Abstract
A partially dominant nuclear gene conferring resistance to the imidazolinone herbicides was previously identified in the cultivated sunflower (Helianthus annuus L.) line CLHA-Plus developed by seed mutagenesis. The objective of this study was to characterize this resistant gene at the phenotypic, biochemical and molecular levels. CLHA-Plus showed a complete susceptibility to sulfonylureas (metsulfuron, tribenuron and chlorsulfuron) but, on the other hand, it showed a complete resistance to imidazolinones (imazamox, imazapyr and imazapic) at two rates of herbicide application. This pattern was in close association with the AHAS-inhibition kinetics of protein extracts of CLHA-Plus challenged with different doses of imazamox and chlorsulfuron. Nucleotide and deduced amino acid sequence comparisons between resistant and susceptible lines indicated that the imidazolinone-resistant AHAS of CLHA-Plus has a threonine codon (ACG) at position 122 (relative to the Arabidopsis thaliana AHAS sequence), whereas the herbicide-susceptible enzyme from BTK47 has an alanine residue (GCG) at this position. Since the resistance genes to AHAS-inhibiting herbicides so far characterized in sunflower code for the catalytic (large) subunit of AHAS, we propose to redesignate the wild type allele as ahasl1 and the incomplete dominant resistant alleles as Ahasl1-1 (previously Imr1 or Ar pur ), Ahasl1-2 (previously Ar kan ) and Ahasl1-3 (for the allele present in CLHA-Plus). The higher tolerance level to imidazolinones and the lack of cross-resistance to other AHAS-inhibiting herbicides of Ahasl1-3 indicate that this induced mutation can be used to develop commercial hybrids with superior levels of tolerance and, at the same time, to assist weed management where control of weedy common sunflower is necessary.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Acetohydroxyacid synthase (AHAS; EC 4.1.3.18), also known as acetolactate synthase (ALS) (Duggleby and Pang 2000), is a key enzyme in the biosynthesis of the branched chain amino acids such as valine, leucine and isoleucine in eukaryotes and prokaryotes (Umbarger 1978; Singh 1999; McCourt and Duggleby 2006). The AHAS enzyme from plants is thought to be an assembly consisting of four catalytic and four regulatory subunits (Lee and Duggleby 2001, 2002; Duggleby et al. 2008).
Five structurally diverse herbicide families including the sulfonylureas (Ray 1984), imidazolinones (Shaner et al. 1984), triazolopyrimidines (Subramanian and Gerwick 1989), pyrimidinyloxybenzoates (Subramanian et al. 1990) and sulfonylaminocarbonyl-triazolinones (Santel et al. 1999) are effective in killing susceptible plants by inhibiting AHAS. Inhibition of AHAS leads to plant death primarily because of amino acid starvation, even though other secondary effects of AHAS inhibition such as build up of 2-ketobutyrate, disruption of protein synthesis and disruption of photosynthate transport, have also been implicated in the mechanism of plant death (Shaner 1991).
A variety of crops including corn (Zea mays L.), canola (Brassica napus L.), sugar beet (Beta vulgaris L.), rice (Oryza sativa L.), cotton (Gossypium hirsutum L.), sunflower (H. annuus), flax (Linum usitatissimum L.), soybean [Glycine max (L.) Merr.] and wheat (Triticum aestivum L.), which are resistant to AHAS-inhibiting herbicides, have been developed by a variety of approaches including somatic cell selection, mutation breeding, plant transformation and interspecific hybridization (Anderson and Georgeson 1989; Croughan 1996; D’Halluin et al. 1992; Newhouse et al. 1991, 1992; Hart et al. 1992; Wright and Penner 1998; Swanson et al. 1989; Subramanian et al. 1990; Rajasekaran et al. 1996; Sebastian et al. 1989; Pozniak and Hucl 2004; Al-Khatib and Miller 2002; Mallory-Smith et al. 1990; McHughen 1989). In most of these cases, resistance is due to a form of AHAS that is less sensitive to herbicide inhibition because of reduced herbicide binding caused by mutations in the genes coding for the catalytic subunit of AHAS. Several authors have reviewed known mutations of the AHAS genes that confer resistance to AHAS-inhibiting herbicides in plants (Preston and Mallory-Smith 2001; Tranel and Wright 2002; Tan et al. 2005). No known amino acid substitutions in the regulatory subunit have been reported to confer herbicide resistance.
Wild sunflower (H. annuus) populations resistant to imidazolinones (IMIs) or sulfonylureas (SUs) have been discovered (Al-Khatib et al. 1998; White et al. 2002). The herbicide resistant trait was introgressed into elite inbred lines of sunflower by conventional breeding methods for the purpose of developing and deploying IMI-resistant and SU-resistant cultivars (Al-Khatib and Miller 2002; Miller and Al-Khatib 2002, 2004). Based on molecular studies, Kolkman et al. (2004) identified and characterized three genes coding for the AHAS catalytic subunits in sunflower (AHAS1, AHAS2 and AHAS3) and demonstrated that the IMI-resistant and SU-resistant genes (Ar pur and Ar kan , respectively) were allelic variants of the same locus (AHAS1). Moreover, they showed that Ar pur harbored a C-to-T mutation in codon 205 (Arabidopsis thaliana nomenclature), whereas Ar kan inbreds harbored a C-to-T mutation in codon 197 (Kolkman et al. 2004). Point mutations resulting in proline (Pro) 197 substitutions confer high levels of sulfonylurea resistance with only small increases in resistance to the imidazolinones and the triazolopyrimidines (Haughn et al. 1988; Lee et al. 1988; Guttieri et al. 1992, 1995; Harms et al. 1992; Mourad and King 1992; Wright et al. 1998). On the other hand, alanine (Ala) 205 substitutions confer moderate resistance to all AHAS inhibitors (Bernasconi et al. 1995; Jander et al. 2003).
Recently, an IMI-resistant sunflower line designated CLHA-Plus was developed through EMS mutagenesis and selection with imazapyr (Sala et al. 2008). Inheritance studies indicated that resistance in CLHA-Plus is conferred by a single nuclear partially dominant gene allelic to the previously described mutant alleles. The objective of this study was to characterize the resistant gene present in CLHA-Plus at the phenotypic, biochemical and molecular levels.
Materials and methods
Plant materials
Four sunflower lines were used: CLHA-Plus, BTK47, BTSu-R1 and RHA426. CLHA-Plus is a mutant IMI-resistant line (Sala et al. 2008), BTK47 is a conventional oilseed line used to develop CLHA-Plus by EMS treatment. BTSu-R1 is a SU-resistant line derived from ANN-Kan that was obtained after three generations of selfing and selection for tribenuron methyl [methyl 2-[4-methoxy-6-methyl-1,3,5-triazin-2-yl(methyl)carbamoylsulfamoyl]benzoate] resistance from the genetic stock SURES-2 developed by Miller and Al-Khatib (2004); and RHA426 is an IMI-resistant public line developed by Miller and Al-Khatib (2002) derived from ANN-Pur.
Greenhouse characterization of herbicide tolerance
Comparisons of CLHA-Plus, BTK47, BTSu-R1 and RHA426 for their relative tolerance level to different herbicides and doses were conducted as separate experiments for each herbicide treatment. Eight seeds of the four lines were sown in 20 × 20 × 30 cm pots. After emergence, plantlets were thinned by hand to leave four plantlets per pot (replication). Five pots of each line were treated with a different herbicide or dose, and an untreated control pot of each line was included in each experiment. Plants were grown in a greenhouse under natural light conditions supplemented with 400 W halide lamps to provide a 14 h day length. Day/night temperatures were 25 and 20°C, respectively. At V4 stage of development (Schneiter and Miller 1981), plants were sprayed with either a single dose of one of three sulfonylurea herbicides (3.6 g a.i. ha−1 of metsulfuron methyl [methyl 2-(4-methoxy-6-methyl-1,3,5-triazin-2-ylcarbamoylsulfamoyl)benzoate]; 15 g a.i. ha−1 tribenuron methyl; and 12.5 g a.i. ha−1 chlorsulfuron [1-(2-chlorophenylsulfonyl)-3-(4-methoxy-6-methyl-1,3,5-triazin-2-yl)urea]), or either of two different doses of one of three imidazolinones (50 and 100 g a.i. ha−1 of imazamox [2-[(RS)-4-isopropyl-4-methyl-5-oxo-2-imidazolin-2-yl]-5-methoxymethylnicotinic acid]; 80 and 160 g a.i. ha−1 of imazapyr [2-[4,5-dihydro-4-methyl-4-(1-methylethyl)-5-oxo-1H-imidazol-2-yl]-3-pyridinecarboxylic acid]; and 50 and 100 g a.i. ha−1 of imazapic [2-[(RS)-4-isopropyl-4-methyl-5-oxo-2-imidazolin-2-yl]-5-methylnicotinic acid]). Fourteen days after treatment, plants were scored phenotypically using a Phytotoxicity Index (PI). PI is a phenotypic scale from 0 to 9 that was assessed for each pot by visual inspection. Plants without any symptoms were recorded as “0”, increasing levels of stunting and yellowing with respect to the untreated control plants were recorded as “1” to “4”; increasing levels of leaf abnormalities and leaf necrosis were recorded from “5” to “8”; dead plants with total necrosis of the apex were recorded as “9”.
Enzyme assay for AHAS activity
An assay for inhibition of AHAS activity was performed on actively growing young leaves approximately 4 weeks after planting. The extraction was performed as follows. Leaves were ground under liquid N2 and extracted with a buffer composed of 100 mM pyruvate, 200 mM KH2PO4, 20 mM MgCl2, 2 mM thiamine pyrophosphate and 20 μM flavin adenine dinucleotide. Homogenate was filtered through two layers of Miracloth (Calbiochem) into a 50 ml conical polypropylene tube. Next, a step to remove leaf polyphenols was added. Briefly, 4–5 ml homogenate was added to a pre-chilled and equilibrated Zeba Desalting Spin column. Columns were then centrifuged cold for 2 min at 1,000×g to obtain flowthrough. Flowthrough samples were assayed immediately. Protein quantitation was determined by using known amounts of bovine serum albumin (BSA) and were used to estimate the amount of protein added to each reaction by the Bradford protein assay (Bradford 1976). The inhibition assay was performed essentially as described in Singh et al. (1988). Briefly, assays were performed in a 96-well format with 50 μl of inhibitor per well to 50 μl of soluble protein extract to give final concentrations of 0.78, 1.56, 3.125, 6.25, 12.5, 25, 50 and 100 μM imazamox or 0.78, 1.56, 3.125, 6.25, 12.5, 25, 50 and 100 nM chlorosulfuron. For negative control wells, 20 μl of 5% H2SO4 was added. To stop the reaction, samples were placed at 37°C for 2 h and 20 μl of 5% H2SO4 was added to each well and the plate was incubated at 60°C for 15 min. For color development, 200 μl of a solution containing creatine (2.5 mg/ml) and γ-naphthol (25 mg/ml) were added to each well. Plates were incubated at 60°C for 15 min then removed and cooled for approximately 5–10 min. Absorbance was measured at 530 nm. Absorbance values for each treatment were expressed as AHAS activity (as estimated by absorbance) as a percentage of the mean of the zero-herbicide controls. Six experiments with three replications of each were conducted for imazamox and one experiment with three replications in the case of chlorsulfuron. Data from each line were fit to a nonlinear regression model by PROC NLIN of SAS (SAS Institute Inc, Cary NC, USA). The nonlinear regression was based on a logistic function mathematically described by Seefeldt et al. 1995.
where β 0 represents the lower asymptote of AHAS activity (%); β 1 represents the mean AHAS activity (%) in the zero-herbicide controls (i.e., upper asymptote); I 50 represents the dose corresponding to AHAS activity midway between the upper and lower asymptotes (50% response); and β 3 [AHAS activity (%) per dose] represents the slope of the curve around the I 50. Dose represents the concentration of inhibitor used in the enzyme assay. The means of each treatment were estimated by PROC GLM of SAS and plotted on the logistic dose response curves.
PCR amplification and direct sequencing of the gene coding for the AHAS1 catalytic subunit from BTK47 and CLHA-Plus genomic DNA
Genomic DNA was extracted from sunflower leaf tissue using Qiagen DNeasy 96 Plant Kit (Qiagen Inc., USA). The AHASL1 gene was PCR amplified in two overlapped fragments and direct-sequenced by DNA LandMarks Inc. (Quebec, Canada). PCR amplification was accomplished with Qiagen HotStart Taq DNA Polymerase and its associated reagents. PCR primers were developed based on the AHASL1 sunflower sequence (Kolkman et al. 2004; GenBank accession no. AY541451) and they are as follows: HA1U254 (5′-CAGACGTGTTGGTGGAAGC-3′); HA1L1195 (5′-CTGTAACGCGACCTTAATATC-3′); HA1U1149 (5′-TGCTGAAATTGGGAAGAATAAG-3′); and HA1L1944 (5′-TTTCGTTCTGCCATCACCC-3′). The primer combination HA1U254/HA1L1195 produced a 963 bp fragment, whereas the primer combination HA1U1149/HA1L1944 produced an 814 bp fragment.
The two PCR fragments cover the majority of mutation sites, which are known to confer resistance to the imidazolinone herbicides (Tranel and Wright 2002). Alignment of the obtained nucleotide sequences was performed and the resulting chromatographs were examined for polymorphisms between the wildtype line, BTK47 and the mutagenized line CLHA-Plus. The AHASL1 nucleotide sequences of the sunflower line HA89 (GenBank accession no. AY541451; Kolkman et al. 2004) and another Compositae Xanthium strumarium L. (XSU16280; Bernasconi et al. 1995) were used as references. Nucleotide and amino acid multiple sequences alignments were generated using ClustalW (http://www.ebi.ac.uk/clustalw), and the output was edited and annotated using GeneDoc software (http://www.psc.edu/biomed/genedoc). Numbering of amino acids followed that of the precursor AHAS from A. thaliana (GenBank accession no. AY124092; Jander et al. 2003). Gene sequences reported herein have been deposited in GenBank with accession numbers EU342348 and EU342349.
Results
Sensitivity of CLHA-Plus to different AHAS inhibiting herbicides
Sunflower is normally sensitive to all the six herbicides used in this study. The susceptible inbred BTK47 was killed 15 days after spraying with all the herbicides tested (Table 1). CLHA-Plus exhibited a complete susceptibility (PI = 9) to metsulfuron, tribenuron and chlorsulfuron and a complete resistance (PI = 0) to imazamox, imazapyr and imazapic at both rates of herbicide application (Table 1). Line BTSu-R1, on the other hand, showed a PI value of zero, when sprayed with the sulfonylurea herbicides metsulfuron methyl, tribenuron methyl and chlorsulfuron, but showed a complete susceptibility (PI = 9) when sprayed with the imidazolinone herbicides imazamox, imazapyr and imazapic at all the assayed doses. Line RHA426 exhibited a variable response depending on the type of herbicide and dose applied (Table 1). It had an average PI score of 5 and 4, when it was sprayed with metsulfuron and tribenuron, respectively; and a score of zero, when it was sprayed with chlorsulfuron. RHA426 showed no symptoms or only a slight chlorosis when sprayed with either imazamox, imazapyr or imazapic in the lower dose (50, 80 and 80 g a.i. ha−1, respectively). However, when the dose of these herbicides increased to 100, 160 and 100 g a.i. ha−1, respectively, RHA426 developed more chlorosis (PI of 3 with imazamox), chlorosis and necrosis (PI = 5 for imazapyr) and even necrosis of the apex (PI = 9 for imazapic).
Biochemical characterization of resistant enzyme
In vitro assays of AHAS were conducted on CLHA-Plus and the susceptible line BTK47 to determine the sensitivity of both lines at the AHAS level, when challenged with imazamox or chlorsulfuron. Specific activities of AHAS in the absence of herbicide ranged from 1.96 ± 0.017 nmol mg−1 min−1 acetoin for CLHA-Plus to 1.93 ± 0.034 nmol mg−1 min−1 acetoin for the susceptible inbred BTK47. No significant differences in specific activity were detected between CLHA-Plus and the susceptible material in the absence of herbicides (P < 0.084), which indicate that resistance in CLHA-Plus was not due to the overexpression of AHAS.
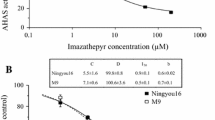
In vitro inhibition curves of AHAS activity at different doses of imazamox in resistant line CLHA-Plus and susceptible line BTK47 are shown in Fig. 1. The log-logistic model accurately described the specific activities of AHAS for the susceptible and the resistant lines. The estimates of the lower asymptote of the logistic regression equation represent the mean activities of AHAS at 100 μM of imazamox (parameter β 0, Fig. 1a). The average value of AHAS activity from extracts of CLHA-Plus was 67.8 ± 4.6%, significantly higher (P < 0.002) than the value of 12.3 ± 2.1% for BTK47. This indicates that much of the AHAS activity from CLHA-Plus was insensitive to imazamox when compared with the inhibition curve for the normal enzyme from BTK47. Average estimated values for β 1 were almost the same for both lines and were not different from 100%. The values for the parameter I 50, which represents the dose corresponding to AHAS activity midway between the upper and lower asymptotes, were also different between both lines (P < 0.005). Since the lower asymptote of AHAS activity inhibition curves were different in CLHA-Plus and BTK47, the estimates of I 50 for both the lines representing the doses which correspond to different AHAS activities (84 and 56% of the untreated control plants for CLHA-Plus and BTK47, respectively). In fact, the concentration of imazamox needed to obtain 50% of inhibition of AHAS activity of the untreated control plants for BTK47 was about 6 μM, whereas for CLHA-Plus this percentage of inhibition was not observed even at the highest herbicide concentration (100 μM). No differences were observed in the AHAS-inhibition kinetics of BTK47 and CLHA-Plus when using chlorsulfuron (Fig. 1b).
In vitro inhibition of AHAS activity in resistant line CLHA-Plus (solid line) and susceptible line BTK47 (dashed line) by imazamox (a) and chlorsulfuron (b). Data points represent the mean at each concentration of herbicide and lines represent the fitted non-linear regression. The regression equation is of the following form: \( {\text{AHAS activity}}\left( {\% {\text{ of the mean of the zero herbicide controls}}} \right) = \beta_{0} + \left( {\beta_{1} - \beta_{0} } \right)/\left[ {1 + \left( {{\text{dose}}/I_{50} } \right)} \right]^{{\beta_{3} }} , \) where β 0 represents the lower asymptote of AHAS activity (%); β 1 represents the upper asymptote; I 50 represents the dose corresponding to AHAS activity midway between the upper and lower asymptotes; and β 3 represents the slope of the curve around I 50. Average values and their standard deviations of each of the four parameters of the equations are provided
Nucleotide sequence comparisons between resistant and susceptible lines
PCR products were sequenced to produce the AHASL1 sequences for CLHA-Plus and BTK47. The alignment of these nucleotide sequences and the nucleotide sequences of the inbred line HA89, Xanthium and Arabidopsis AHAS genes revealed that the AHASL1 gene from CLHA-Plus has a G-to-A transition relative to the AHASL1 of BTK47 and the other sequences.
An alignment of the predicted amino acid sequences of the AHASL1 nucleotide sequences of CLHA-Plus, BTK47, Xanthium, sunflower and A. thaliana is provided in Fig. 2. In relative to the AHASL1 amino acid sequence of BTK47, the AHASL1 amino acid sequence of CLHA-Plus has an alanine-to-threonine substitution at amino acid position 122 relative to the A. thaliana sequence.
Partial alignment of deduced amino acid sequences of AHAS catalytic subunit of sunflower, cocklebur and arabidopsis. Sunflower sequences are represented as EU342348 (BTK47, susceptible), EU342349 (CLHA-Plus, resistant), and AY541451 (USDA line HA89, wildtype; Kolkman et al. 2004). Genbank accession no. XSU16280 refers to Xanthium (cocklebur, Bernasconi et al. 1995) and AY124092 refers to Arabidopsis (Jander et al. 2003). The nucleotide alteration generating the codon 122 substitution is denoted in bold-face type with an asterisk above the mutation
Discussion
Biochemical mechanisms endowing herbicide resistance in plants can include increased metabolism, sequestration, reduced uptake and/or translocation and modification of the herbicide target site (Powles and Holtum 1994). For AHAS-inhibiting herbicides, both target site and non target site resistance mechanisms have been reported (Saari et al. 1994). The results obtained in this study with respect to AHAS activity inhibition kinetics indicated that IMI-resistance in CLHA-Plus is target site based and that is not the result of AHAS overexpression (Fig. 1).
The whole plant response pattern of tolerance of CLHA-Plus to several sulfonylurea and imidazolinone herbicides differed with respect to the other tolerant lines tested. It has a high tolerance level to imidazolinones but it is susceptible to all the sulfonylureas used. This pattern is in close association with the AHAS-inhibition kinetics of protein extracts of CLHA-Plus. The enzyme is resistant to imidazolinone herbicides as shown by the high β 0 value for imazamox when compared with BTK47, but it is susceptible to sulfonylureas as shown by the low I 50 value for chlorsulfuron (Fig. 1).
Sequencing results indicate that the imidazolinone-resistant AHAS of CLHA-Plus has a threonine residue (ACG) at position 122, whereas the herbicide-susceptible enzyme from BTK47 has an alanine residue (GCG) at this position (Fig. 2). This mutation (A122T) has been identified as the basis for AHAS-inhibition resistance in several species: X. strumarium (Bernasconi et al. 1995), Solanum ptycanthum Dun. (Milliman et al. 2003), Amaranthus retroflexus (McNaughton et al. 2005), A. hybridus L. (Trucco et al. 2006), Nicotiana tabacum L. (Chong and Choi 2000), maize (Zea mays L., Bernasconi et al. 1995), sugarbeet (B. vulgaris, Wright and Penner 1998) and A. thaliana (Jander et al. 2003).
Several AHAS substitutions will result in resistance to AHAS inhibitors; however, the magnitude of resistance to different AHAS inhibiting herbicides varies widely among substitutions (Saari et al. 1994; Tranel and Wright 2002). Although exceptions exist, resistance caused by an altered AHAS can be generally classified into three types on the basis of cross resistance: (1) SU-specific resistance, (2) IMI-specific resistance, and (3) broad cross resistance. Substitutions of Pro 197 usually result in SU- but not IMI-resistance, whereas substitutions of Ala 122 result in IMI- but not SU-resistance. On the other hand, substitutions of Ala 205 display broad cross resistance; however, the levels of resistance are moderate (Saari et al. 1994; Tranel and Wright 2002). Our results were in close agreement with this generalization. Line BTSu-R1 (P197L) was resistant to all the sulfonylureas assayed; RHA426 (A205 V) was moderately resistant to both SU and IMI herbicides; and CLHA-Plus (A122T) was highly resistant solely to imidazolinones. Both SU and IMI herbicides, which have partially overlapping binding sites, inhibit AHAS by binding within and obstructing the channel leading to the active site of the enzyme. A122T substitution makes important hydrophobic contacts to the isopropyl and methyl substituents of the dihydroimidazolone ring of the IMI molecule, and the mutation to a larger polar residue such as threonine would tend to preclude the herbicide from its binding site. On the other hand, this threonine substitution would not seriously compromise sulfonylurea binding (McCourt et al. 2006).
Resistance genes to AHAS-inhibiting herbicides in sunflower have received different names. Imr1 was proposed by Bruniard and Miller (2001) while studying the inheritance of the imidazolinone resistance. AHAS1 as the locus name was proposed by Kolkman et al. (2004), together with Ar kan and Ar pur for the SU- and IMI-resistance alleles, respectively. Since the resistant genes so far characterized code for the catalytic (large) subunit of AHAS and to distinguish these genes from the regulatory (small) subunit genes, we propose to redesignate the susceptible wild type allele as ahasl1 and the incomplete dominant resistant alleles as Ahasl1-1 (previously Imr1 or Ar pur ), Ahasl1-2 (previously Ar kan ) and Ahasl1-3 (for the allele present in CLHA-Plus).
CLHA-Plus was the only resistant plant that could be recovered after imazapyr selection from a population of ca. 590,000 M2 plants (Sala et al. 2008). Seed mutagenesis of BTK47 with EMS has been effective in generating a single base pair mutation in the gene coding for the AHAS1 catalytic subunit in sunflower. Most EMS induced mutations result in a single base pair GC → AT transition (Ashburner 1990), most likely the results of O-6-ethylguanine adducts that mispair with thymine during DNA replication (Snow et al. 1984) and occur at a 5′-purine-guanine-3′ motif (Bently et al. 2000; Inukai et al. 2000). Jander et al. (2003) isolated 12 EMS induced mutations in A. thaliana resulting in imidazolinone resistance and noted that all identified mutations were single GC → AT transitions in the genes coding for the catalytic subunit of AHAS. The EMS induced mutation identified in AHASL1 of CLHA-Plus also follows this general pattern.
Apart from the fact that it is the only member of the family of AHAS genes in sunflower where all the induced and natural mutations for herbicide resistance where described thus far, AHASL1 is also the most polymorphic (Kolkman et al. 2004) and the most preponderant in the sunflower EST database (Kozik et al. 2002). This suggest that the AHASL1 gene is the gene family member that encodes the AHAS enzyme with essential housekeeping functions in sunflower and that it is predominantly expressed in tissues affected by herbicide treatment. Divergent patterns of expression of different members of the AHAS multigene family were also demonstrated in other species like Brassica napus (Ouellet et al. 1992) and Gossypium hirsutum (Grula et al. 1995).
The possibility of gene flow from IMI-resistant domesticated sunflower to wild relatives (Faure et al. 2002; Massinga et al. 2003) and the lack of a competitive penalty associated with the Ahasl1-1 mutation in common sunflower (Marshall et al. 2001) suggests that widespread use of the hybrids could result in emergence of new herbicide-resistant weedy sunflower biotypes by both selection and gene flow. As was argued by Kolkman et al. (2004), the discovery and careful management of different resistance genes, especially mutations that lack cross-resistance to different classes of AHAS-inhibiting herbicides, is needed for better management of domesticated sunflower and other crops where control of weedy common sunflower is necessary. Phenotypic, biochemical and molecular characterization of Ahasl1-3 from CLHA-Plus provided here indicate that this induced mutation would fulfill these requirements.
References
Al-Khatib K, Baumgartner JR, Peterson DE, Currie RS (1998) Imazethapyr resistance in common sunflower (Helianthus annuus). Weed Sci 46:403–407
Al-Khatib K, Miller JF (2002) Registration of four genetic stocks of sunflower resistant to imidazolinone herbicides. Crop Sci 40:869–870
Anderson PC, Georgeson M (1989) Herbicide-tolerant mutants of corn. Genome 34:994–999
Ashburner M (1990) Drosophila: a Laboratory Handbook. Cold Spring Harbor Laboratory Press, Cold Spring Harbor
Bently A, Mac Lennan B, Calvo J, Dearolf CR (2000) Targeted recovery of mutations in Drosophila. Genetics 156:1169–1173
Bernasconi P, Woodworth AR, Rosen BA, Subramanian MV, Siehl DL (1995) A naturally occurring point mutation confers broad range tolerance to herbicides that target acetolactate synthase. J Biol Chem 270:17381–17385
Bradford MM (1976) A rapid and sensitive method for the quantification of microgram quantities of protein utilizing the principle of protein-dye binding. Ann Biochem 72:248–254
Bruniard JM, Miller JF (2001) Inheritance of imidazolinone herbicide resistance in sunflower. Helia 24:11–16
Chong CK, Choi JD (2000) Amino acid residues conferring herbicide tolerance in tobacco acetolactate synthase. Biochem Biophys Res Commun 279:462–467
Croughan TP (1996) Herbicide resistant rice. US Patent 5,545,822
D’Halluin KM, Bossut M, Bonne E, Mazur B, Leemans J, Botterman J (1992) Transformation of sugarbeet (Beta vulgaris L.) and evaluation of herbicide resistance in transgenic plants. Biotechnology 10:309–314
Duggleby RG, Pang SS (2000) Acetohydroxyacid synthase. J Biochem Mol Biol 33:1–36
Duggleby RG, McCourt JA, Guddat L (2008) Structure and mechanism of inhibition of plant acetohydroxiacid synthase. Plant Physiol Biochem 46:309–324
Faure N, Serieys H, Bervillé A (2002) Potential gene flow from cultivated sunflower to volunteer, wild Helianthus species in Europe. Agric Ecosyst Environ 89:183–190
Grula JW, Hudspeth RL, Hobbs SL, Anderson DM (1995) Organization, inheritance and expression of acetohydroxyacid synthase genes in the cotton allotetraploid Gossypium hirsutum. Plant Mol Biol 28:837–846
Guttieri MJ, Eberlein CV, Mallory-Smith CA, Thill DC, Hoffman DL (1992) DNA sequence variation in domain A of the acetolactate synthase genes of herbicide-resistant and -susceptible weed biotypes. Weed Sci 40:670–676
Guttieri MJ, Eberlein CV, Thill DC (1995) Diverse mutations in the acetolactate synthase allele confer chlorsulfuron resistance in Kochia scoparia biotypes. Weed Sci 43:175–178
Harms CT, Armour SL, DiMaio JJ, Middlesteadt LA, Murray D, Negrotto DV, Thompson-Tyler H, Weymann K, Montoya AL, Shillito RD, Jen GC (1992) Herbicide resistance due to amplification of a mutant acetohydroxyacid synthase allele. Mol Gen Genet 233:427–435
Hart SE, Saunders JW, Penner D (1992) Chlorsulfuron resistant sugar beet: cross-resistance and physiological basis of resistance. Weed Sci 40:378–383
Haughn GW, Smith J, Mazur B, Somerville C (1988) Transformation with a mutant Arabidopsis acetolactate synthase gene renders tobacco resistant to sulfonylureas. Mol Gen Genet 211:266–271
Inukai T, Sako A, Hirano HY, Sano Y (2000) Analysis of intragenic recombination at wx in rice: correlation between molecular and genetic maps within the locus. Genome 43:589–596
Jander G, Baerson SR, Hudak JA, Gonzalez KA, Gruys KJ, Last RL (2003) Ethylmethanesulfonate saturation mutagenesis in Arabidopsis to determine frequency of herbicide resistance. Plant Physiol 131:139–146
Kolkman JM, Slabaugh MB, Bruniard JM, Berry S, Bushman BS, Olungu C, Maes N, Abratti G, Zambelli A, Miller JF, Leon A, Knapp SJ (2004) Acetohydroxyacid synthase mutations conferring resistance to imidazolinone or sulfonylurea herbicides in sunflower. Theor Appl Genet 109:1147–1159
Kozik A, Michelmore RW, Knapp SJ, Matvienko MS, Rieseberg L, Lin H, van Damme M, Lavelle D, Chevalier P, Ziegle J, Ellison P, Kolkman JM, Slabaugh MB, Livingston K, Zhou LZ, Church S, Edberg S, Jackson L, Kesseli R, Bradford K (2002) Sunflower and lettuce ESTs from the compositae genome project. http://compgenomics.ucdavis.edu
Lee YT, Duggleby RG (2001) Identification of the regulatory subunit of Arabidopsis thaliana acetohydroxyacid synthase and reconstitution with its catalytic subunit. Biochem 40:6836–6844
Lee YT, Duggleby RG (2002) Regulatory interactions in Arabidopsis thaliana acetohydroxyacid synthase. FEBS Lett 512:180–184
Lee KY, Townsend J, Tepperman J, Black M, Chui CF, Mazur B, Dunsmuir P, Bedbrook J (1988) The molecular basis of sulfonylurea resistance in tobacco. EMBO J 7:1241–1248
Mallory-Smith CA, Thill DC, Dial MJ (1990) Identification of sulfonylurea herbicide-resistant prickly lettuce (Lactuca serriola). Weed Technol 4:163–168
Marshall MW, Al-Khatib K, Loughin T (2001) Gene flow, growth, and competitiveness of imazethapyr-resistant common sunflower. Weed Sci 49:14–21
Massinga RA, Al-Khatib K, Amand PSt, Miller JF (2003) Gene flow from imidazolinone-resistant domesticated sunflower to wild relatives. Weed Sci 51:854–862
McCourt JA, Duggleby RG (2006) Acetohydroxyacid synthase and its role en the biosynthetic pathway for branched-chain amino acids. Amino Acids 31:173
McCourt JA, Pang SS, King-Scott J, Guddat LW, Duggleby RG (2006) Herbicide-binding sites revealed in the structure of plant acetohydroxyacid synthase. Proc Natl Acad Sci USA 103:569–573
McHughen A (1989) Agrobacterium mediated transfer of chlorsulfuron resistance to commercial flax cultivars. Plant Cell Rep 8:445–449
McNaughton KE, Letarte J, Lee EA, Tardif FJ (2005) Mutations in ALS confer herbicide resistance in redroot pigweed (Amaranthus retroflexus) and powell amaranth (Amaranthus powellii). Weed Sci 53:17–22
Miller JF, Al-Khatib K (2002) Registration of imidazolinone herbicide-resistant sunflower maintainer (HA425) and fertility restorer (RHA426 and RHA427) germplasms. Crop Sci 42:988–989
Miller JF, Al-Khatib K (2004) Registration of two oilseed sunflower genetic stocks, SURES-1 and SURES-2, resistant to tribenuron herbicide. Crop Sci 44:1037–1038
Milliman LD, Riechers DE, Wax LM, Simmons FW (2003) Characterization of two biotypes of imidazolinone-resistant eastern black nightshade (Solanum ptycanthum). Weed Sci 51:139–144
Mourad G, King J (1992) Effect of four classes of herbicides on growth and acetolactate synthase activity in several variants of Arabidopsis thaliana. Planta 188:491–497
Newhouse K, Singh BK, Shaner DL, Stidham M (1991) Mutations in corn (Zea mays L.) conferring resistance to imidazolinones. Theor Appl Genet 83:65–70
Newhouse K, Smith W, Starrett M, Schaefer T, Singh BK (1992) Tolerance to imidazolinone herbicides in wheat. Plant Physiol 100:882–886
Ouellet T, Rutledge RG, Miki BL (1992) Members of the acetohydroxyacid synthase multigene family of Brassica napus have divergent patterns of expression. Plant J 2:321–330
Powles SB, Holtum JAM (1994) Herbicide resistance in plants: biology and biochemistry. Lewis Publishers, Boca Ratón
Pozniak CJ, Hucl PJ (2004) Genetic analysis of imidazolinone resistance in mutation-derived lines of common wheat. Crop Sci 44:23–30
Preston C, Mallory-Smith CA (2001) Biochemical mechanisms, inheritance, and molecular genetics of herbicide resistance in weeds. In: Powles SB, Shaner DL (eds) Herbicide resistance and world grains. CRC Press, Boca Raton, pp 23–60
Rajasekaran K, Grula JW, Anderson DM (1996) Selection and characterization of mutant cotton (Gossypium hirsutum L.) cell lines resistant to sulfonylurea and imidazolinone herbicides. Plant Sci 199:115–124
Ray TB (1984) Site of action of chlorsulfuron. Inhibition of valine and isoleucine biosynthesis in plants. Plant Physiol 75:827–831
Saari LL, Cotterman JC, Thill DC (1994) Resistance to acetolactate synthase inhibiting herbicides. In: Powles SB, Holtum JAM (eds) Herbicide resistance in plants: biology and biochemistry. Lewis Publishers, Boca Ratón, pp 83–139
Sala CA, Bulos M, Echarte AM (2008) Genetic analysis of an induced mutation conferring imidazolinone resistance in sunflower. Crop Sci (in press)
Santel HJ, Bowden BA, Sorenson VM, Mueller KH, Reynolds J (1999) Flucarbazone-sodium—a new herbicide for grass control in wheat. Proc West Soc Weed Sci 52:124–125
Schneiter AA, Miller JF (1981) Description of sunflower growth stages. Crop Sci 21:901–903
Sebastian SA, Fader GM, Ulrich JF, Forney DR, Chaleff RS (1989) Semi-dominant soybean mutation for resistance to sulfonylurea herbicides. Crop Sci 29:1403–1408
Seefeldt SS, Jensen JE, Fuerst EP (1995) Log-logistic analysis of herbicide dose response relationships. Weed Technol 9:218–227
Shaner DL (1991) Physiological effects of the imidazolinone herbicides. In: Shaner DL, O’Connor SL (eds) The imidazolinone herbicides. Lewis, Ann Arbor, pp 129–138
Shaner DL, Anderson PC, Stidham MA (1984) Imidazolinones: potent inhibitors of acetohydroxyacid synthase. Plant Physiol 76:545–546
Singh BK (1999) Biosynthesis of valine, leucine and isoleucine. In: Singh BK (ed) Plant amino acids. Marcel Dekker Inc, New York, pp 227–247
Singh BK, Stidham MA, Shaner DL (1988) Assay of acetohydroxyacid synthase. Ann Biochem 171:173–179
Snow ET, Foote RS, Mitra S (1984) Base-pairing properties of O-6-methylguanine in template DNA during in vitro DNA replication. J Biol Chem 259:8095–8100
Subramanian MV, Gerwick BC (1989) Inhibition of acetolactate synthase by triazolopyrimidines: a review of recent developments. In: Whitaker JR, Sonnet PE (eds) Biocatalysis in agricultural biotechnology. American Chemical Society, Washington, pp 277–288
Subramanian MV, Hung HY, Dias JM, Miner VW, Butler JH, Jachetta JJ (1990) Properties of mutant acetolactate synthases resistant to triazolopyrimidine sulfonanilide. Plant Physiol 94:239–244
Swanson EB, Hergesell MJ, Arnoldo M, Sippell DW, Wong RSC (1989) Microspore mutagenesis and selection: canola plants with field tolerance to imidazolinones. Theor Appl Genet 78:525–530
Tan S, Evans RR, Dahmer ML, Singh BK, Shaner DL (2005) Imidazolinone-tolerant crops: history, current status and future. Pest Manag Sci 61:246–257
Tranel PJ, Wright TR (2002) Resistance of weeds to AHAS inhibiting herbicides: what have we learned? Weed Sci 50:700–712
Trucco F, Hager AG, Tranel PJ (2006) Acetolactate synthase mutation conferring imidazolinone-specific herbicide resistance in Amaranthus hybridus. J Plant Physiol 163:475–479
Umbarger HE (1978) Amino acid biosynthesis and its regulation. Annu Rev Biochem 47:533–606
White AD, Owen MD, Hartzler RG, Cardina J (2002) Common sunflower resistance to acetolactate-inhibiting herbicides. Weed Sci 50:432–437
Wright TR, Bascomb NF, Sturner SF, Penner D (1998) Biochemical mechanism and molecular basis for ALS-inhibiting herbicide resistance in sugar beet (Beta vulgaris) somatic cell selections. Weed Sci 46:13–23
Wright TR, Penner D (1998) Cell selection and inheritance of imidazolinone resistance in sugar beet (Beta vulgaris). Theor Appl Genet 96:612–620
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by A. Bervillé.
Rights and permissions
About this article
Cite this article
Sala, C.A., Bulos, M., Echarte, M. et al. Molecular and biochemical characterization of an induced mutation conferring imidazolinone resistance in sunflower. Theor Appl Genet 118, 105–112 (2008). https://doi.org/10.1007/s00122-008-0880-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00122-008-0880-6