Abstract
Blackleg (stem canker) caused by the fungus Leptosphaeria maculans is one of the most damaging diseases of oilseed rape (Brassica napus). Crop relatives represent a valuable source of “new” resistance genes that could be used to diversify cultivar resistance. B. rapa, one of the progenitors of B. napus, is a potential source of new resistance genes. However, most of the accessions are heterozygous so it is impossible to directly detect the plant genes conferring specific resistance due to the complex patterns of avirulence genes in L. maculans isolates. We developed a strategy to simultaneously characterize and introgress resistance genes from B. rapa, by homologous recombination, into B. napus. One B. rapa plant resistant to one L. maculans isolate was used to produce B. rapa backcross progeny and a resynthesized B. napus plant from which a population of doubled haploid lines was derived after crossing with natural B. napus. We then used molecular analyses and resistance tests on these populations to identify and map the resistance genes and to characterize their introgression from B. rapa into B. napus. Three specific genes conferring resistance to L. maculans (Rlm1, Rlm2 and Rlm7) were identified in B. rapa. Comparisons of genetic maps showed that two of these genes were located on the R7 linkage group, in a region homologous to the region on linkage group N7 in B. napus, where these genes have been reported previously. The results of our study offer new perspectives for gene introgression and cloning in Brassicas.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Stem canker (Blackleg) caused by the fungal pathogen Leptosphaeria maculans Desm. (Ces & de Not), affects all cultivated Brassica species and is responsible for severe yield losses worldwide in Brassica napus L. production (Fitt et al. 2006; Gugel and Petrie 1992). The most effective way to control stem canker is by genetic resistance (Rimmer and vandenBerg 1992). Two types of resistance are commonly used. On the one hand, quantitative resistance is partial, race non-specific and under polygenic control (Pilet et al. 1998) and is usually evaluated in adult plants in the field. Quantitative resistance reduces the severity of symptoms and has been suggested to be durable. On the other hand, qualitative seedling resistance is controlled by a single gene (Ansan-Melayah et al. 1998), leads to complete resistance and is race specific. Qualitative resistance may be evaluated in seedlings from the cotyledon stage to the adult stage. Several specific resistance genes (Rlm) have been identified in Brassica species (Delourme et al. 2006a). Each specific resistance gene only confers resistance against races of the fungus, which have the corresponding avirulence (AvrLm) gene and therefore this type of resistance gene exerts a high selection pressure on the pathogen population. L. maculans populations evolve rapidly by genetic recombination during sexual reproduction, coupled with large-scale dispersal of ascospores (West et al. 2001), and adapt to major resistance genes. For example, Rlm1, which was used in a large proportion of French oilseed rape production areas, was overcome by pathogen populations within 3 years (Rouxel et al. 2003). More recently, in Australia, the efficacy of the “Surpass 400” specific resistance in the ‘Surpass 400’ cultivar was lost after only two to three growing seasons (Li et al. 2003; Sprague et al. 2006). Finally, several specific resistance genes are no longer effective when corresponding virulence genes become more common in the local pathogen population (Balesdent et al. 2005). In order to improve the durability of resistance, different specific resistance genes could be managed in time and space and used in combination with quantitative resistance and therefore, new sources of resistance are needed.
Different sources of specific resistance have already been identified. By screening a collection of oilseed rape varieties with a differential set of L. maculans isolates, six resistance genes were detected in B. napus: Rlm1 was identified in ‘Quinta’ (Ansan-Melayah et al. 1998), Rlm2 and Rlm3 in ‘Glacier’ (Ansan-Melayah et al. 1998; Balesdent et al. 2002), Rlm4 in ‘Jet Neuf’ (Balesdent et al. 2001), Rlm7 in non-commercial lines (Balesdent et al. 2002), and Rlm9 in ‘Darmor’ (Delourme et al. 2004). Five of these genes (Rlm1, Rlm3, Rlm4, Rlm7 and Rlm9) are on the N7 linkage group, and only Rlm2 is on a different linkage group (Delourme et al. 2004; 2006a). Some specific resistance genes were also identified in other Brassica species. Cultivated Brassica species are organized into three homeologous genomes, named A, B and C, of which A and C are present in B. napus. All the resistance genes detected in B. napus are on the A genome, and four resistance genes were also described on the A genome in B. rapa (AA): Rlm8 (Balesdent et al. 2002), LepR1, LepR2 (Yu et al. 2005) and LepR3 (Li and Cowling 2003). The majority of Brassica species containing the B genome also carry specific resistance to L. maculans. Rlm5 and Rlm6 were identified on the B genome of B. juncea (AABB) (Balesdent et al. 2002; Rimmer and vandenBerg 1992) and Rlm10 was found in B. nigra (BB) (Chevre et al. 1996; Chevre, Personal Communication). To date, no specific resistance genes were detected on the C genome, in either diploid or polyploid species (Rimmer and vandenBerg 1992).
A few genes have been introduced from the B genome of B. juncea into B. napus on the DY717 linkage group (Barret et al. 1998; Chevre et al. 1997; Saal et al. 2004), but only large introgressions were obtained and no recombination was observed between the B genome introgressed fragment and its homeologous region in B. napus, because of the low level of homology between the genomes. Resistance genes were also introduced into B. napus from B. rapa (Crouch et al. 1994) and their introgression, via synthetic B. napus, was successful because B. rapa is a progenitor of B. napus. We assume that these B. rapa genes were introgressed by homologous recombination, but they were only mapped in B. napus, without information on their original position in B. rapa (Yu et al. 2004, 2005).
The aim of this study was to assess a method to carry out a concurrent introgression in B. napus and characterization of the specific resistance genes present in B. rapa, in spite of the heterozygosity of most B. rapa accessions and the complex AvrLm patterns of the fungus isolates. One resistant plant was selected from an old French forage B. rapa variety, which is a heterozygous plant population, by screening seedlings for resistance with several L. maculans isolates. The avirulence patterns of the isolates used were mostly representative of the most common AvrLm genes in the French L. maculans populations (Balesdent et al. 2006). Specific resistance genes carried by this plant were mapped simultaneously in B. rapa and in B. napus upon their introgression via resynthesized B. napus. Therefore, this is the first report of a map of the linkage group carrying resistance genes in B. rapa. In addition, we identified the Rlm genes derived from B. rapa and characterized their introgression in B. napus, using several races of L. maculans with known patterns of avirulence genes (AvrLm) and by molecular analyses.
Materials and methods
Fungal material
Ten L. maculans isolates were used to test the plant material for resistance genes. PHW1223 (Balesdent et al. 2005), IBCN14 (Balesdent et al. 2005), S7 (Brun et al. 2001), R2 (Somda et al. 1999), 929 (kindly provided by Dr. J.P. Tewari, Edmonton, Canada, in 1993) and R1 M1.3 (obtained at Le Rheu in 2001 from a single ascospore sampled from an infected ‘SMX’ plant, which harbors Rlm1 and Rlm6) are natural isolates, whereas v45.57, v33.6.3, v29.3.1 and v48.2.6 are progeny isolates obtained following in vitro crosses between L. maculans isolates. In brief, v33.6.3 is an isolate derived from the cross 33 (Balesdent et al. 2002); v29.3.1 and 19.2.1 (Balesdent et al. 2002) were crossed to generate isolate v45.57 (Balesdent, unpublished data); and v48.2.6 was recovered from the cross 48 between 19.4.45 and v34.4.4 (Balesdent et al. 2002). The pattern of avirulence alleles (Avrlm), for most of the ten known AvrLm genes, in each isolate was previously established on an appropriate differential host set under controlled conditions (Balesdent et al. 2002; Balesdent et al. 2005; Balesdent, Personal Communication; Brun, Personal Communication). In this paper, we described isolate avirulence patterns using the nomenclature proposed by Balesdent et al. (2005); the avirulence alleles present in the isolate are listed, only the avirulence alleles in parentheses have not been characterized.
Plant material
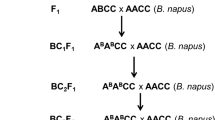
Seedlings of an old non homogeneous French forage B. rapa variety, ‘Chicon’, were screened with several isolates and one plant, ‘C1.3’, was selected for its resistance, at the cotyledon stage, to isolate S7 (Av1-5-6-7-(8)). This plant was crossed to a susceptible B. rapa doubled haploid line, ‘Z1’ (kindly provided by AAFC, Canada), to produce F1 hybrids. One F1 plant was then backcrossed to ‘Z1’ to produce 164 BC1 progeny, named CZB1 (Fig. 1). The same ‘C1.3’ plant was crossed to ‘RC’, a B. oleracea ssp. alboglabra doubled haploid line, and the F1 plant was colchicine-doubled to produce a resynthesized B. napus (RCC). The resynthesized B. napus was crossed to ‘Darmor’ to produce eight hybrids, RCCD. ‘Darmor’ is a winter oilseed rape variety that contains both quantitative adult-stage resistance to L. maculans and the specific resistance gene Rlm9. A population of doubled haploid lines (RCCD.DH) was produced from these hybrids using microspore cultures (Polsoni et al. 1988). More than 300 doubled haploid (DH) lines were produced either by spontaneous chromosome doubling or colchicine treatment. Thirty-one DH lines (between one to ten lines per F1 hybrid) with good seed set and showing differential reaction to isolates S7, 929 and v45.57 were used in this study (Fig. 1). In addition, 150 DH lines obtained from a B. napus ‘Darmor-bzh’ × ‘Yudal’ hybrid and previously described by Foisset et al. (1996) were also used in this study.
Resistance tests at the seedling stage
A cotyledon test derived from the test of Williams and Delwiche (1979) was performed following the protocol described by Somda et al. (1999). A plastic cover was placed over the inoculated plants to create an atmosphere with 100% relative humidity (RH) and the plants were incubated in the dark at 20°C for 24 h in a growth chamber. Inoculated plants were then placed in an atmosphere with 80–90% RH (without a plastic cover) and a 16-h photoperiod at 20°C (±1°C). Symptoms on cotyledons were scored 14 and 21 days after inoculation using the 0 (no visible reaction) to 11 (total collapse of the tissue) scale. Plants were considered to be susceptible to an isolate when all replicates had a score higher than or equal to 9, at 21 days after inoculation, and resistant when all replicates had a score less than or equal to 3. When intermediate scores were obtained, we considered that the genotype had an intermediate behavior. In B. rapa BC progeny, each plant was inoculated with two isolates, one per cotyledon, and each cotyledon lobe was treated as a replicate, therefore, making two inoculation sites per plant and per isolate. For doubled haploid lines, each isolate was inoculated on three plants of each line and on the four cotyledon lobes of each plant. Each cotyledon lobe was treated as a replicate; therefore, 12 inoculation sites per isolate and per line were used.
Molecular analyses
Genomic DNA was extracted from the young leaves of all individual plants according to the method of Doyle and Doyle (1990). The DNA concentration was adjusted to 10, 50 and 1 ng/μl for RAPD, AFLP and SSR assays or specific markers, respectively.
Bulked segregant analysis
Sixteen plants showing resistance and 11 plants showing susceptibility to both R2 and S7 isolates were selected from the CZB1 population and their genomic DNA pooled to form the resistant (RB1) and the susceptible (SB1) bulks, respectively (Michelmore et al. 1991). These bulks were used to determine the linkage group carrying the resistance genes. As a second step, two additional bulks, RB2 and SB2, were constituted from the CZB1 population in order to cover a larger genomic region with molecular markers: the RB2 bulk was composed of the pooled genomic DNA from 12 plants showing resistance to the R2 isolate, and carrying the ‘C1.3’ allele for four molecular markers that were detected as linked to the resistance in the first bulked segregant analysis, or that were carried by the same linkage group as these markers; the SB2 bulk was composed of seven plants showing susceptibility to the R2 isolate and the ‘Z1’ allele at these four loci.
Random amplified polymorphic DNA (RAPD) markers
The RAPD primers (Operon Technologies) covering several A genome chromosomes were selected from published reference maps (Lombard and Delourme 2001). The PCRs were performed using the protocol of Foisset et al. (1996). Amplified products were separated on 1.8% agarose gels and images acquired after staining with ethidium bromide. The RAPD markers were named by combining the primer code and the size of the amplified DNA fragments (for example, W11.1690).
Simple sequence repeat (SSR) markers
The SSR primers used in this study were obtained from the UK Cropnet Database (prefixed with Ol; http://www.ukcrop.net/perl/ace/search/BrassicaDB) or were developed by Celera AgGen (prefixed with CB or Bras) and are publicly available (Piquemal et al. 2005). The PCRs were performed in a total volume of 5 μl, containing M13 universal fluorescent tag to label the amplified DNA fragments. The amplified products were electrophoresed through 6.5% denaturing polyacrylamide gels and detected with an automated DNA sequencer (LI-COR Biosciences).
Amplified fragment length polymorphism (AFLP) and sequence-specific amplification polymorphism (SSAP) markers
Amplified fragment length polymorphism assays were conducted essentially as described in a previous study (Vos et al. 1995). For AFLP and SSAP, DNA was digested by EcoRI plus MseI. After ligation of double-stranded adaptors to the ends of the restriction fragments, pre-amplification was performed with EcoRI and MseI primer pairs with one selective nucleotide at their 3′-end. For AFLP, the selective amplification was performed with EcoRI and MseI primer pairs with three selective nucleotides at their 3′-end. The EcoRI + 3 primers were labeled by IRD700 or IRD800 fluorochrome (MWG Biotech). For SSAP, the selective amplification was performed with one unlabeled EcoRI primer with three selective nucleotides at its 3′-end and one primer designed in the nucleotide binding site (NBS) conserved region of NBS-LRR resistance genes (Meyers et al. 1999) and labeled by IRD700 or IRD800 fluorochrome (MWG Biotech). Three different NBS primers designed by Rocherieux (2003) were used, NBS1 (GGGGGGGTAGTGGGAAAGACGAC), NBS2 (GGCGGTTCAGGGAA-AGACATAC) and NBS1c (GTCGTCTTCCCAGCTACCCCAGTCCC). For AFLP and SSAP, the amplified products were separated on 5.5% denaturing acrylamide gels and detected with the same sequencer as SSR markers. AFLP and SSAP markers were named using the international code for each primer pair and the size of the amplified fragment. AFLP markers that revealed polymorphisms between the RB2 and SB2 bulks were then checked on all the 19 individuals making up the bulks. Markers, which showed a co-segregation ratio with the resistance phenotype equal to or greater than 15 out of 19 were retained and tested on 116 additional plants from the BC1 B. rapa population.
Specific markers
Specific molecular markers (prefixed with At) were developed from Arabidopsis sequences in this study, using the published alignment between N7 and chromosome 1 of Arabidopsis (Mayerhofer et al. 2005). Two pairs of primers, designed on At1g80400 (F: AGTCCTTGA-GGCCTTAGAGAAGA and R: CGTTTGAGGACTGTGTTGTCC) and At1g80530 (F: GACGTTCCCGCTTTACTCC and R: CTCATAGGAAAATTCCTCATATTGGT) genes, showed polymorphic loci between ‘C1.3’ and ‘Darmor’ on R7/N7. PCR was performed in a final volume of 8 μl containing 2 mM MgCl2, 310 μM dNTP, 0.62 μM of each primer, 0.062 U/μl Taq polymerase (Promega), 1× Taq polymerase buffer (Promega) and 1.8 ng/μl genomic DNA. The DNA was amplified using a cycling profile of 5 min at 94°C followed by 30 cycles of 30 s at 94°C, 1 min at 55°C (for primers designed on At1g80040) or at 60°C (for primers designed on At1g80530) and 45 s at 72°C. At the end of 30 cycles, the reactions were held at 72°C for 5 min. Amplified products were separated on 2% agarose gels and images acquired after staining with ethidium bromide.
Statistical analyses and construction of genetic linkage maps
The genetic control of resistance and the segregation of molecular markers in the B. rapa BC progeny (CZB1) were analyzed using Chi-square tests. Segregating markers were scored for each plant and linkage analysis performed with MAPMAKER/EXP version 3.0 (Lincoln et al. 1992). Linkage groups were established with a LOD-score threshold of 4.0 and a maximum recombination frequency of 0.4. The Kosambi function (Kosambi 1944) was used to evaluate the genetic distances (in centiMorgans; cM) between linked markers.
Results
Genetic analyses of resistance in B. rapa progeny
As a first step in our study, a BC1 progeny was derived from ‘C1.3’ to determine the genetic control of resistance.
Resistance tests were performed on all 164 plants of the B. rapa BC1 progeny (CZB1) using the fungal isolates R2 (Av5-7-(8)-10) and S7 (Av1-5-6-7-(8)). These two isolates were chosen for their particular patterns of AvrLm alleles (virulent on mustard resistance genes Rlm6 (R2) and Rlm10 (S7)), and because they differed at the AvrLm1 allele. Each seedling was inoculated with both isolates, one isolate on each cotyledon (Table 1). We found that 81 plants were resistant and 47 plants were susceptible to both the isolates, and 17 plants were resistant to S7 and susceptible to R2. The reaction to one of the two isolates was unclear in 19 plants. Among these, five plants were resistant and one susceptible to S7, and five plants were resistant and eight susceptible to R2. Based on the resistance segregation ratios, Chi-square test confirmed that resistance to R2 is compatible with a single dominant gene, rR2 (86R:72S; P = 0.26), whereas resistance to S7 is conferred by two dominant genes (103R:48S), separated by a genetic distance of at least 21 cM (α = 0.05). Furthermore, because no plants were both resistant to R2 and susceptible to S7, it suggested that rR2 was closely linked or may be one of the genes governing resistance to S7 (rS7.1). This gene could be one of Rlm5, Rlm7 or Rlm8, because the two fungal isolates shared AvrLm5, AvrLm7, and possibly AvrLm8. The second gene conferring resistance to S7, rS7.2, appears to also be linked to rR2 because only 17 out of the 64 plants susceptible to R2 were also resistant to S7. If we assume that rR2 and rS7.1 are the same gene, the genetic distance between rR2 and rS7.2 is therefore estimated to be 26.6 cM, based on a sample of 64 plants.
Mapping of resistance genes in the B. rapa progeny
For the second step in our study, we mapped the genes conferring resistances to S7 and R2 in the B. rapa BC1 progeny.
First, we searched for polymorphic markers linked to the resistance genes by using 34 RAPD primers in a bulked segregant analysis. Only four markers were polymorphic in our progeny and among these, two markers, W11.1690, which was previously mapped to DY10 (corresponding to N7; Delourme et al. 2006b) in B. napus (Lombard and Delourme 2001), and Y13.1500, which remains unmapped in B. napus, were present in the resistant bulk RB1 and absent in the susceptible bulk SB1. The linkage between these markers and the resistance genes was confirmed by genotyping 135 CZB1 progeny plants.
Second, we checked that the resistance genes are located on linkage group R7, the chromosome homologous to B. napus N7 (Parkin and Lydiate 1997), because this position was suggested by the linkage between resistance genes and W11.1690. The 135 CZB1 plants were therefore genotyped with CB10450 and CB10439 SSR markers that were previously mapped to N7 linkage group in B. napus according to Delourme et al. (2006b) and Piquemal et al. (2005). The linkage between CB10439 and rR2/rS7.1 confirmed the location of the resistance genes on R7 (Fig. 2).
Genetic map of the R7 linkage group in B. rapa BC progeny and its homologue N7, in the B. napus ‘Darmor-bzh’ × ‘Yudal’ DH population. The locus conferring resistance to the L. maculans isolate R2 was designated rR2 while rS7.1 and rS7.2 correspond to the putative position of the loci conferring resistance to the S7 isolate. The molecular markers which are underlined were polymorphic between ‘C1.3’ and ‘Darmor’ and were used in RCCD.DH characterization
Third we identified additional molecular markers in the targeted genomic region, by using the second set of bulks (RB2 and SB2) built on the phenotypic resistance data and genotypic data at W11.1690, Y13.1500, CB10439 and CB10450 markers. Of the 170 AFLP and 36 SSAP primer combinations analyzed, 54 had at least one marker present in ‘C1.3’ and in the RB2 resistant bulk, and absent in ‘Z1’ and in the SB2 susceptible bulk. These 54 AFLP or SSAP primer combinations were then used to genotype the 19 plants making up the bulks. Thirty-five of these combinations generated markers that were linked to the resistance genes and finally, 12 markers were tested on the 135 B. rapa BC plants to establish the genetic map of the R7 linkage group (Fig. 2). The genes rR2 and rS7.1 were located at a distance of 4 and 3 cM from the markers E35M69.146 and E36M70.292, respectively. The position of rS7.2 was estimated by only including plants susceptible to the R2 isolate in the mapping population (48 plants). The rS7.2 gene was found to be located in a 9.9-cM interval between E44M66.277 and E36M73.403.
Identification of resistance genes in doubled haploid lines after introgression in B. napus
In the third step of our study, the resistant plant ‘C1.3’ was used to produce a resynthesized B. napus. This resynthesized B. napus was then crossed to the B. napus variety ‘Darmor’ to generate a doubled haploid segregating population, RCCD.DH (Fig. 1). As opposed to the genetically unique B.rapa BC1 plants, which only allowed us to test a maximum of two isolates per genotype (one per cotyledon), the use of selfed progeny of RCC.DH lines provided an opportunity to test several isolates per genotype, which was necessary to fully characterize the resistance genes carried by ‘C1.3’. Therefore, selfed progeny of 31 doubled haploid lines from the RCCD.DH population was used to perform the subsequent tests.
None of the lines tested were resistant to fungal isolates v45.57 (Av3-6-8-10), v48.2.6 (Av3-5) or IBCN14 (Av5-6-(10)) (Table 2), which indicated that the DH lines did not contain the genes Rlm3, Rlm5, Rlm6, Rlm8 or Rlm10. By knowing that these genes were absent in the lines tested, it was possible to determine the resistance genes present in the DH line using the other isolates. The detection of lines resistant to isolates R2 (Av5-7-(8)-10), v33.6.3 (Av1-5-6-8-(10)), or PHW1223 (Av5-6-8-9-(10)) indicated that Rlm7, Rlm1 and Rlm9, respectively, were segregating in the population. The four lines resistant to the isolate 929 (Av2-3-5-6-9-10), but susceptible to PHW1223 (Av5-6-8-9-(10)), showed that Rlm2 was also present in the population. Finally, all the DH lines without Rlm7 were susceptible to R1M1.3 isolate (Av4-5-7-(8)-(10)), suggesting that Rlm4 may be absent in these lines. However, we did not have an appropriate isolate to determine whether Rlm4 was in fact absent in the population or may have been lost due to linkage to Rlm7.
By combining the results of all the resistance tests (Table 2), we could deduce that Rlm1 was present in five lines (lines 18–22) and absent in 23 lines, Rlm2 was present in five lines (lines 20–21 and lines 28–30) and absent in 19 lines, Rlm7 was present in 10 lines (lines 11–20) and absent in 20 lines and Rlm9 was present in 7 lines (lines 22–28) and absent in 23 lines. Six lines carried more than one resistance gene.
According to these results, resistance to the R2 (Av5-7-(8)-10) isolate, observed in the B. rapa BC progeny, was conferred by Rlm7, which also conferred resistance to the S7 isolate (Av1-5-6-7-(8)). The rR2 and rS7.1 loci mapped in the BC progeny thus corresponded to the Rlm7 locus. The Rlm1 gene, detected in ‘C1.3’, also conferred resistance to the S7 isolate, and was mapped as the rS7.2 locus in B. rapa BC progeny.
Localization of specific resistance genes in doubled haploid lines after introgression into B. napus
In the last step of our study, we positioned resistance genes on the R7/N7 linkage group in the RCCD.DH lines, in order to assess the efficiency of resistance gene transfer from B. rapa to B. napus. The map of the N7 linkage group developed in ‘Darmor-bzh’ × ‘Yudal’ (Delourme et al. 2006b; Foisset et al. 1996; Lombard and Delourme 2001; Piquemal et al. 2005) was improved for this study by using AFLP markers (Fig. 2) and five markers common to both B. rapa and B. napus were used to align the R7 and N7 linkage groups (Fig. 2). The 31 RCCD.DH lines were genotyped by 19 molecular markers polymorphic between ‘C1.3’ and ‘Darmor’ (Fig. 3). Nine of these markers were mapped to the N7 B. napus linkage group (Fig. 2), among these: two were SSR markers already mapped in B. napus (Delourme et al. 2006b; Piquemal et al. 2005) and seven were AFLP markers added to N7 in B. napus in this study; eight markers were mapped on B. rapa R7 linkage group (Fig. 2); and two specific markers were mapped to the R7/N7 linkage group in a ‘Darmor’ × ‘C1.3’ interspecific hybrid (unpublished data). This mapping effort thus allowed us to cover, in the DH lines, a region corresponding to the B. rapa R7 region between E32M71.237 and Y13.1500, i.e., 64% of the R7 linkage group. The markers were ordered according to their relative position in B. rapa and B. napus and to the recombination observed in the RCCD.DH lines (Fig. 3), but genetic distances between markers were not evaluated in this population because of the limited number of lines.
The putative positions of Rlm1, Rlm2, Rlm7 and Rlm9 in the RCCD.DH lines were deduced from the comparison of the R7/N7 genotypes of lines with or without the Rlm genes (Fig. 3). Because all the RCCD.DH lines were derived from the same ‘RCC’ synthetic plant, produced by colchicine doubling of a single F1 hybrid (‘RC’ × ‘C1.3’), ‘RCC’ was homozygous at the Rlm1, Rlm2 and Rlm7 loci. Rlm1, Rlm2 and Rlm7 originated from ‘C1.3’ and should be linked to ‘C1.3’ alleles. Rlm9 was present in ‘Darmor’ and should be linked to ‘Darmor’ alleles.
The ten lines carrying Rlm7 differed from the 22 lines without Rlm7 only on the At1g80400-E35M73.200 interval, with all resistant lines containing ‘C1.3’ alleles on this interval and all susceptible lines containing ‘Darmor’ alleles. This indicated that Rlm7 was localized in this interval (Fig. 3).
For Rlm1, Rlm2 and Rlm9, no interval seemed to differentiate resistant from susceptible lines, in spite of regions in the linkage group common to all resistant lines either for Rlm1, or Rlm9. Indeed, for each interval included in these regions, we found some susceptible lines containing alleles from the resistant parent (Fig. 3).
Discussion
This study showed that in order to characterize three L. maculans specific resistance genes (Rlm1, Rlm2 and Rlm7) present in the B. rapa ‘C1.3’ plant it was necessary to analyze two segregating populations, one BC1 B. rapa population and a B. napus doubled haploid population, derived from the same B. rapa resistant genotype. To identify resistance genes in B. rapa genetic resources, which are generally heterozygous, several fungal isolates must be tested on each plant because the fungal isolates used possess complex patterns of avirulence genes. In BC1 plants, however, only two isolates can be analyzed at one time, so plant material was produced that permitted us to simultaneously identify resistance genes and assess the efficiency of their introgression in B. napus. Doubled haploid lines were produced from resynthesized B. napus using ‘C1.3’ as the B. rapa progenitor, and by crossing it to a B. napus cultivar which carries a resistance gene on N7. This strategy had the added benefit of determining the respective positions of both B. rapa and B. napus resistance genes. The number of lines (31) was sufficient to segregate out all the resistance genes from ‘C1.3’ and to obtain various sizes of introgression.
In the B. rapa progeny, Rlm1 was mapped to the R7 linkage group, between the markers E44M66.277 and E36M73.403, but due to the lack of polymorphic molecular markers between ‘C1.3’ and ‘Darmor’ in this region, it was not possible to pin point Rlm1 in RCCD.DH lines. The position of Rlm7 was mapped in both the B. rapa and the RCCD.DH populations. The comparison of the two maps indicated that the Rlm7 locus is in the At1g80040-E36M70.292 interval. Interestingly, Rlm1 and Rlm7 are two resistance genes naturally present in B. napus and have been mapped to DY10, which corresponds to the N7 linkage group (Delourme et al. 2004, 2006b), on intervals homologous to the B. rapa regions in this present study. Thus our results suggest that these resistance genes have the same position in B. rapa and B. napus. Either these genes are orthologous and were present on the B. rapa A genome prior to the formation of B. napus, and were present in one of the B. rapa plants at the origin of B. napus, or these genes were transferred from one species to the other through interspecific hybridization.
In B. napus, other L. maculans specific resistance genes have been suggested to be located on the N7 linkage group, in different cultivars (Delourme et al. 2006a, 2004; Dion et al. 1995; Ferreira et al. 1995; Mayerhofer et al. 2005; Rimmer 2006). Hence, there appears to be a cluster of resistance genes on N7 in B. napus. If these B. napus genes have orthologs in B. rapa, as it seems to be the case for Rlm1 and Rlm7, then a cluster of resistance genes may also be present in B. rapa, and other resistance genes may be found on R7 among B. rapa species. Another hypothesis is that some resistance genes appeared in B. napus after its formation. Indeed, clusters of resistance genes are known to evolve rapidly through unequal crossing over, gene conversion or recombination, which may generate new alleles or new resistance genes (Noel et al. 1999).
In this study, the Rlm9 gene could not be mapped, however in a previous study (Delourme et al. 2004) it was positioned in B. napus on the N7 linkage group between the markers A14.880 and C02.1375. Two of our findings appear to confirm the position of Rlm9 on N7: first, Rlm7 and Rlm9 genes were never found simultaneously in the same plant, and second, all RCCD.DH plants carrying Rlm9 possessed ‘Darmor’ alleles on N7 between the CB10439 and E46M70.134 markers. However, since plants were found that lacked Rlm9 although they had the ‘Darmor’ alleles in this interval, Rlm9 may be a major resistance gene whose expression is regulated by other genetic factors.
In our study, the Rlm genes detected in B. rapa were introgressed into B. napus by homologous recombination via the creation of a synthetic B. napus. Homologous recombination occurred at a high frequency and along the entire linkage group, because our molecular analysis of the RCCD.DH lines showed large variations in the sizes of introgressed fragments. This observation agrees with the results reported by Parkin and Lydiate (1997), which showed that R7 and N7 linkage groups are essentially the same. This may explain why some resistance genes introduced from B. rapa could be used successfully in B. napus commercial lines, such as the ‘Surpass 400’ resistance gene (Li and Cowling 2003), whereas resistance genes found on the Brassica B genome showed a lack of recombination when they were introgressed into B. napus (Barret et al. 1998; Chevre et al. 1997).
Three specific resistance genes, Rlm1, Rlm2 and Rlm7, were detected in a single B. rapa plant. Further screening of the same or of other B. rapa cultivars with L. maculans isolates may detect other resistance genes and perhaps even resistance genes that do not exist naturally in B. napus. Resistance genes have been detected in B. rapa: among 555 accessions, 4.5% contained Rlm1, Rlm2 or Rlm4 (Delourme et al. 2006a). Both Rlm7 and Rlm8 were detected in at least one B. rapa accession (Balesdent et al. 2002; Delourme et al. 2006a). The Rlm8 gene was recently described and has not yet been observed in B. napus. Three resistance genes, LepR1, LepR2 and LepR3, were introduced from B. rapa into B. napus (Crouch et al. 1994). The LepR1 gene was mapped to N2, and LepR2 and LepR3 to N10 (Yu et al. 2004; Yu et al. 2005), clearly indicating that they are different from Rlm1 and Rlm7 that are present in ‘C1.3’. In our study, we did not map the Rlm2 gene, but if its position is conserved between B. rapa and B. napus, it is expected to map to R10 which corresponds to B. napus LG16-N10 (Delourme et al. 2004, 2006b), so in a similar position to LepR2 or LepR3. This suggests either that Rlm2 corresponds to LepR2 or to LepR3, or that another cluster of L. maculans resistance genes exists on N10/R10 in B. rapa.
We have produced plant material that may be used to clone different Rlm genes identified in this study. In spite of the presence of different resistance genes on the same linkage group (N7) and the complex pattern of AvrLm genes in the L. maculans isolates, the resistant doubled haploid lines with very small introgression should allow the identification of molecular markers closely linked to each Rlm gene. Subsequent fine mapping would then be possible by screening the 300 DH lines produced with molecular markers and an appropriate isolate of L. maculans. For example, isolate R2 could be used for Rlm7, because Rlm5, Rlm8 and Rlm10 were not present in this material.
Last, the material characterized was produced in a ‘Darmor’ genetic background, which carries polygenic resistance conferred by various quantitative trait loci (Pilet et al. 1998). It has been suggested that the combination of quantitative and qualitative resistance may improve the durability of resistance (Brun et al. 2000). In the future, therefore, it should be possible to choose DH lines carrying different combinations of quantitative trait loci and Rlm genes effective against the local pathogen population, and to assess the genetic structure that provides the most effective and durable resistance under field conditions.
References
Ansan-Melayah D, Balesdent MH, Delourme R, Pilet ML, Tanguy X, Renard M, Rouxel T (1998) Genes for race-specific resistance against blackleg disease in Brassica napus L. Plant Breed 117:373–378
Balesdent MH, Attard A, Ansan-Melayah D, Delourme R, Renard M, Rouxel T (2001) Genetic control and host range of avirulence toward Brassica napus cultivars Quinta and Jet Neuf in Leptosphaeria maculans. Phytopathology 91:70–76
Balesdent MH, Attard A, Kuhn AL, Rouxel T (2002) New avirulence genes in the phytopathogenic fungus Leptosphaeria maculans. Phytopathology 92:1122–1133
Balesdent MH, Barbetti MJ, Li H, Sivasithamparam K, Gout L, Rouxel T (2005) Analysis of Leptosphaeria maculans race structure in a worldwide collection of isolates. Phytopathology 95:1061–1071
Balesdent MH, Louvard K, Pinochet X, Rouxel T (2006) A large-scale survey of races of Leptosphaeria maculans occurring on oilseed rape in France. Eur J Plant Pathol 114:53–65
Barret P, Guerif J, Reynoird JP, Delourme R, Eber F, Renard M, Chevre AM (1998) Selection of stable Brassica napus Brassica juncea recombinant lines resistant to blackleg (Leptosphaeria maculans). 2. A ‘to and fro’ strategy to localise and characterise interspecific introgressions on the B. napus genome. Theor Appl Genet 96:1097–1103
Brun H, Levivier S, Somda I, Ruer D, Renard M, Chevre AM (2000) A field method for evaluating the potential durability of new resistance sources: application to the Leptosphaeria maculans Brassica napus pathosystem. Phytopathology 90:961–966
Brun H, Ruer D, Levivier S, Somda I, Renard M, Chevre AM (2001) Presence in Leptosphaeria maculans populations of isolates virulent on resistance introgressed into Brassica napus from the B. nigra B genome. Plant Pathol 50:69–74
Chevre AM, Eber F, This P, Barret P, Tanguy X, Brun H, Delseny M, Renard M (1996) Characterization of Brassica nigra chromosomes and of blackleg resistance in B. napus-B. nigra addition lines. Plant Breed 115:113–118
Chevre AM, Barret P, Eber F, Dupuy P, Brun H, Tanguy X, Renard M (1997) Selection of stable Brassica napus B. juncea recombinant lines resistant to blackleg (Leptosphaeria maculans).1. Identification of molecular markers, chromosomal and genomic origin of the introgression. Theor Appl Genet 95:1104–1111
Crouch JH, Lewis BG, Mithen RF (1994) The effect of A-genome substitution on the resistance of Brassica napus to infection by Leptosphaeria maculans. Plant Breed 112:265–278
Delourme R, Pilet-Nayel ML, Archipiano M, Horvais R, Tanguy X, Rouxel T, Brun H, Renard M, Balesdent MH (2004) A cluster of major specific resistance genes to Leptosphaeria maculans in Brassica napus. Phytopathology 94:578–583
Delourme R, Chevre AM, Brun H, Rouxel T, Balesdent MH, Dias JS, Salisbury P, Renard M, Rimmer SR (2006a) Major gene and polygenic resistance to Leptosphaeria maculans in oilseed rape (Brassica napus). Eur J Plant Pathol 114:41–52
Delourme R, Falentin C, Huteau V, Clouet V, Horvais R, Gandon B, Specel S, Hanneton L, Dheu JE, Deschamps M, Margale E, Vincourt P, Renard M (2006b) Genetic control of oil content in oilseed rape (Brassica napus L.). Theor Appl Genet 113:1331–1345
Dion Y, Gugel RK, Rakow GFW, SeguinSwartz G, Landry BS (1995) RFLP mapping of resistance to the blackleg disease [causal agent, Leptosphaeria maculans (Desm) Ces, et de Not] in canola (Brassica napus L). Theor Appl Genet 91:1190–1194
Doyle JJ, Doyle JL (1990) Isolation of plant DNA from fresh tissue. Focus 12:13–15
Ferreira ME, Rimmer SR, Williams PH, Osborn TC (1995) Mapping loci controlling Brassica napus resistance to Leptosphaeria maculans under different screening conditions. Phytopathology 85:213–217
Fitt BDL, Brun H, Barbetti MJ, Rimmer SR (2006) World-wide importance of phoma stem canker (Leptosphaeria maculans and L. biglobosa) on oilseed rape (Brassica napus). Eur J Plant Pathol 114:3–15
Foisset N, Delourme R, Barret P, Hubert N, Landry BS, Renard M (1996) Molecular-mapping analysis in Brassica napus using isozyme, RAPD and RFLP markers on a doubled haploid progeny. Theor Appl Genet 93:1017–1025
Gugel RK, Petrie GA (1992) History, occurrence, impact, and control of blackleg of rapeseed. Can J Plant Pathol 14:36–45
Kosambi DD (1944) The estimation of map distances from recombination values. Ann Eugen 12:172–175
Li CX, Cowling WA (2003) Identification of a single dominant allele for resistance to blackleg in Brassica napus ‘Surpass 400’. Plant Breed 122:485–488
Li H, Sivasithamparam K, Barbetti MJ (2003) Breakdown of a Brassica rapa subsp. sylvestris single dominant blackleg resistance gene in B. napus rapeseed by Leptosphaeria maculans field isolates in Australia. Plant Dis 87: 752–752
Lincoln SE, Daly MJ, Lander ES (1992) Constructing genetic linkage maps with MAPMAKER/EXP 3.0: a tutorial and reference manual, 3rd edn. Whitehead Institute Technical Report, Cambridge
Lombard V, Delourme R (2001) A consensus linkage map for rapeseed (Brassica napus L.): construction and integration of three individual maps from DH populations. Theor Appl Genet 103:491–507
Mayerhofer R, Wilde K, Mayerhofer M, Lydiate D, Bansal VK, Good AG, Parkin IAP (2005) Complexities of chromosome landing in a highly duplicated genome: toward map-based cloning of a gene controlling blackleg resistance in Brassica napus. Genetics 171:1977–1988
Meyers BC, Dickerman AW, Michelmore RW, Sivaramakrishnan S, Sobral BW, Young ND (1999) Plant disease resistance genes encode members of an ancient and diverse protein family within the nucleotide-binding superfamily. Plant J 20:317–332
Michelmore RW, Paran I, Kesseli RV (1991) Identification of markers linked to disease-resistance genes by bulked segregant analysis: a rapid method to detect markers in specific genomic regions by using segregating populations. Proc Natl Acad Sci USA 88:9828–9832
Noel L, Moores TL, van der Biezen EA, Parniske M, Daniels MJ, Parker JE, Jones JDG (1999) Pronounced intraspecific haplotype divergence at the RPP5 complex disease resistance locus of Arabidopsis. Plant Cell 11:2099–2111
Parkin IAP, Lydiate DJ (1997) Conserved patterns of chromosome pairing and recombination in Brassica napus crosses. Genome 40:496–504
Pilet ML, Delourme R, Foisset N, Renard R (1998) Identification of loci contributing to quantitative field resistance to blackleg disease, causal agent Leptosphaeria maculans (Desm.) Ces. et de Not., in winter rapeseed (Brassica napus L.). Theor Appl Genet 96:23–30
Piquemal J, Cinquin E, Couton F, Rondeau C, Seignoret E, Doucet I, Perret D, Villeger MJ, Vincourt P, Blanchard P (2005) Construction of an oilseed rape (Brassica napus L.) genetic map with SSR markers. Theor Appl Genet 111:1514–1523
Polsoni L, Kott S, Beversdorf WD (1988) Large-scale microspore culture technique for mutation-selection studies in Brassica napus. Can J Bot 66:1681–1685
Rimmer SR (2006) Resistance genes to Leptosphaeria maculans in Brassica napus. Can J Plant Pathol 28:288–297
Rimmer SR, vandenBerg CGJ (1992) Resistance of oilseed Brassica spp. blackleg caused by Leptosphaeria maculans. Can J Plant Pathol 14:56–66
Rocherieux J (2003) Analyse génétique structurale et fonctionnelle de la résistance à la hernie chez Brassica oleracea. Ph D thesis, Ecole nationale supérieure agronomique de Rennes
Rouxel T, Penaud A, Pinochet X, Brun H, Gout L, Delourme R, Schmit J, Balesdent MH (2003) A 10-year survey of populations of Leptosphaeria maculans in France indicates a rapid adaptation towards the Rlm1 resistance gene of oilseed rape. Eur J Plant Pathol 109:871–881
Saal B, Brun H, Glais I, Struss A (2004) Identification of a Brassica juncea-derived recessive gene conferring resistance to Leptosphaeria maculans in oilseed rape. Plant Breed 123:505–511
Somda I, Delourme R, Renard M, Brun H (1999) Pathogenicity of Leptosphaeria maculans isolates on a Brassica napus-B. juncea recombinant line. Phytopathology 89:169–175
Sprague SJ, Marcroft SJ, Hayden HL, Howlett BJ (2006) Major gene resistance to blackleg in Brassica napus overcome within three years of commercial production in southeastern Australia. Plant Dis 90:190–198
Vos P, Hogers R, Bleeker M, Reijans M, Vandelee T, Hornes M, Frijters A, Pot J, Peleman J, Kuiper M, Zabeau M (1995) AFLP—a new technique for DNA-fingerprinting. Nucleic Acids Res 23:4407–4414
West JS, Kharbanda PD, Barbetti MJ, Fitt BDL (2001) Epidemiology and management of Leptosphaeria maculans (phoma stem canker) on oilseed rape in Australia, Canada and Europe. Plant Pathol 50:10–27
Williams PH, Delwiche PA (1979) Screening for resistance to blackleg of crucifers in the seedling stage. In: Proceedings Eucarpia conference, breeding of Cruciferous crops, Wageningen, pp 164–170
Yu FQ, Rimmer SR, Lydiate DJ (2004) Identification and mapping of a third blackleg resistance locus in Brassica napus derived from B. rapa subsp. sylvestris. Plant and animal genome XII conference Abstract, p 520
Yu F, Lydiate DJ, Rimmer SR (2005) Identification of two novel genes for blackleg resistance in Brassica napus. Theor Appl Genet 110:969–979
Acknowledgments
Our colleagues of OUEST-Génopole® (Le Rheu, France) are gratefully acknowledged for their technical assistance. We thank R. Philippe and J. C. Letanneur (INRA Rennes, France) for producing plant material, O. Quenez (INRA Rennes, France) for generating specific molecular markers and E. Jenczewski (INRA Versailles, France) and R. Kutcher (AAFC Melfort, Canada) for their critical reading of the manuscript. This work was funded by INRA and CETIOM.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by I. Rajcan.
Rights and permissions
About this article
Cite this article
Leflon, M., Brun, H., Eber, F. et al. Detection, introgression and localization of genes conferring specific resistance to Leptosphaeria maculans from Brassica rapa into B. napus . Theor Appl Genet 115, 897–906 (2007). https://doi.org/10.1007/s00122-007-0616-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00122-007-0616-z