ITs) on each accession were assessed using a 0–4 scale. The IT scale used for barley is a modification of the one developed for wheat by Stakman et al. (1962) and is based primarily on uredinia size as described by Miller and Lambert (1955). Mesothetic reactions (i.e. those that include more than one IT on a single leaf) are common on barley infected with Pgt and Pgs. Thus, all of the ITs observed in each host accession-rust culture combination were recorded in order of their prevalence on the leaf. The symbols + and − denote more and less sporulation of uredinia, respectively, as compared to the original scale description. Thus, a 23-1 reaction denotes that IT 2 was most common, followed by IT 3 (with slightly less sporulation than described in the scale), and finally IT 1 in lowest frequency.
Plant materials and RNA isolation
The three necrotic mutants and cv. Steptoe were grown in a growth chamber maintained at 21°C (16-h light) and 16°C (8-h dark). The primary leaves from ten 10-day-old seedlings were pooled, frozen in liquid nitrogen, and stored at −80°C until needed for RNA isolation. Total RNA was isolated using the hot (60°C) phenol/guanidinium thiocyanate method. Trizol-like reagent is made from 38% saturated phenol, pH 4.3 (Fisher Scientific, Pittsburg, PA), 1 M guanidine thiocyanate (Fisher Scientific), 0.1 M sodium acetate, pH 5.0 and 5% glycerol (Fisher Scientific). RNA was purified further using the RNeasy Midi kit (Qiagen, Valencia, CA). Three independent biological replicates were performed for each genotype.
Microarray analysis
Target synthesis and GeneChip hybridization, washing, staining, and scanning were performed at the Molecular Biology Core at Washington State University. Microarray output was examined visually for excessive background noise and physical anomalies. The default MAS (Microarray Suite 5.0) statistical values were used for all analyses. All probe sets on each array were scaled to a mean target signal intensity of 125, with the signal correlating to the amount of transcript in the sample. An absolute analysis using MAS was performed to assess the relative abundance of the 22,792 represented transcripts based on signal and detection (present, absent, or marginal). The resulting data from the absolute analysis were exported into Microsoft EXCEL and then imported into GeneSifter software (GeneSifter.net, Seattle, WA). Transcripts expressed differentially at a statistically significant level were determined using the Welch t test with variances not assumed equal, a P value cutoff of 0.05 and Benjamini and Hochberg False discovery rate under 5%. Gene expression changes were considered to be significant if the change was greater than twofold.
Allelism tests
Crosses were made among the necS1-1, necS1-2, and necS1-3 mutants in the field at Spillman farm in Pullman, WA during the summer of 2005. Ten F1 seeds from each cross were planted in the greenhouse where the necrosis phenotype was noted at both seedling and adult plant stages. Twenty-five F2 seeds from each cross were planted in the greenhouse and the phenotype noted at the seedling stage.
Southern analysis and genetic mapping
Plant genomic DNA was extracted as previously described (Kleinhofs et al. 1993). DNA probes were labeled with [α-32P] dCTP (New England Nuclear, Boston, MA) using the ALL-IN-ONE random prime kit (SIGMA, St Louis, MO) and hybridized to barley genomic DNA blots. DNA was digested with restriction enzymes following the manufacturer’s recommendations and transferred to nylon membranes (New England Nuclear) by the alkaline-transfer procedure. Hybridizations were at 65°C and a final wash at 65°C with 0.5X SSC, 1% SDS. The Steptoe/Morex “mini-mapper” population, consisting of 35 selected doubled-haploid lines (DHL), was used for placement of markers to the barley Bin map (Kleinhofs and Graner 2002; Kleinhofs et al. 1993). The Contig4211_at and Contig4212_s_at probes for Southern analysis and genetic mapping were RT-PCR products from Steptoe cDNA using primer sets 4211F/4211R and 4212F/4212R (Table 2).
The gene Contig10850_s_at was non-polymorphic with RFLP markers; therefore, primer sets 10850F2 (5′-GAAGGTGGCAAACAACAATACC-3′) and 10850R2 (5′-GTCACATTGTTCCACTCACTTG-3′) were designed to amplify Steptoe and Morex genomic DNA. PCR reactions of 50 μl contained 100 ng of genomic DNA, 0.2mMdNTP mix, 25 pmol of each primer, 2.5 μl of REDTaq DNA polymerase (Sigma), and 5 μl of 10xRedTaq reaction buffer. Amplification was performed in a PTC-100 programmable thermal controller (MJ Research, Cambridge, MA, USA) at 95°C for 4 min, followed by 35 cycles of 95°C for 1 min, 60°C for 1 min, and 72°C for 1 min; this was followed by 7 min at 72°C. Amplification products were sequenced with the BigDye terminator system on ABI Prizm 377 DNA sequencer (Applied Biosystems, Foster City, CA) at the Bioanalytical Center, Washington State University, Pullman. Nine single nucleotide polymorphic sites identified between Morex and Steptoe were separated in a 5% denaturing polyacrylamide gel as described (Yan et al. 2003).
A nec1j (Morex FN338) x necS1-3 (Steptoe) population consisting of 180 F2 plants was used to map the necS1 phenotype and demonstrate cosegregation with molecular markers for contigs 4211_at, 4212_s_at, and 10850_s_at. Plants with the nec1j phenotype have small black pin-point lesions and are very distinct from the necS1-3 phenotype (Rostoks et al. 2003, 2006). The population was examined and identified 75 necrotic plants, which fit the expected 9:7 ratio for two independently segregating genes (X 2 = 0.28). The necrotic plants were then examined visually and using a nec1 specific molecular marker (Rostoks et al. 2006) to eliminate the nec1j homozygous group. The remaining 41 necrotic plants were tested with molecular markers for contigs 4211_at, 4212_s_at, and 10850_at to demonstrate cosegregation with necS1-3 phenotype.
BSMV constructs and cloning of cDNAs
For VIGS experiments, the BSMV vector (Holzberg et al. 2002) was used to silence candidate NecS1 genes in barley. BSMV is a tripartite RNA virus consisting of α, β, and γ genomes. VIGS constructs were engineered by cloning candidate NecS1 gene fragments using gene-specific primers harboring NotI and PacI restriction sites at their extremity into the γ genome. The 359-bp fragment 5′-end of the gene represented by Contig4211_at was cloned with 5′-ATATTAATTAACCTTAGCCATGGATAGTCACTCCGC-3′ forward primer and 5′-TATGCGGCCGCATCAGACTAAGCGCAAACACCCATA-3′ reverse primer. Primer sequences used to clone the 301-bp portion of Contig10850_s_at were forward primer 5′-ATATTAATTAAGGTAGCAACTGATGGTCTTTGGGA-3′ and reverse primer 5′-TATGCGGCCGCATGAAATCCAAATTCCTACTGAT-3′. PCR-amplified fragments from cDNA clones HVSMEm0012D10 (acc. # EF446604) and HVSMEm0006g13 (acc. # EF446603), for Contig4211_at and Contig10850_s_at, respectively, were digested with PacI-NotI and inserted in antisense orientation in the γ.bPDS2-as vector (Holzberg et al. 2002) to generate constructs BSMV.4211as and BSMV.10850as. The BSMV.MCS construct was generated from the multiple cloning site of pBluescript, for use as a virus inoculation control (R. Brueggeman unpublished). Generation of BSMV infectious RNAs from cDNA clones and inoculation procedures were as previously described (Holzberg et al. 2002).
PCR and semi-quantitative RT-PCR
Total RNA samples generated for the microarray experiment as described above were also used for RT-PCR after DNase I digestion (Ambion, Austin, TX, USA). Single-strand cDNA was synthesized with the Superscript First Strand Synthesis System (Invitrogen, Carlsbad, CA, USA) using oligo(dT)12–18 as primer. PCR was performed using RedTaq DNA polymerase (Sigma, St Louis, MO, USA) as described with primers listed in Table 2. The barley ubiquitin gene (GenBank accession M60175) was used as a control with primer sequences described by Rostoks et al. (2003). PCR was carried out with 20–50 ng genomic DNA using primers listed in Table 2. Five primer pairs covering the Contig4211_at gene open reading frame are 4211F/4211R, 4212F/4212R, 4211F1/4211R1, 4211F3/4211R3, and 4212F1/4211R4 (Table 2).
Data access
All detailed data and protocols from these experiments have been deposited in the Plant Expression Database (www.plexdb.org). Files are categorized under accession number BB54 and can be downloaded as individual CEL, CHP, DAT, or EXP files under “Downloads”.
Results
Barley lesion mimic mutants have enhanced resistance to stem rust
We reported previously the isolation of several fast-neutron mutant lines derived from cv. Steptoe that displayed a disease lesion mimic phenotype (Rostoks et al. 2003). Mutants necS1-1, necS1-2, and necS1-3 were similar in their general appearances and in the development of necrotic lesions, except that necS1-3 plants were taller and more vigorous than the other two mutant plants. Some lesion mimic mutants exhibit extensive necrotic areas across their leaf surface (e.g. >60%). This is certain to affect the infection process by biotrophic pathogens like the rusts, which require healthy cells to infect and ramify in the host. The three mutants described here had relatively low (about 20–30%) areas of leaf necrosis; thus, there were sufficiently large areas of non-necrotic (healthy) tissue for the rust fungus to infect and to observe the resulting infection types (ITs). When infected with Pgt pathotypes MCC and QCC and Pgs isolate 92-MN-90, all three mutants exhibited enhanced resistance compared to wild type cv. Steptoe. The enhanced resistance was manifested by markedly smaller and fewer uredinia (Table 1, Fig. 1). ITs on the mutants ranged from 0 to 23-1, with an occasional three type, compared to ITs of 3-2 to 3 for Steptoe (Sun and Steffenson 2005; see “Materials and methods”). Other lesion mimic mutants tested (12 total) showed infection patterns identical to the parent cv. Steptoe (data not shown). These results suggest that the necS1 lesion mimic mutants necS1-1, necS1-2, and necS1-3 carry broad-spectrum resistance to not only different races of Pgt, but also to Pgs, another forma specialis of P. graminis. Interestingly, when infected with barley stripe rust, P. striiformis f. sp. hordei races PSH-14, PSH-48, and PSH-54, the necS1 mutants showed susceptibility comparable to the parent cv. Steptoe (data not shown).
Allelism tests
Because the necS1-1, necS1-2, and necS1-3 mutants have similar necrotic phenotypes and increased resistance to stem rust, we conducted allelism tests. All ten F1 plants from each of three crosses necS1-1/necS1-2, necS1-1/necS1-3, and necS1-2/necS1-3 showed necrotic phenotypes, suggesting that these three mutants are allelic. The allelism tests were confirmed in the F2 generation and were in accordance with previous observations that these three necS1 mutations are inherited in a recessive manner.
Transcript-based cloning identified the candidate NecS1 gene
Fast neutron bombardment induces deletion mutations (Li et al. 2001). Since the FN mutants were induced with fast neutrons, we expected that they would be due to deletions. We hypothesized that mutations abolishing transcript presence or stability may be identified by using microarray analysis, which examines transcript abundance. We compared basal gene expression profiles in wild type cv. Steptoe and necS1 mutant lines using the Affymetrix Barley1 GeneChip (Close et al. 2004) that contains >22,000 expressed genes. The expression profiles of necS1-1, necS1-2, and necS1-3 were very similar, which is consistent with the results of the allelism tests. At significance level of P = 0.05, 1558, 1081, and 1108 genes were down-regulated and 2184, 1590, and 1583 genes were up-regulated in necS1-1, necS1-2, and necS1-3 mutants, respectively, compared with wild type cv. Steptoe. All three necS1 mutants shared the same 544 down-regulated genes and 841 up-regulated genes compared with Steptoe (Supplemental Table 1; Fig. 2).
Venn diagram for down-regulated genes (a) and up-regulated genes (b) in necS1-1, necS1-2 and necS1-3 mutants compared to wild type Steptoe. a There are 544 overlapping genes that were down-regulated in necS1-1 (Left), necS1-2 (Right) and necS1-3 (Bottom). b There are 841 overlapping genes that were up-regulated in necS1-1 (Left), necS1-2 (Right) and necS1-3 (Bottom)
In order to identify the deleted gene in the necS1 mutants, 30 of the most highly down-regulated genes were tested one by one using genomic PCR, RT-PCR (data not shown) and Southern blot hybridization (data not shown). These analyses identified two genes, represented by three probesets (Contig4211_at, Contig4212_s_at and Contig10850_s_at) that were apparently deleted in all three necS1 mutants (Fig. 3). Lack of PCR amplification from necS1 mutants’ genomic DNA and RNA using the primer pairs 4211F/4211R, 4212F/4212R, and 10850F/R (Fig. 3; Table 2) confirmed that the genes corresponding to Contig4211_at, Contig4212_s_at and Contig10850_s_at were deleted and transcriptionally silent in these necS1 mutant lines. These data were in agreement with the microarray data. Both Contig4211_at and Contig4212_s_at represent a single gene, encoding a Ca2+/H+ exchanging protein. Contig10850_s_at encodes a protein phosphatase 2C (PP2C). Contig4601_s_at, encoding a putative ubiquitin-specific protein, was deleted only in necS1-1 and necS1-2, but not in the necS1-3 mutant. Two other unknown genes, detected by probesets Contig4845_at and HW03M24T_s_at, were deleted only in the necS1-3 mutant line (Fig. 3, Table 2). Therefore, Contig4601_s_at, Contig4845_at and HW03M24T_s_at were removed from further candidacy for the NecS1 gene. The remaining 25 highly down-regulated genes are still present in the necS1 mutants’ genome as demonstrated by PCR-amplification with the corresponding primer sets (Table 2).
Genetic mapping of Contig4211_at, Contig4212_s_at, and Contig10850_at
In the nec1j (Morex) x necS1-3 (Steptoe) 180 F2 population 75 were identified as necrotic. Of these, 34 were identified as nec1j homozygotes. The remaining 41 necrotic plants with necS1 phenotype were examined with molecular markers for Contig4211_at, Contig4212_s_at, and Contig10850_s_at and showed absolute co-segregation. This indicated very close linkage, which can not be calculated directly due to absence of recombinants. However, using Muller’s formulae (Muller 1923) the chance of failure to detect at least one recombinant in this population at the 5% recombination level is 1%. The chance of failure to detect at least one recombinant at 1% recombination increases to 40%.
Contig4211_at, Contig4212_s_at, and Contig10850_s_at mapped to chromosome 3(3H) bin 6, co-segregating with the marker ABG399. Thus, the deletions in this genetic region encompass the putative NecS1 gene. We hypothesize that one, or both of these genes may be responsible for the necrosis and defense response phenotype of the necS1-1, necS1-2, and necS1-3 mutants.
Necrotic phenotype on BSMV-Silenced barley leaves
In order to assess the function of candidate NecS1 genes in the cell death-signaling pathway, we used the BSMV-VIGS strategy. If silencing of a candidate gene could cause the lesion mimic phenotype in a barley line, then this candidate gene is likely to be the NecS1 gene. The barley line Q21861 was selected from several different barley cultivars tested under our experiment condition. It tolerates substantial levels of BSMV accumulation required to elicit a significant VIGS response (Zhang et al. 2007; Brueggeman et al. 2007). In the case of Rpr1 gene, the reduction of endogenous Rpr1 mRNA was 50–75% measured by quantitative real-time PCR in our Q21861 system (unpublished data). Q21861 was used in this BSMV-VIGS experiment because it harbors both Contig4211_at and Contig10850_s_at based on the fact that both genes can be amplified with the five primer pairs spanning the entire open reading frame of these genes from Morex, Steptoe and Q21861 genomic DNA (Fig. 4).
The gene represented by Contig4211_at appears to be present and intact in cvs. Morex, Steptoe and Q21861. Five Contig4211_at gene fragments (lanes 1–5) were amplified with primer pairs covering the open reading frame. Lane 1 4211F/4211R, Lane 2 4211F1/4211R1, Lane 3 4211F3/4211R3, Lane 4 4211F4/4211R4, Lane 5 4212F/4212R. The primer sequences are given in Table 2
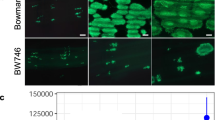
VIGS constructs with antisense cDNA fragments from Contig4211_at and Contig10850_s_at (BSMV.4211as, BSMV.10850as) were tested. First and second leaves of 12-day-old Q21861 seedlings were inoculated with each of these constructs. As the plants grew, mild viral symptoms such as yellow or white streaks were typically observed in many BSMV-infected leaves (Fig. 5). As early as 7 days post virus infection, the leaves infected with BSMV.4211as started to develop necrotic spots similar to the original necrotic FN mutants, necS1-1, necS1-2, and necS1-3. At 14 days post virus infection, more prominent necrotic spots developed in different leaves of BSMV.4211as infected plants (Fig. 5). The necrotic spots were easily distinguishable from viral symptoms by their color and discrete size. No necrotic spots were observed in BSMV.10850as and BSMV.MCS infected control plants. The Q21861 seedlings without virus inoculation grown along with BSMV-infected plants were green and healthy, ruling out environmental stress as a possible cause of the necrosis. The necS1-1, necS1-2, and necS1-3 mutants were planted and grown at the same time in the same growth chamber for comparison. They displayed necrotic symptoms 7 days after planting and the necrotic area became more extensive as they grew older. Two repetitions of this experiment produced the same results. A total of 10 out of 40 BSMV.4211as infected plants developed necrotic spots. In contrast, none of the 40 plants infected each with BSMV.10850as or BSMV.MCS showed necrotic spots. These results suggest that Contig4211_at is a strong candidate for the NecS1 gene. We realize that VIGS experiments are subject to artifacts and off-target effects, therefore further research with a different target sequence within the gene or an entirely different experimental approach may be required for positive proof that Contig4211_at is the NecS1 gene.
The candidate barley NecS1 gene (Contig4211_at), silenced by BSMV-VIGS, produced a phenotype with necrotic spots while the candidate gene contig 10850_s_at and controls did not. Three biological replicates for each VIGS construct are presented. First and second leaves of 12-day-old Q21861 seedlings were inoculated with each of these constructs. Starting 7-day post infection, new leaves were scored for necrotic lesions. Seedling leaves infected with BSMV.4211as construct showed large necrotic spots 14 days post infection, while seedlings inoculated with BSMV.10850as did not. Infection with BSMV.MCS, used as a negative control, also failed to elicit a necrosis response. Forty seedlings were used for each of two repetitions for the two candidate gene tests while 20 seedlings were used for two repetitions of the BSMV.MCS negative control. FN044, FN211 and FN303 are original necrotic mutants necS1-1, necS1-2 and necS1-3, respectively. Un-inoculated Q21861 seedlings were green and healthy, growing under the same growth condition as the BSMV infected plants. All photographs were taken of 26-day-old plants grown under growth chamber condition
Contig4211_at, the NecS1 gene, encodes a cation/proton exchanging protein (CAX1) in barley
A search for Contig4211_at and Contig4212_s_at specific cDNA clones in the HarvEST database (http://138.23.191.142/hweb) identified several in the barley libraries (http://www.genome.arizona.edu/genome/barley.html). The longest cDNA clone HVSMEm0012D10 (acc. # EF446604) was sequenced. Sequence analysis of this 1,869-bp cDNA clone revealed that it contains a 1,389-bp open reading frame (ORF), and translation of this ORF would produce a protein of 462 amino acids. A search of current databases with the deduced amino acid sequence revealed significant similarity to various plant CAX1 proteins (See Supplemental Fig. 1). The deduced amino acid sequence is 81, 73, and 64% identical to Oryza sativa, Zea mays, and Arabidopsis thaliana cation/proton exchanger 1 (CAX1) proteins, respectively; therefore, we named the protein HvCAX1. The AtCAX1 and OsCAX1a have been localized to vacuolar membranes and predicted to have 11 trans-membrane domains (TMs) (Cheng et al. 2003; Kamiya and Maeshima 2004; Shigaki et al. 2006). The same 11 TMs were found in HvCAX1 by the TMpred program (http://www.ch.embnet.org/software/TMPRED_form.html) (See Supplemental Fig. 1). AtCAX1 is a high capacity Ca2+-specific transporter, while OsCAX1a transports Ca2+ into vacuoles and is involved in Ca2+ homeostasis in cells that suffer from high concentrations of Ca2+. Therefore, we have identified a candidate NecS1 gene as encoding the first CAX1 protein described in barley. The NecS1 gene can be amplified from barley cvs. Morex, Steptoe and Q21861 (Fig. 4), suggesting the gene sequence is conserved among these barley cultivars.
Defense-related barley genes are induced in necS1 mutants
Many lesion mimic mutants exhibit a state of increased disease resistance and show high, constitutive levels of pathogenesis-related (PR) gene expression (Lorrain et al. 2003). Microarray analysis showed that these three necrotic mutants display enhanced expression of an array of defense-related genes. Among 841 overlapping up-regulated genes in necS1-1, necS1-2, and necS1-3 compared to wild type Steptoe (Fig. 2), there are several classes of pathogenesis-related genes. The highly up-regulated genes include glucan endo-1,3-beta-glucanase, pathogenesis-related proteins 1, 2, 4, 5, PR-10a, thaumatin-like protein TLP8, and others. To validate the microarray data, we conducted semi-quantitative RT-PCR on five up-regulated defense-related genes (Fig. 6), Contig4405_x_at (pathogenesis-related protein PR-10a), Contig2212_s_at (pathogenesis-related protein prb1-3 precursor) Contig2214_s_at (pathogenesis-related protein 1a), Contig2550_x_at (pathogenesis-related protein 4), and HVSMEm0003C15r2_s_at (endo-1,3-beta-glucanase) with the corresponding primer sets (Table 2). Our results show that each of the five genes tested exhibited increased transcript accumulation in necS1-1, necS1-2, and necS1-3 plants compared to wild type Steptoe, confirming the microarray data.
Discussion
We identified a novel lesion-mimic mutant necS1 that rendered barley cv. Steptoe resistant to several stem rust pathotypes/cultures in an Rpg1-independent manner. Although lesion mimic mutants could result from mutations that affect plant cell physiology and therefore may be unrelated to disease defense responses, at least some of them are thought to represent defects in genes that regulate HR cell death programs directly. A series of Arabidopsis mutants that exhibit constitutive initiation of HR-like cell death in the absence of a pathogen have been isolated, and the mutants showed heightened resistance to virulent bacterial and oomycete pathogens when lesions were present (Dietrich et al. 1994; Greenberg et al. 1994). In addition, mutations that result in the constitutive expression of defense mechanisms, such as the cpr mutations cpr1 (Bowling et al. 1994), cpr5 (Bowling et al. 1997), cpr20 and cpr21 (Silva et al. 1999), and cpr22 (Yoshioka et al. 2001), cause spontaneous lesion phenotypes.
Although the lesion mimic mutants necS1-1, necS1-2, and necS1-3 exhibited enhanced resistance to several pathotypes/isolates of P. graminis, they did not show the same effect against barley stripe rust (P. striiformis f. sp. hordei, Psh). Like barley mlo mutants, the resistance trait conferred by necS1 is recessive and probably non-race specific. Homozygous mutant (mlo) alleles of the Mlo gene confer broad-spectrum disease resistance to the biotrophic powdery mildew fungus, Blumeria graminis f. sp. hordei. However, barley mlo mutants do not differ in their infection phenotype to a range of other phytopathogens, such as barley leaf rust (P. hordei), and stripe rust (Psh) (Jorgensen 1977). The mlo mutants exhibit enhanced susceptibility to the fungal pathogens Magnaporthe grisea and Bipolaris sorokiniana (Jarosch et al. 1999; Kumar et al. 2001). It is plausible that each pathogen species evolved its own specific means to suppress and overcome general or specialized host-defense mechanisms (Panstruga 2005).
Fast neutron generates deletions in the plant genome. In Arabidopsis the size of these deletions can range from 0.8 to 12 kb (Li et al. 2001). In barley, deletions up to 100 kb have been observed (Zhang et al. unpublished data). In one study of wheat fast-neutron mutants, the largest deletions span 70 cM of genetic distance, and the smallest span 6 cM, which may correspond to 2 Mb (Faris et al. 2003). Transcript-based cloning (Mitra et al. 2004; Zhang et al. 2006) is a gene cloning method based on expression-level polymorphism between a wild type plant and mutant by a microarray approach. In this study, microarray analysis quickly identified candidate NecS1 genes based on deletions present in the necS1 mutants. At the same time, many up regulated genes in the necS1 mutants, compared to wild type Steptoe, were defense-related and highly expressed, which may explain the disease resistance to several stem rust pathotypes. However, since Barley1 GeneChip does not include all the barley genes and fast-neutron generated deletion in barley can be very large, microarray analysis may not be able to identify all the deleted genes. In a search of rice genome sequences (http://www.tigr.org/tigr-scripts/osa1_web/gbrowse/rice/), both Contig4211_at and Contig10850_s_at are located in chromosome 1 in rice, containing about 40 genes in a ~668 kb region. It is possible that other genes in the deletions might influence the necrotic phenotype.
Our results suggested that Contig4211_at is a strong candidate for the NecS1 gene. The NecS1 gene is predicted to encode a cation/proton-exchanging protein, HvCAX1. CAXs (for CAtion eXchangers) are a group of proteins that export cations from the cytosol to maintain optimal ionic concentrations in the cell. CAXs are thought to play important roles in signal transduction. In signaling events, the basal Ca2+ concentration in the cytosol is restored by sequestering the transient increase of free calcium into the vacuole (Sanders et al. 1999). CAXs are a multigene family in Arabidopsis (Maser et al. 2001) and rice (Kamiya et al. 2005). Arabidopsis CAX1 is localized on the vacuolar membrane (Cheng et al. 2003). The Arabidopsis cax1 mutant showed increased tolerance to a variety of ionic stresses, suggesting that the disruption in Ca2+ loading into the vacuole affects some signaling pathways. Five cation/H+ exchangers (CAX) were identified from rice, four isoforms of OsCAX conferred tolerance to calcium (Kamiya et al. 2005). This candidate NecS1 is the first CAX gene function identified from barley, and deletion of this gene resulted in a lesion mimic phenotype and increased disease resistance. One hypothesis for this effect is that the sequestration of Ca2+ in the vacuole is blocked so that cytosolic Ca2+ ions are extremely high, thereby perturbing Ca2+-dependent processes. Various abiotic stresses induce Ca2+ release from different stores, such as the vacuole. In response to pathogen attack, the influx of calcium is one of the earliest events (Atkinson et al. 1990). Calcium influx and the transient increase in cytosolic calcium levels after elicitor treatment are necessary for the induction of the oxidative burst, one of the signaling events associated with HR. In the necS1 mutant, cytosolic Ca2+ is probably constitutively high even in the absence of pathogen because of the disruption of HvCAX1, leading to the induction of oxidative burst and spontaneous HR.
Recent studies have demonstrated that vectors based on BSMV can be effective reverse-genetics tools for the analysis of gene function in monocots. BSMV-mediated silencing of phytoene desaturase (PDS) caused a robust photobleaching phenotype in barley (Holzberg et al. 2002). In another study, plants inoculated with BSMV carrying PDS fragment gave rise to a 70 to 84% reduction in PDS mRNA (Bruun-Rasmussen et al. 2007). Furthermore, Hein et al. (2005) and Scofield et al. (2005) have demonstrated functional characterization of genes associated with disease resistance in barley and wheat using BSMV-VIGS. In our study, silencing of the barley candidate NecS1 gene led to a visual necrotic phenotype, milder, but nevertheless similar to the original fast neutron generated mutants. This result strongly suggested that the mutation of the NecS1 gene might lead to spontaneous HR and consequently increased stem rust resistance in barley. In Arabidopsis, a T-DNA insertion mutant, cax1, exhibited altered plant development, perturbed ion homeostasis and hormone sensitivity, as well as altered expression of an auxin-regulated promoter-reporter gene fusion (Cheng et al. 2003). Although necrosis was not reported in the cax1 mutant, it would be interesting to test expression of some defense-related genes and their corresponding disease reaction to a variety of pathogens in Arabidopsis.
In summary, the appearance of necrotic lesions suggests that HR is affected in the necS1 mutants. Certain defense-related genes are highly expressed and are constitutive, suggesting that they may have contributed to broad-spectrum stem rust resistance in these mutants. Transcript-based cloning was used to isolate the barley NecS1 gene, and to identify a strong candidate as encoding a predicted CAX1 protein. The identification of this gene was facilitated by inhibition of the wild-type gene expression by BSMV-VIGS delivered antisense gene fragments generating a phenotype similar to the necS1 phenotypes. The presented results implicating the gene as a putative component in the HR pathway, should allow us to address questions regarding the molecular mechanisms leading to a necrotic phenotype and to advance our understanding of plant HR, calcium signaling and disease resistance.
References
Atkinson MM, Keppler LD, Orlandi EW, Baker CJ, Mischke CF (1990) Involvement of plasma membrane calcium influx in bacterial induction of the K+/H+ and hypersensitive response in tobacco. Plant Physiol 92:215–221
Balague C, Lin B, Alcon C, Flottes G, Malmstrom S, Kohler C, Neuhaus G, Pelletier G, Gaymard F, Roby D (2003) HLM1, an essential signaling component in the hypersensitive response, is a member of the cyclic nucleotide-gated channel ion channel family. Plant Cell 15:365–379
Bowler C, Neuhaus G, Yamagata H, Chua NH (1994) Cyclic GMP and calcium mediate phytochrome phototransduction. Cell 77:73–81
Bowling SA, Guo A, Cao H, Gordon AS, Klessig DF, Dong X (1994) A mutation in Arabidopsis that leads to constitutive expression of systemic acquired resistance. Plant Cell 6:1845–1857
Bowling SA, Clarke JD, Liu Y, Klessig DF, Dong X (1997) The crp5 mutant of Arabidopsis expresses both NPR1-dependent and NPR1-independent resistance. Plant Cell 9:1573–1584
Brueggeman R, Rostoks N, Kudrna D, Kilian A, Han F, Chen J, Druka A, Steffenson B, Kleinhofs A (2002) The barley stem rust-resistance gene Rpg1 is a novel disease-resistance gene with homology to receptor kinases. Proc Natl Acad Sci USA 99:9328–9333
Brueggeman R, Druka A, Nirmala J, Drader T, Cavileer T, Bennypaul H, Gill K, Steffenson B, Kleinhofs A (2007) The barley stem rust resistance gene Rpg5 encodes the NBS-LRR and protein kinase domains in a single gene. Keystone symposia abstract, Plant Cell Biology, Coeur d’Alene, ID, p 109
Bruun-Rasmussen M, Madsen CT, Jessing S, Albrechtsen M (2007) Stability of Barley stripe mosaic virus-induced gene silencing in barley. Mol Plant Microbe Interact 20:1323–1331
Cheng NH, Pittman JK, Barkla BJ, Shigaki T, Hirschi KD (2003) The Arabidopsis cax1 mutant exhibits impaired ion hemeostasis, development, and hormonal responses and reveals interplay among vacuolar transporters. Plant Cell 15:347–364
Close TJ, Wanamaker S, Caldo RA, Turner SM, Ashlock DA, Dickerson JA, Wing RA, Muehlbauer GJ, Kleinhofs A, Wise RP (2004) A new resource for cereal genomics: 22 K barley GeneChip comes of age. Plant Physiol 134:960–968
Clough SJ, Fengler KA, Yu IC, Lippok B, Smith RKJ, Bent AF (2000) The Arabidopsis dnd1 “defense no death” gene encodes a mutated cyclic nucleotide-gated ion channel. Proc Natl Acad Sci USA 97:9323–9328
Dietrich RA, Delaney TP, Uknes SJ, Ward ER, Ryals JA, Dangl JL (1994) Arabidopsis mutants simulate disease resistance response. Cell 77:565–577
Evans DE, Williams LE (1998) P-type calcium ATPases in higher plants- biochemical, molecular and functional properties. Biochim Biophys Acta 1376:1–25
Faris JD, Fellers JP, Brooks SA, Gill BS (2003) A bacterial artificial chromosome contig spanning the major domestication locus Q in wheat and identification of a candidate gene. Genetics 164:311–321
Gray J, Close PS, Briggs SP, Johal GS (1997) A novel suppressor of cell death in plants by the Lls1 gene of maize. Cell 89:25–31
Greenberg JT, Guo A, Klessig DF, Ausubel FM (1994) Programmed cell death in plants: a pathogen-triggered response activated coordinately with multiple defense functions. Cell 77:551–564
Hein I, Barciszewska-Pacak M, Hrubikova K, Williamson S, Dinesen M, Soenderby IE, Sundar S, Jarmolowski A, Shirasu K, Lacomme C (2005) Virus-induced gene silencing-based functional characterization of genes associated with powdery mildew resistance in barley. Plant Physiol 138:2155–2164
Hirschi KD, Zhen RG, Cunningham KW, Rea PA, Fink GR (1996) CAX1, an H+/Ca2+ antiporter from Arabidopsis. Proc Natl Acad Sci USA 93:8782–8786
Holzberg S, Brosio P, Gross C, Pogue GP (2002) Barley stripe mosaic virus-induced gene silencing in a monocot plant. Plant J 30:315–327
Jambunathan N, Siani JM, McNellis TW (2001) A humidity-sensitive Arabidopsis copine mutant exhibits prcocious cell death and increased disease resistance. Plant Cell 13:2225–2240
Jarosch B, Kogel KH, Schaffrath U (1999) The ambivalence of the barley Mlo locus: Mutations conferring resistance against powdery mildew (Blumeria graminis f. sp. hordei) enhance susceptibility to the rice blast fungus Magnaporthe grisea. Mol Plant Microbe Interact 12:508–514
Johal GS, Hulbert S, Briggs SP (1995) Disease lesion mimic mutations of maize: a model for cell death in plants. BioEssays 17:685–692
Jorgensen JH (1977) Spectrum of resistance conferred by ML-O powdery mildew resistance genes in barley. Euphytica 26:55–62
Kamiya T, Maeshima M (2004) Residues in internal repeats of the rice cation/H+ exchanger are involved in the transport and selection of cations. J Biol Chem 279:812–819
Kamiya T, Akahori T, Maeshima M (2005) Expression profile of the genes for rice cation/H+ exchanger family and functional analysis in yeast. Plant Cell Physiol 46:1735–1740
Kleinhofs A, Graner A (2002) An integrated map of the barley genome. In: Vasil I, Phillips RL (ed) DNA-Based Markers in Plants. Kluwer, Boston, pp 187–199
Kleinhofs A, Kilian A, Saghai Maroof MA, Biyashev RM, Hayes P, Chen FQ, Lapitan N, Fenwick A, Blake TK, Kanazin V, Ananiev E, Dahleen L, Kudrna D, Bollinger J, Knapp SJ, Liu B, Sorrells M, Heun M, Franckowiak JD, Hoffman D, Skadsen R, Steffenson BJ (1993) A molecular, isozyme and morphological map of the barley (Hordeum vulgare) genome. Theor Appl Genet 86:705–712
Knight MR, Sampbell AK, Smith SM, Trewavas AJ (1991) Transgenic plant aequorin reports the effects of touch and cold-shock and elicitors on cytoplasmic calcium. Nature 352:524–526
Knight H, Trewavas AJ, Knight MR (1996) Cold calcium signaling in Arabidopsis involves two cellular pools and change in calcium signature after acclimation. Plant Cell 8:489–503
Kumar J, Huckelhoven R, Beckhove U, Nagarajan S, Kogel KH (2001) A compromised Mlo pathway affects the response of barley to the necrotrophic fungus Bipolaris sorokiniana (Telemorph: Cochliobolus sativus) and its toxins. Phytopathology 91:127–133
Li X, Song Y, Century K, Straight S, Ronald PC, Dong X, Lassner M, Zhang Y (2001) A fast neutron deletion mutagenesis-based reverse genetics system for plants. Plant J 27:235–242
Lorrain S, Vailleau F, Balague C, Roby D (2003) Lesion mimic mutants: keys for deciphering cell death and defense pathways in plants? Trends Plant Sci 8:263–271
Maeshima M (2001) Tonoplast Transporters: Organization and Function. Ann Rev Plant Physiol Plant Mol Biol 52:469–497
Maser P, Thomine S, Schroeder JI, Ward JM, Hirschi KD, Sze H, Talke IN, Amtmann A, Maathuis FJ, Sanders D, Harper JF, Tchieu J, Gribskov M, Persans MW, Salt DE, Kim SA, Guerinot ML (2001) Phylogenetic relationships within cation transporter families of Arabidopsis. Plant Physiol 126:1646–1667
Miller JD, Lambert JW (1955) Variability and inheritance of reaction of barley to race 15B of stem rust. Agron J 47:373–377
Mitra RM, Gleason CA, Edwards A, Hadfield J, Downie JA, Oldroyd GE, Long SR (2004) A Ca2+/calmodulin-dependent protein kinase required for symbiotic nodule development: Gene identification by transcript-based cloning. Proc Natl Acad Sci USA 101:4701–4705
Muller HJ (1923) A simple formula giving the number of individuals required for obtaining one of a given frequency. Am Nat 57:66–73
Nimchuk Z, Eulgem T, Holt BFIII, Dangl JL (2003) Recognition and response in the plant immune system. Ann Rev Genet 37:579–609
Panstruga R (2005) Serpentine plant MLO proteins as entry portals for powdery mildew fungi. Biochem Soc Trans 33:389–392
Pooviah BW, Reddy AS (1993) Calcium and signal transduction in plants. CRC Crit Rev Plant Sci 12:185–211
Rostoks N, Schmierer D, Kudrna D, Kleinhofs A (2003) Barley putative hypersensitive induced reaction gene: genetic mapping, sequence analyses and differential expression in disease lesion mimic mutants. Theor Appl Genet 107:1094–1101
Rostoks N, Schmierer D, Mudie S, Drader T, Brueggeman R, Caldwell DG, Waugh R, Kleinhofs A (2006) Barley necrotic locus nec1 encodes the cyclic nucleotide-gated ion channel 4 homologous to the Arabidopsis HLM1. Mol Gen Genomics 275:159–168
Sanders D, Brownlee C, Harper JF (1999) Communicating with calcium. Plant Cell 11:691–706
Scofield SR, Huang L, Brandt AS, Gill BS (2005) Development of a virus-induced gene-silencing system for hexaploid wheat and its use in functional analysis of the Lr21-mediated leaf rust resistance pathway. Plant Physiol 138:2165–2173
Shigaki T, Rees I, Nakhleh L, Hirschi KD (2006) Identification of three distinct phylogenetic groups of CAX cation/proton antiporters. J Mol Evol 63:815–825
Silva H, Yoshioka K, Dooner HK, Klessig DF (1999) Characterization of a new Arabidopsis mutant exhibiting enhanced disease resistance. Mol Plant Microbe Interact 12:1053–1063
Stakman EC, Stewart DM, Loegering WQ (1962) Identification of physiological races of Puccinia graminis f. sp. tritici. USDA/Agricultural Research Service Publication No. E617 (Revised 1962) pp 53
Sun Y, Steffenson BJ (2005) Reaction of barley seedlings with different stem rust resistance genes to Puccinia graminis f. sp. tritici and P. g. f. sp. secalis. Can J Plant Pathol 27:80–89
Yan GP, Chen XM, Line RF, Wellings CR (2003) Resistance gene-analog polymorphism markers co-segregating with the Yr5 gene for resistance to wheat stripe rust. Theor Appl Genet 106:636–643
Yoshioka K, Kachroo P, Tsui F, Sharma SB, Shah J, Klessig DF (2001) Environmentally sensitive, SA-dependent defense responses in the cpr22 mutant of Arabidopsis. Plant J 26:447–459
Zhang L, Fetch T, Nirmala J, Schmierer D, Brueggeman R, Steffenson B, Kleinhofs A (2006) Rpr1, a gene required for Rpg1-dependent resistance to stem rust in barley. Theor Appl Genet 113:847–855
Zhang L, Nirmala J, Drader T, Brueggeman R, Gill U, Gill K, Kleinhofs A (2007) Characterization and functional analysis of Rpr1 gene in barley. Keystone symposia abstract, Plant Cell Biology, Coeur d’Alene, ID, p 321
Acknowledgments
This is Scientific Paper No. 0801-07 from the College of Agricultural, Human, and Natural Sciences Research Center, Washington State University, Pullman, WA 99164, Project 0196. Research was supported by the National Research Initiative of the USDA Cooperative State Research, Education and Extension Service grant number #2004-35301-14635 and by the U.S. Barley Genome Project. We thank Derek Pouchnik, Stephanie Dahl and Tamas Szinyei for excellent technical assistance.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by R. Waugh.
Electronic supplementary material
Rights and permissions
About this article
Cite this article
Zhang, L., Lavery, L., Gill, U. et al. A cation/proton-exchanging protein is a candidate for the barley NecS1 gene controlling necrosis and enhanced defense response to stem rust. Theor Appl Genet 118, 385–397 (2009). https://doi.org/10.1007/s00122-008-0910-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00122-008-0910-4