Abstract
Tan spot, caused by Pyrenophora tritici-repentis, is a destructive foliar disease of wheat causing significant yield reduction in major wheat growing areas throughout the world. The objective of this study was to identify quantitative trait loci (QTL) conferring resistance to tan spot in the synthetic hexaploid wheat (SHW) line TA4152-60. A doubled haploid (DH) mapping population derived from TA4152-60 × ND495 was inoculated with conidia produced by isolates of each of four virulent races of P. tritici-repentis found in North America. QTL analysis revealed a total of five genomic regions significantly associated with tan spot resistance, all of which were contributed by the SHW line. Among them, two novel QTLs located on chromosome arms 2AS and 5BL conferred resistance to all isolates tested. Another novel QTL on chromosome arm 5AL conferred resistance to isolates of races 1, 2 and 5, and a QTL specific to a race 3 isolate was detected on chromosome arm 4AL. None of these QTLs corresponded to known host selective toxin (HST) insensitivity loci, but a second QTL on chromosome arm 5BL conferred resistance to the Ptr ToxA producing isolates of races 1 and 2 and corresponded to the Tsn1 (Ptr ToxA sensitivity) locus. This indicates that the wheat-P. tritici-repentis pathosystem is much more complex than previously thought and that selecting for toxin insensitivity alone will not necessarily lead to tan spot resistance. The markers associated with the QTLs identified in this work will be useful for deploying the SHW line as a tan spot resistance source in wheat breeding.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Tan spot, caused by the fungus Pyrenophora tritici-repentis (Died.) Drechs. [anamorph Drechslera tritici-repentis (Died.) Shoem.], is a destructive foliar disease of common wheat (Triticum aestivum L.) and durum wheat (T. turgidum L. var. durum), as well as other grass species (Ali and Francl 2003; Hosford 1971; Krupinsky 1992). Riede et al. (1996) reported that yield losses caused by tan spot were capable of approaching 50%. Disease incidence has become more common in recent years due in part to the application of reduced tillage practices in cereal growing regions of the world (Strelkov and Lamari 2003).
Isolates of P. tritici-repentis differ in virulence. Susceptibility of wheat to P. tritici-repentis is manifested by the development of necrosis and/or chlorosis depending on the race of the pathogen. Isolates of P. tritici-repentis have been grouped into eight races based on their virulence pattern on five differential wheat lines: Glenlea, Katepwa, Salamouni, 6B365, and 6B662 (Lamari et al. 1995, 2003; Strelkov et al. 2002; Strelkov and Lamari, 2003) and on the presence of genes coding for specific host-selective toxins (HSTs) (Andrie et al. 2007). Races 1–5 have all been found in North America (Ali et al. 1999; Lamari et al. 2003), with races 1 and 2 being the most prevalent (Ali and Francl 2003; Lamari et al. 1998; Lamari and Bernier 1989a).
Reports of tan spot resistance in wheat range from qualitative (Gamba and Lamari 1998; Gamba et al. 1998; Lee and Gough 1984; Lamari and Bernier 1989b, 1991; Sykes and Bernier 1991; Singh et al. 2006a; Tadesse et al. 2007) to quantitative (Cheong et al. 2004; Elias et al. 1989; Faris et al. 1997, 1999; Friesen et al. 2003; Faris and Friesen 2005; Nagle et al. 1982). P. tritici-repentis is known to produce at least three HSTs (Effertz et al. 2002; Lamari and Bernier 1989b; Orolaza et al. 1995; Tomás and Bockus 1987; Tuori et al. 1995) that interact with specific host sensitivity genes to cause disease (Effertz et al. 2002; Friesen et al. 2003; Friesen and Faris 2004; Lamari and Bernier 1989b). Among the identified HSTs, Ptr ToxA, a well-characterized HST produced by races 1 and 2 (Tomás and Bockus 1987; Tuori et al. 1995), was shown to be responsible for the development of necrosis (Lamari and Bernier 1989b). Sensitivity to Ptr ToxA is conditioned by a single dominant gene named Tsn1 (Lamari and Bernier 1989b) on chromosome arm 5BL (Faris et al. 1996; Anderson et al. 1999). Insensitivity to Ptr ToxA is highly associated with resistance (Cheong et al. 2004; Friesen et al. 2003; Lamari and Bernier 1989b).
Ptr ToxB (Orolaza et al. 1995) and Ptr ToxC (Effertz et al. 2002) are both chlorosis-inducing HSTs. The Ptr ToxB-sensitivity gene (Tsc2) was mapped to chromosome arm 2BS and was shown to confer susceptibility to the race 5 isolate DW5 (Friesen and Faris 2004). Sensitivity to Ptr ToxC is conditioned by the gene Tsc1, found on the short arm of chromosome 1A (Effertz et al. 2002). Sensitivity to Ptr ToxC has also been shown to be associated with tan spot susceptibility (Effertz et al. 1998, 2002; Faris et al. 1997, 1999). Therefore, several major resistance genes and quantitative trait loci (QTLs) identified have been due to insensitivity to HSTs produced by the various races of P. tritici-repentis.
In addition to the known toxin sensitivity genes, Faris and Friesen (2005) identified resistance QTLs on chromosome arms 1BS and 3BL in a recombinant inbred population derived from Grandin × BR34. Neither of these QTLs has been shown to be associated with toxin insensitivity genes. They also found that the Tsn1–Ptr ToxA interaction was not a significant factor in tan spot development in that population. This result, along with other recent studies (Andrie et al. 2007; Friesen et al. 2002, 2003; Riede et al. 1996; Singh et al. 2006a, b; Tadesse et al. 2007) has indicated the potential for additional complexity in the wheat-P. tritici-repentis pathosystem.
Synthetic hexaploid wheat (SHW) (×Aegilotriticum spp., 2n = 6x = 42, AABBDD) is the induced amphiploid from the hybrid between tetraploid wheat (Triticum turgidum L., 2n = 4x = 28, AABB) and Aegilops tauschii Coss. (2n = 2x = 14, DD), and is commonly used as bridging germplasm in the introgression of desirable genes from Ae. tauschii to common wheat (reviewed by Cox, 1998). Xu et al. (2004) identified 41 SHW lines resistant to a tan spot race 1 isolate, and among them, TA4152-60 showed a high level of resistance. In this study, we evaluated a doubled haploid (DH) population derived from the cross between TA4152-60 and an elite North Dakota hard red spring wheat line (ND495) for reaction to races 1, 2, 3 and 5 of P. tritici-repentis and used QTL analysis to identify genomic regions and molecular markers associated with resistance.
Materials and methods
Plant materials
A mapping population consisting of 120 DH lines derived from the SHW line TA4152-60 and the North Dakota breeding line ND495, which was used for developing whole genome linkage maps (Chu et al. 2008), was used for the identification of tan spot resistance QTLs. TA4152-60 was developed at the International Maize and Wheat Improvement Center (CIMMYT) from a cross between the durum wheat variety Scoop 1 and the Ae. tauschii accession WPI358 (TA2516). TA4152-60 is insensitive to Ptr ToxA and resistant to Pti2, a race 1 isolate of P. tritici-repentis (Xu et al. 2004). ND495 is a selection from ‘Justin*2/3/ND 259/Conley//ND 112’, and is sensitive to Ptr ToxA and susceptible to Pti2 (race 1). Our tests also indicated that ND495 and TA4152-60 showed different reactions to isolates of race 2 (86–124), race 3 (OH99) and race 5 (DW5) (Table 1).
Fungal isolates, inoculation, and rating
Because races 1, 2, 3 and 5 are all found in North America and isolates of race 4 are avirulent on wheat (Ali et al. 1999; Lamari et al. 2003), we selected isolates Pti2 (race 1) (Friesen et al. 2002), 86–124 (race 2) (Friesen et al. 2003), OH99 (race 3) (Engle et al. 2006; Faris and Friesen 2005), and DW5 (race 5) (Ali et al. 1999; Friesen and Faris 2004) for this study. All isolates have been race characterized based on the standard differential set as proposed by Lamari et al. (2003). In addition, the presence of ToxA has been evaluated in each isolate used, with ToxA being present in races 1 and 2 and absent in races 3 and 5 (data not shown). For evaluation of disease reaction, all plants were inoculated with conidia of each isolate separately. Disease inoculation was conducted in three experiments under controlled conditions using procedures described in Friesen et al. (2003). For each experiment, nine seeds of each line were planted in three super-cell cones (Stuewe and Sons, Inc., Corvallis, OR, USA) with three seeds per cone. Cones were placed in RL98 trays (Stuewe and Sons, Inc., Corvallis, OR, USA). To eliminate any edge effect, the susceptible cultivar Grandin was planted in all the cones around the borders on each RL98 tray except for six cones, which were used for planting the parents. Therefore, a total of 27 plants were used for each line with each isolate. Fungi were grown and conidia were harvested as described by Lamari and Bernier (1989a). Spore inoculum was adjusted to 3,000 spores/ml, and two drops of Tween-20 were added per 100 ml of inoculum. Plants were inoculated until runoff and placed in 100% relative humidity in the light at 21°C for 24 h, and then placed in a growth chamber under a 12-h photoperiod at 21°C. Reaction types were rated 7-day post-inoculation using the 1–5 reaction type rating scale developed by Lamari and Bernier (1989a), where 1 = resistant, 2 = moderately resistant, 3 = moderately resistant to moderately susceptible, 4 = moderately susceptible, and 5 = susceptible. Lines showing equal numbers of two reaction types were given an intermediate score (e.g. lines showing equal numbers of reaction type 1 and 2 were scored as 1.5). Ptr ToxA sensitivity of each DH line was obtained previously (Chu et al. 2008). Purified Ptr ToxB and Ptr ToxC were not available for this research.
Molecular mapping and QTL analysis
The linkage maps developed for this DH population were previously reported and consisted of 632 markers and spanned 3,811.5 cM with an average density of one marker per 6.03 cM (Chu et al. 2008). A subset of 449 markers spaced approximately 5–20 cM apart and giving the most complete genome coverage was selected and used for QTL detection.
Bartlett’s χ 2 was calculated to test the homogeneity of variances among different experiments using the SAS program (SAS Institute 1999), and data from homogeneous experiments were combined for QTL analysis. Individual markers significantly (P < 0.001) associated with tan spot resistance were identified through single-factor regression analysis using the computer program QGENE (Nelson 1997). Simple interval mapping (SIM) and composite interval-regression mapping (CIM) were performed using the computer program Map Manager QTX (Manly et al. 2001) to evaluate marker intervals putatively associated with trait phenotypes. A permutation test with 5,000 permutations was conducted to determine that the critical LOD threshold of 3.0 in this DH population yields an experiment-wise significance level of 0.05. Markers with significant (P < 0.001) main effects were tested against all other markers to identify significant (P < 0.000001) interactions (Manly et al. 2001). Markers with the most significant effect for each QTL and the interactions were assembled into multiple regression models to determine the coefficient of determination (R 2), which is the total amount of variation explained by the model.
Results
Evaluation of tan spot in the DH population
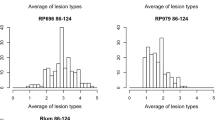
Tan spot reaction types caused by the four isolates of P. tritici-repentis were recorded 7-day post-inoculation for each DH line. Homogeneity tests indicated the data from the three replicated experiments for each isolate were homogeneous. Bartlett’s χ 2 was 2.67, 1.87, 3.84 and 4.61, and the associated P values with 2 degrees of freedom were 0.26, 0.39, 0.15 and 0.10 for reaction to isolates Pti2 (race 1), 86–124 (race 2), OH99 (race 3), and DW5 (race 5), respectively. Thus, the reaction type data for each isolate were combined and the means for each isolate were used for QTL analysis. The SHW line TA4152-60 was highly resistant to all isolates and had average reaction types of 1.0–1.3, whereas ND495 showed average reaction types of 3.0 for reaction to OH99 and 4.0–4.3 for Pti2, 86–124, and DW5 (Table 1, Fig. 1). Therefore, ND495 was moderately susceptible to the race 3 isolate and susceptible to the isolates of races 1, 2 and 5.
Histograms of average reaction type distribution in the doubled haploid population after inoculation with the four isolates/races of P. tritici-repentis. For isolates representing races 1 and 2, Ptr ToxA insensitive and sensitive lines are indicated in gray and black, respectively. White bars for isolates representing races 3 and 5 refer to the whole population
Distribution of the reaction type means in the DH population indicated that resistance to all four isolates was quantitatively controlled, and little transgressive segregation was observed (Fig. 1). Because races 1 and 2 produce Ptr ToxA (Tomás and Bockus 1987; Tuori et al. 1995), and Ptr ToxA insensitivity is known to be associated with resistance (Friesen et al. 2003; Lamari and Bernier 1989b), we compared the previously obtained Ptr ToxA sensitivity data (Chu et al. 2008) with the distribution of Pti2 and 86–124 reaction types among Ptr ToxA sensitive and insensitive DH lines (Fig. 1). Of the 57 Ptr ToxA insensitive DH lines, 32 (56%) and 44 (77%) showed reaction type means less than 2.5 for reaction to Pti2 and 86–124, respectively, and among the 63 Ptr ToxA sensitive lines, 47 (75%) and 33 (52%) had mean reaction types greater than 3.0 for reaction to Pti2 and 86–124, respectively. This suggests that a compatible Tsn1–Ptr ToxA interaction plays a significant role in disease development in this population, which corroborates the results of the QTL analysis (see below).
QTL Identification
A total of five genomic regions harboring QTL were revealed through single-factor regression, SIM, and CIM, and they were all significantly associated with the resistance contributed by TA4152-60 (Tables 2, 3; Fig. 2). Two of the QTL were located on chromosome arm 5BL (designated as QTs.fcu-5BL.1 and QTs.fcu-5BL.2), and the remaining three were on chromosome arms 2AS (QTs.fcu-2AS), 4AL (QTs.fcu-4AL) and 5AL (QTs.fcu-5AL) (Fig. 2).
Composite interval regression maps of QTLs associated with the resistance contributed by TA4152-60. The positions of marker loci are shown to the left of the linkage groups and centiMorgan (cM) distances between loci are shown along the right. Red, green, purple, and blue lines indicate QTLs for resistance to isolates Pti2 (race 1), 86–124 (race 2), OH99 (race 3), and DW5 (race 5), respectively. The vertical dotted line represents the logarithm of the odds (LOD) significance threshold of 3.0. The LOD and R 2 values for each QTL are listed in Table 3
Four of the five QTLs, QTs.fcu-2AS, QTs.fcu-5AL, QTs.fcu-5BL.1 and QTs.fcu-5BL.2, were significantly associated with the resistance to Pti2 (race 1) of P. tritici-repentis (Table 3; Fig. 2). The interval between Xgwm71.2 and Xfcp526 defined the peak of QTs.fcu-2AS, which explained 14% of the phenotypic variation using CIM (Table 3). Xbarc10 was the most significant marker within the interval explaining 12% of the variation (Table 2). QTs.fcu-5AL peaked within the interval defined by markers Xbarc1061 and Xcfa2163, and it explained 10% of the phenotypic variation in CIM (Table 3). Xfcp412 was the marker most significantly associated with QTs.fcu-5AL for resistance to Pti2, and it explained 14% of the variation in single-factor regression (Table 2). The two QTLs on chromosome arm 5BL, QTs.fcu-5BL.1 and QTs.fcu-5BL.2, explained 22 and 17% of the variation in CIM, respectively (Table 3, Fig. 2). QTs.fcu-5BL.1 was located in the proximal region of 5BL and peaked at the interval defined by Xbarc138 and Xfcp505 (Table 3, Fig. 2). Xbarc128.2 was the most significant marker within the QTs.fcu-5BL.1 interval and explained 20% of the variation in single-factor regression (Table 2). QTs.fcu-5BL.2 was located more distal and peaked in the interval between Tsn1 and Xbarc142 (Fig. 2). In single-factor regression, the Tsn1 locus explained 17% of the variation in disease caused by Pti2 (Table 2). A significant interaction between the marker Xfcp547 located on chromosome arm 2BL and Tsn1 on 5BL was also identified, and the interaction explained 10% of the variation (Table 2). The most significant marker associated with each of the four QTLs as well as the interaction were assembled into a multiple regression model, which explained a total of 49% of the variation for resistance to Pti2 (Table 2).
Resistance to isolate 86–124 (race 2) was governed by the same four QTLs associated with resistance to Pti2 (race 1) (Table 3, Fig. 2). QTs.fcu-2AS peaked between Xgwm515 and Xfcp526 and explained 22% of phenotypic variation in CIM (Table 3, Fig. 2). The most significant marker within the interval was Xbarc10, and it explained 20% of the variation in single-factor regression (Table 2). QTs.fcu-5BL.1 also explained 22% of phenotypic variation in CIM (Table 3) and peaked between markers Xgwm66 and Xfcp505 (Fig. 2). The most significant marker within the interval was Xbarc128.2, which explained 20% of the variation in single-factor regression (Table 2). QTs.fcu-5AL peaked between Xbarc1061 and Xcfa2163 on 5AL, and it explained 9% of the phenotypic variation in CIM (Table 3). The marker Xfcp412 was the most significant and explained 11% of the variation in single-factor regression (Table 2). QTs.fcu-5BL.2 explained 14% of the variation in CIM (Table 3), and peaked between Tsn1 and Xbarc142, the interval similar to that for resistance to Pti2. The Tsn1 locus had the most significant effect within the QTs.fcu-5BL.2 region and explained 15% of the variation in single-factor regression (Table 2). The interaction between Xfcp547 and Tsn1, which was significantly associated with resistance to Pti2 was also significantly associated with resistance to 86–124, and it explained 9% of the trait variation (Table 2). Markers associated with the four significant QTLs and the interaction were all significant in a multiple regression model, which explained a total of 54% of the phenotypic variation for resistance to 86–124 (race 2) (Table 2).
Resistance to isolate OH99 (race 3) was conditioned by QTs.fcu-2AS, QTs.fcu-4AL and QTs.fcu-5BL.1, and they explained 14, 10 and 26% of the phenotypic variation in CIM, respectively (Table 3, Fig. 2). QTs.fcu-2AS peaked at the same marker interval as that for resistance to 86–124, and the most significant marker within the interval was Xbarc10, which explained 12% of the variation in single-factor regression (Table 2). QTs.fcu-4AL was located near the middle of chromosome arm 4AL and peaked within the interval defined by markers Xbarc236 and Xgwm644 (Fig. 2). The marker Xbarc1047 was the most significant and explained 9% of the variation in single-factor regression (Table 2). QTs.fcu-5BL.1 was the most significant QTL for resistance to OH99, and was flanked by markers Xbarc138 and Xfcp505, an interval similar to that observed for resistance to Pti2 and 86–124. As with resistance to Pti2 and 86–124, Xbarc128.2 was the most significant marker associated with QTs.fcu-5BL.1 for resistance to OH99, and it explained 24% of the variation in single-factor regression (Table 2). No significant interactions between markers with significant main effects and other loci were found. Markers Xbarc10, Xbarc1047, and Xbarc128.2 were assembled into a multiple regression model and explained a total of 45% of the variation for resistance to isolate OH99 (race 3) (Table 2).
For isolate DW5 (race 5), three QTLs, QTs.fcu-2AS, QTs.fcu-5AL and QTs.fcu-5BL.1 were all significantly associated with the resistance, and they explained 19, 14, and 14% of the trait variation in CIM, respectively (Table 3, Fig. 2). QTs.fcu-2AS peaked in the same region as observed for resistance to isolates Pti2, 86–124, and OH99 (Fig. 2), and the most significant marker was again Xbarc10 explaining 19% of the variation in single-factor regression (Table 2). QTs.fcu-5AL was within the interval defined by markers Xbarc1061 and Xcfa2185, which is the same region as observed for resistance to Pti2 and 86–124 (Fig. 2). Xfcp412 was the most significant marker within the interval and explained 18% variation in single-factor regression (Table 2). QTs.fcu-5BL.1 peaked within the same genomic region as observed for resistance to Pti2, 86–124, and OH99, and again, Xbarc128.2 was the most significant marker and explained 12% of the variation in single-factor regression (Table 2). A significant interaction between the marker Xcfa2106 located on chromosome arm 7BS and Xbarc128.2 was also identified, and the interaction explained 13% of the variation in single-factor regression (Table 2). The three QTLs and the interaction were all significant in the multiple regression model and together they explained 48% (Table 2) of the phenotypic variation.
Discussion
In this research, we identified a total of five QTLs that conferred resistance to tan spot contributed by TA4152-60. QTs.fcu-2AS and QTs.fcu-5BL.1 were significantly associated with resistance to isolates representing all four races. QTs.fcu-5AL was significantly associated with resistance to the isolates representing races 1, 2 and 5. QTs.fcu-5BL.2, which harbors the Tsn1 locus governing reaction to Ptr ToxA, was associated with resistance to the isolates representing races 1 and 2, which are the only two races that produce Ptr ToxA (Lamari and Bernier 1989b, Tomás and Bockus 1987; Tuori et al. 1995). QTs.fcu-4AL was significantly associated with resistance to the race 3 isolate only (Table 3).
Friesen and Faris (2004) identified a QTL on chromosome arm 2AS conferring resistance to the same race 5 isolate used here and the marker Xcdo447 showed the most significant effects. In our research, QTs.fcu-2AS was located in a more proximal position of the arm and Xbarc10 had the most significant effects. Xcdo447 and Xbarc10 are located in different deletion bins (Erayman et al. 2004; Sourdille et al. 2004), which suggests that QTs.fcu-2AS is not the same as the one identified by Friesen and Faris (2004). The QTL QTs.fcu-4AL identified in this study conferred resistance only to the race 3 isolate OH99 (Fig. 2). Based on the position of the common marker Xbarc206 among the maps of Faris et al. (1997, 1999), Sourdille et al. (2004), and ours, QTs.fcu-4AL is probably the same as that previously identified (Faris et al. 1997, 1999; Faris and Friesen 2005; Friesen and Faris 2004). However, this QTL was reported to confer resistance to a race 1 isolate (Faris et al. 1997, 1999) and a race 5 isolate (Friesen and Faris 2004; Faris and Friesen 2005). Although closely linked, different genes conferring tan spot resistance might exist in this region, or different alleles of the same locus might have the ability to recognize different races/isolates. From the position of common marker Xcfa2163 on maps of Liu et al. (2005) and ours, QTs.fcu-5AL identified in this research was different from the 5AL QTL reported in Faris and Friesen (2005). As for the genomic region containing QTs.fcu-5BL.1, there were no previous reports regarding its association with either disease resistance or insensitivity to HSTs. Therefore, QTs.fcu-2AS, QTs.fcu-5AL and QTs.fcu-5BL.1 are probably novel tan spot resistance QTLs derived from the SHW line.
By definition, races 3 and 5 produce Ptr ToxC and Ptr ToxB, respectively, and the Tsc1 and Tsc2 genes govern sensitivity to Ptr ToxC and Ptr ToxB, respectively (Effertz et al. 2002; Friesen and Faris 2004). We identified no QTLs within the genomic regions of 1AS and 2BS, which are known to harbor the Tsc1 and Tsc2 genes. Purified Ptr ToxC and Ptr ToxB were not available for this project, but the fact that no QTLs corresponding to the known genomic regions of Tsc1 and Tsc2 were identified indicates that either these genes are not segregating in our population, or Ptr ToxB and Ptr ToxC were not significant factors in causing disease. In either case, our results indicate that isolates OH99 and DW5 produce virulence factors in addition to Ptr ToxC and Ptr ToxB.
Four out of the five resistance QTLs detected in this study were not associated with any known toxin insensitivity locus. It is possible that underlying the four QTLs are toxin insensitivity genes which have yet to be discovered. Two independent research groups have reported the finding of a fourth and fifth toxin, which were both referred to as Ptr ToxD (Manning et al. 2002, Meinhardt et al. 2003). Lines containing host sensitivity to these two toxins have been reported but chromosomal locations of the sensitivity loci have not been identified. Therefore, other toxins probably exist and might be associated with any of these four QTLs. It is also possible that these QTLs are non-toxin associated, and that some other mechanism of resistance other than toxin insensitivity is conferred by the SHW line similar to that proposed by Faris and Friesen (2005). It is also possible that a combination of these scenarios exist. Regardless, it is apparent that each of the isolates used in this work harbor virulence factors in addition to the known toxins. This fact may have implications on the race classification system. Andrie et al. (2007) reported that the current differential set may need to be expanded due to its insufficiency in classification based on the additional complexity beyond the HSTs described. It was also noted that genotyping of the isolates for the known HST-producing genes (i.e. ToxA and ToxB) is also needed to increase confidence in race classification.
All the resistance QTLs identified in this study were located in the A and B genome chromosomes of the SHW line. It was somewhat surprising that no QTLs were detected in the D genome because the tetraploid parent (Scoop 1), which donated the A and B genomes, showed a mean reaction type of 2.8 in response to isolate Pti2 (race 1) (Xu et al. 2004) indicating Scoop 1 was moderately resistant to moderately susceptible. Tadesse et al. (2007) reported resistance to the race 1 isolate ASC1b on chromosome arm 3DS derived from three SHW lines. Therefore, we expected the high level of resistance in TA4152-60 to be due to genes in the D genome donated by the Ae. tauschii accession WPI358 (TA2516). However, after inoculating the Ae. tauschii accession with Pti2, it showed a reaction type of 3–4 indicating it was also susceptible to tan spot (data not shown). This substantiates the lack of resistance identified on the D genome in our population.
He et al. (2003) compared global gene expression of a SHW line with its diploid and tetraploid parents and found that a significant number of genes had altered expression levels in the hexaploid. Islam et al. (2003) compared the proteome patterns of diploid, tetraploid and hexaploid wheat and found the expression of proteins in hexaploid wheat is affected by interactions among the diploid genomes when they coincide within the hexaploid nucleus. The durum cultivar Scoop 1 carries the resistance genes that underlie the QTLs identified in this research, but it is possible that these genes are not active or are expressed at lower levels in the tetraploid. Once the A and B genomes of the tetraploid are combined with the D genome from Ae. tauschii, the resistance genes may be activated or up regulated leading to higher levels of resistance in TA4152-60. Therefore, comparing the expression level of resistance genes in the tetraploid parent with that of the SHW line might shed light on the issue.
In conclusion, our research revealed five QTLs conferring resistance to tan spot in the SHW line, three of which are novel. Four of the QTLs do not correspond to known toxin insensitivity loci that were previously shown to play significant roles in disease development for the corresponding races. This finding indicates that the wheat-tan spot system is more complex than previously thought, and that isolates harbor numerous virulence factors (possibly unidentified HSTs) in addition to those HSTs currently used to define races. The results of this research and that of Faris and Friesen (2005) indicate that genotypes insensitive to the known tan spot toxins are not always resistant to the disease and that additional and/or alternate factors are involved. Although the currently identified toxins, which define the P. tritici-repentis race classification system, have been shown to be highly important in disease and should be used in selection of the most resistant wheat varieties, our previous (Faris and Friesen 2005) and current research shows that wheat breeders should not rely solely on these toxins, since other virulence factors are highly important in disease development. This research may have implications for the tan spot race classification system but additional research on the pathogen side to define these new virulence factors will be necessary. The molecular markers identified in this research will be useful for deploying the SHW-derived tan spot resistance QTLs in wheat breeding.
References
Ali S, Francl LJ (2003) Population structure of Pyrenophora tritici-repentis prevalent on wheat and noncereal grasses in the Great Plains. Plant Dis 87:418–422
Ali S, Francl LJ, Dewolf ED (1999) First report of Pyrenophora tritici-repentis race 5 from North America. Plant Dis 83:591
Anderson JA, Effertz RJ, Faris JD, Francl LJ, Meinhardt SW, Gill BS (1999) Genetic analysis of sensitivity to Pyrenophora tritici-repentis necrosis-inducing toxin in durum and common wheat. Phytopathology 89:293–297
Andrie RM, Pandelova I, Ciuffetti LM (2007) A combination of phenotypic and genotypic characterization strengthens Pyrenophora tritici-repentis race identification. Phytopathology 97:694–701
Cheong J, Wallwork H, Williams KJ (2004) Identification of a major QTL for yellow leaf spot resistance in the wheat varieties Brookton and Cranbrook. Aust J Agric Res 55:315–319
Chu CG, Xu SS, Friesen TL, Faris JD (2008) Whole genome mapping in a wheat doubled haploid population using SSRs and TRAPs and the identification of QTL for agronomic traits. Mol Breed [online]. doi:10.1007/s11032-008-9171-9
Cox TS (1998) Deepening the wheat gene pool. J Crop Prod 1:1–25
Effertz RJ, Anderson JA, Francl LJ (1998) QTLs associated with resistance to chlorosis induction by in adult wheat. Can J Plant Pathol 20:438–439
Effertz RJ, Meinhardt SW, Anderson JA, Jordahl JG, Francl LJ (2002) Identification of a chlorosis-inducing toxin from Pyrenophora tritici-repentis and the chromosomal location of an insensitivity locus in wheat. Phytopathology 92:527–533
Elias E, Cantrell RG, Hosford RM Jr (1989) Heritability of resistance to tan spot in durum wheat and its association with other agronomic traits. Crop Sci 29:299–304
Engle JS, Madden LV, Lipps PE (2006) Distribution and pathogenic characterization of Pyrenophora tritici-repentis and Stagonospora nodorum in Ohio. Phytopathology 96:1355–1362
Erayman M, Sandhu D, Sidhu D, Dilbirligi M, Baenziger PS, Gill KS (2004) Demarcating the gene-rich regions of the wheat genome. Nucleic Acids Res 32:3546–3565
Faris JD, Anderson JA, Francl LJ, Jordahl JG (1996) Chromosomal location of a gene conditioning insensitivity in wheat to a necrosis-inducing culture filtrate from Pyrenophora tritici-repentis. Phytopathology 86:459–463
Faris JD, Anderson JA, Francl LJ, Jordahl JG (1997) RFLP mapping of resistance to chlorosis induction by Pyrenophora tritici-repentis in wheat. Theor Appl Genet 94:98–103
Faris JD, Friesen TL (2005) Identification of quantitative trait loci for race-nonspecific resistance to tan spot in wheat. Theor Appl Genet 111:386–392
Faris JD, Li WL, Liu DJ, Chen PD, Gill BS (1999) Candidate gene analysis of quantitative disease resistance in wheat. Theor Appl Genet 98:219–225
Friesen TL, Ali S, Kianian S, Francl LJ, Rasmussen JB (2003) Role of host sensitivity to Ptr ToxA in development of tan spot of wheat. Phytopathology 93:397–401
Friesen TL, Faris JD (2004) Molecular mapping of resistance to Pyrenophora tritici-repentis race 5 and sensitivity to Ptr ToxB in wheat. Theor Appl Genet 109:464–471
Friesen TL, Rasmussen JB, Ali S, Kwon CY, Francl LJ, Meinhardt SW (2002) Reaction to Pyrenophora tritici-repentis race 1 by wheat mutants insensitive to Ptr ToxA. Phytopathology 92:38–42
Gamba FM, Lamari L (1998) Mendelian inheritance of resistance to tan spot (Pyrenophora tritici-repentis) in selected genotypes of durum wheat (Triticum turgidum). Can J Plant Pathol 20:408–414
Gamba FM, Lamari L, Brule-Babel A (1998) Inheritance of race-specific necrotic and chlorotic reactions induced by Pyrenophora tritici-repentis in hexaploid wheats. Can J Plant Pathol 20:401–407
He P, Friebe BR, Gill BS, Zhou JM (2003) Allopolyploidy alters gene expression in the highly stable hexaploid wheat. Plant Mol Biol 52:401–414
Hosford RM Jr (1971) A form of Pyrenophora trichostoma pathogenic to wheat and other grasses. Phytopathology 62:28–32
Islam N, Tsujimoto H, Hirano H (2003) Proteome analysis of diploid, tetraploid and hexaploid wheat: towards understanding genome interaction in protein expression. Proteomics 3:549–557
Krupinsky JM (1992) Grass hosts of Pyrenophora tritici-repentis. Plant Dis 76:92–95
Lamari L, Bernier CC (1989a) Evaluation of wheat lines and cultivars for reaction to tan spot Pyrenophora tritici-repentis based on lesion size. Can J Plant Pathol 11:49–56
Lamari L, Bernier CC (1989b) Toxin of Pyrenophora tritici-repentis: host-specificity, significance in disease, and inheritance of host reaction. Phytopathology 79:740–744
Lamari L, Bernier CC (1991) Genetics of tan necrosis and extensive chlorosis in tan spot of wheat caused by Pyrenophora tritici-repentis. Phytopathology 81:1092–1095
Lamari L, Gilbert J, Tekauz A (1998) Race differentiation in Pyrenophora tritici-repentis and survey of physiologic variation in western Canada. Can J Plant Pathol 20:396–400
Lamari L, Sayoud R, Boulif M, Bernier CC (1995) Identification of a new race of Pyrenophora tritici-repentis: implications for the current pathotype classification system. Can J Plant Pathol 17:312–318
Lamari L, Strelkov SE, Yahyaoui A, Orabi J, Smith RB (2003) The identification of two new races of Pyrenophora tritici-repentis from the host center of diversity confirms a one-to-one relationship in tan spot of wheat. Phytopathology 93:391–396
Lee TS, Gough FJ (1984) Inheritance of Septoria leaf blotch (S. tritici) and Pyrenophora tan spot (P. tritici-repentis) resistance in Triticum aestivum cv. Carifen 12. Plant Dis 68:848–851
Liu ZH, Anderson JA, Hu J, Friesen TL, Rasmussen JB, Faris JD (2005) A wheat intervarietal genetic linkage map based on microsatellite and target region amplified polymorphism markers and its utility for detecting quantitative trait loci. Theor Appl Genet 111:782–794
Manning VA, Pandelova I, Ciuffetti LM (2002) A race for a novel host selective toxin. Phytopathology 92:S51
Manly KK, Cudmore RH Jr, Meer JM (2001) Map Manager QTX, cross platform software for genetic mapping. Mamm Genome 12:930–932
Meinhardt S, Ali S, Ling H, Francl L (2003) A new race of Pyrenophora tritici-repentis that produces a putative host-selective toxin. In: Rasmussen JB, Friesen TL, Ali S (eds) Proceedings of the fourth international wheat tan spot and spot blotch workshop. North Dakota Agric. Exp. Station, Fargo, pp 117–121
Nagle BJ, Frohberg RC, Hosford RM Jr (1982) Inheritance of resistance to tan spot of wheat. In: Hosford RM Jr (ed) Tan spot of wheat and related diseases workshop. North Dakota Agricultural Experimental Station, Fargo, pp 40–45
Nelson JC (1997) QGENE: software for marker-based genomic analysis and breeding. Mol Breed 3:239–245
Orolaza NP, Lamari L, Balance GM (1995) Evidence of a host specific chlorosis toxin from Pyrenophora tritici-repentis, the causal agent of tan spot of wheat. Phytopathology 85:1282–1287
Riede CR, Francl LJ, Anderson JA, Jordahl JG, Meinhardt SW (1996) Additional source of resistance to tan spot of wheat. Crop Sci 36:771–777
SAS Institute (1999) SAS/STAT user’s guide, releases: 8.2, 8.1, 8.0. SAS Institute, Inc., Cary
Singh PK, Gonzalez-Hernandez JL, Mergoum M, Ali S, Adhikari TB, Kianian SF, Elias EM, Hughes GR (2006a) Identification and molecular mapping of a gene conferring resistance to Pyrenophora tritici-repentis race 3 in tetraploid wheat. Phytopathology 96:885–889
Singh PK, Mergoum M, Ali S, Adhikari TB, Elias EM, Anderson JA, Glover KD, Berzonsky WA (2006b) Evaluation of elite wheat germplasm for resistance to tan spot. Plant Dis 90:1320–1325
Sourdille P, Singh S, Cadalen T, Brown-Guedira GL, Gay G, Qi L, Gill BS, Dufour P, Murigneux A, Bernard M (2004) Microsatellite-based deletion bin system for the establishment of genetic-physical map relationships in wheat (Triticum aestivum L.). Funct Integr Genomics 4:12–25
Strelkov SE, Lamari L, Sayoud R, Smith RB (2002) Comparative virulence of chlorosis-inducing races of Pyrenophora triticirepentis. Can J Plant Pathol 24:29–35
Strelkov SE, Lamari L (2003) Host-parasite interactions in tan spot [Pyrenophora tritici-repentis] of wheat. Can J Plant Pathol 25:339–349
Sykes EE, Bernier CC (1991) Qualitative inheritance of tan spot resistance in hexaploid, tetraploid, and diploid wheat. Can J Plant Pathol 13:38–44
Tadesse W, Schmolke M, Hsam SLK, Mohler V, Wenzel G, Zeller FJ (2007) Molecular mapping of resistance genes to tan spot [Pyrenophora tritici-repentis race 1] in synthetic wheat lines. Theor Appl Genet 114:855–862
Tomás A, Bockus WW (1987) Cultivar specific toxicity of culture filtrate of Pyrenophora tritici-repentis. Phytopathology 77:1337–1366
Tuori RP, Wolpert TJ, Ciuffetti LM (1995) Purification and immunological characterization of toxic components from cultures of Pyrenophora tritici-repentis. Mol Plant Microbe Interact 8:41–48
Xu SS, Friesen TL, Mujeeb-Kazi A (2004) Seedling resistance to tan spot and Stagonospora nodorum blotch in synthetic hexaploid wheats. Crop Sci 44:2238–2245
Acknowledgments
The authors thank P. E. Lipps and S. Ali for providing P. tritici-repentis isolates, J. G. Hu and G. T. Yu for critical review. This research was supported by USDA-ARS CRIS Projects 5442-22000-037-00D and 5442-22000-030-00D.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by J. W. Snape.
Mention of trade names or commercial products in this article is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the U.S. Department of Agriculture.
Rights and permissions
About this article
Cite this article
Chu, CG., Friesen, T.L., Xu, S.S. et al. Identification of novel tan spot resistance loci beyond the known host-selective toxin insensitivity genes in wheat. Theor Appl Genet 117, 873–881 (2008). https://doi.org/10.1007/s00122-008-0826-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00122-008-0826-z