Abstract
Forage quality of maize is influenced by both the content and structure of lignin in the cell wall. Phenylalanine Ammonia-Lyase (PAL) catalyzes the first step in lignin biosynthesis in plants; the deamination of l-phenylalanine to cinnamic acid. Successive enzymatic steps lead to the formation of three monolignols, constituting the complex structure of lignin. We have cloned and sequenced a PAL genomic sequence from 32 maize inbred lines currently employed in forage maize breeding programs in Europe. Low nucleotide diversity and excessive linkage disequilibrium (LD) was identified at this PAL locus, possibly reflecting selective constrains resulting from PAL being the first enzyme in the monolignol, and other, pathways. While the association analysis was affected by extended LD and population structure, several individual polymorphisms were associated with neutral detergent fiber (not considering population structure) and a single polymorphism was associated with in vitro digestibility of organic matter (considering population structure).
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Maize (Zea mays L.) is widely used as a forage crop in European agriculture. During recent decades, breeding efforts have led to a substantial increase in the whole plant yield, facilitated by improved maize stalk standability, and stalk rot and lodging resistance (Barrière et al. 2004). However, during the same period of time there has been a steady decrease in cell wall digestibility, and, consequently, in feeding value of elite maize hybrids (Barrière et al. 2005).
Cell wall digestibility in forage crops is influenced by both lignin content and lignin structure (reviewed by Barrière et al. 2003). The first step in lignin biosynthesis in plants is the deamination of l-phenylalanine by Phenylalanine Ammonia-Lyase (PAL) to cinnamic acid. PAL also catalyzes the first step of several other phenylpropanoid pathways, leading to the formation of a variety of secondary metabolites (reviewed by Winkel 2004). A full-length PAL cDNA has been isolated from maize and the encoded enzyme has been shown to catalyze the deamination of both l-phenylalanine and l-tyrosine (Rosler et al. 1997). Successive enzymatic steps in the monolignol pathway lead to the formation of three monolignols (p-hydroxycinnamyl alcohols); p-coumaryl-, coniferyl-, and sinapyl alcohols from which p-hydroxyphenyl- (H), guaiacyl- (G), and syringyl units (S), respectively, are derived. Subsequently, G, S, and H undergo polymerization by oxidases to form lignin (reviewed by Boerjan et al. 2003; Grabber et al. 2004). Not unexpectedly, given that PAL is the first enzyme in lignin biosynthesis, impaired expression of PAL results in defective lignin formation in tobacco (Nicotiana tabbacum L.) and Arabidopsis (Sewalt et al. 1997; Raes et al. 2003; Rohde et al. 2004).
The brown-midrib (bm) mutants of maize are characterized by decreased lignin content, altered cell wall composition, and a brown-reddish color of leaf midribs. Of the four known bm mutants, bm3 exhibits the strongest effect on plant phenotype, and several feeding studies in dairy cattle have shown the positive impact of bm3 mutants on intake and digestibility of forage maize (reviewed by Barrière et al. 2003). This phenotype is caused by a knock-out mutation in the caffeic acid O-methyl transferase (COMT) gene (Collazo et al. 1992; Vignols et al. 1995; Morrow et al. 1997). However, this phenotype also results in inferior agronomic performance such as lodging and lower biomass yield, restricting the use of bm3 mutants in maize hybrid breeding programs (Ballard et al. 2001; Cherney et al. 1991; reviewed by Pedersen et al. 2005). Thus, the characterization of genetic diversity in other genes involved in lignin biosynthesis could facilitate identification of allelic variation more applicable to breeding programs.
Recently, reports have emerged on nucleotide diversity and extent of linkage disequlibrium (LD) at the COMT locus in maize genotypes currently employed in breeding programs (Fontaine and Barrière 2003; Guillet-Claude et al. 2004a; Zein et al. 2006). Knowledge on the extent of LD is relevant when estimating the marker saturation necessary for high-resolution association analysis at a given locus. Following the pioneering study in plants, associating individual Dwarf8 polymorphisms with flowering time in maize (Thornsberry et al. 2001), association analysis has been applied in a number of species (reviewed by Gupta et al. 2005). In maize, association analyses have been employed to associate individual candidate gene polymorphisms to phenotypic variation in flowering time, endosperm color, starch production, and maysin and chlorogenic acid accumulation (Palaisa et al. 2003; Wilson et al. 2004; Andersen et al. 2005; Szalma et al. 2005). In addition, associations between cell wall digestibility and genetic variation in COMT and other “lignin genes” have been reported (Guillet-Claude et al. 2004a, b; Lübberstedt et al. 2005). Thus, by association analysis causative polymorphisms can be identified from which functional markers can be derived (Andersen and Lübberstedt 2003).
Further characterization of genes affecting fodder quality would facilitate targeted identification, maintenance, and utilization of genetic variation for this trait in maize breeding lines. Three putative PAL genes have been mapped to unlinked positions in the maize genome (http://www.maizegdb.org) by RFLP based on a partial PAL cDNA sequence (GenBank no. M95077; Keith et al. 1993). Thus, it is likely that several PAL genes are present in maize, as is the case in Arabidopsis where four PAL genes have been identified (Raes et al. 2003). The genomic sequence analyzed in the present study has been obtained based on a full-length cDNA sequence (GenBank no. L77912) identified by Rosler et al. (1997), and is throughout the text denoted PAL. Given that PAL is the first enzyme in the biosynthesis of lignin and the phenotypic consequences of impairing PAL expression in tobacco and Arabidopsis, the aims of the present study were to (1) examine the sequence diversity at the PAL locus in European inbred lines of maize, (2) study the extent of LD at this locus in these lines, and (3) test associations between individual PAL polymorphisms and four different phenotypic traits related to forage quality.
Materials and methods
Plant materials and phenotypic analyses
A collection of 32 maize inbred lines consisting of 19 Flints and 13 Dents were included in the analysis. Twenty-nine lines were elite inbreds from the current breeding program of KWS Saat AG and three lines were from the public domain (AS01, AS02, and AS03 identical to F7, F2, and EP1, respectively; Table 1). The 32 lines were selected based on digestibility of neutral detergent fiber (DNDF) values to represent a broad range of variability for this trait in central European germplasm employed in forage maize breeding. The included lines were derived from several breeding populations of Flint and Dent, respectively, and are not related apart from lines AS20 and AS21 which are an isogenic line pair included in the analysis based on contrasting DNDF values. The inbred lines were evaluated in Grucking (sandy loam) in 2002, 2003, and 2004, and in Bernburg (sandy loam) in 2003 and 2004. The experiments included 49 entries in a 7 × 7 lattice design with two replications. Plots consisted of single rows, 0.75 m apart and 3 m long with a total of 20 plants. About 50 days after flowering, the ears were manually removed and the stover was chopped. Approximately 1 kg of the material was collected and dried at 40°C after which the stover was ground to pass through a 1 mm sieve. Quality analyses were performed with near infrared reflectance spectroscopy (NIRS) based on previous calibrations on the data of 300 inbred lines (unpublished results). The following data were recorded: % water soluble carbohydrates (WSC) (Luff Schoorl 1929), % neutral detergent fiber (NDF) (VanSoest 1963), in vitro digestibility in % of organic matter (IVDOM) (Tilley and Terry 1963), and DNDF given by the formula DNDF = 100−(100−IVDOM)/(NDF × DM/OM/100) where DM is dry matter content and OM is organic matter content of the sample (Tilley and Terry 1963; VanSoest 1963).
DNA isolation, PCR amplification, and DNA sequencing
Plants were grown for DNA isolation in the greenhouse and leaves were harvested at 3 weeks after germination. Genomic DNA was extracted from the leaves using the Maxi CTAB method (Saghai-Maroof et al. 1984). DNA templates for sequencing were obtained by polymerase chain reaction (PCR) to produce two overlapping fragments using Taq DNA polymerase and primers based on the sequence of a full-length PAL cDNA of maize (GenBank no. L77912; Rosler et al. 1997). The combination of the forward and reverse primers PAL_F1 (5′- ACT CCT CCG GCT CTT CTT CTC) and PAL_R1 (5′- CTT GTG GGT CAG GTG GTC CGT) produced a 2,200 bp fragment spanning the full first exon, the intron, and the 5′ end of the second exon. The combination of the forward and reverse primers PAL_F2 (5′- CGC CGA GGC GTT CAA GAT C) and PAL_R2 (5′- GTG GCA GGG CAC AGC TAC) produced a 1,640 bp fragment, overlapping with the PAL_F1 – PAL_R1 fragment, spanning the second exon, and part of the 3′-UTR.
DNA amplification was performed in a 50 μl reaction mixture containing 20 ng genomic DNA, primers (200 nM), dNTPs (200 μM), 1 M Betain and two units of Taq polymerase (Peqlab, Erlangen, Germany). Touchdown PCR was applied as follows: an initial denaturation step at 95°C for 2 min, seven amplification cycles: 45 s at 95°C; 45 s at 68°C (minus 1°C per cycle), 2 min at 72°C, followed by 30 amplification cycles: 30 s at 94°C; 45 s at 60°C, 2 min at 72°C, and a final extension step at 72°C for 10 min. Products were separated by gel electrophoresis on 1.5% agarose gels, visualized by ethidium bromide staining and photographed using an eagle eye apparatus (Herolab, Wiesloch, Germany).
Fragments were purified using QiaQuick spin columns (Qiagen, Valencia, USA) according to the manufacturer’s instructions, and sequenced directly using internal sequence specific primers and the Big Dye1.1 dye-terminator sequencing kit on an ABI 377 (PE Biosystems, Foster City, USA). Electropherograms of overlapping sequencing fragments were manually edited using the software package Sequence Navigator version 1.1 from PE Biosystems. Final full alignment was built up using default settings of the Clustal program version 1.8 (Thompson et al. 1994) followed by manual refinement to minimize the number of gaps.
Analysis of sequence data
DNA sequences were analyzed for the complete sample and within individual subpopulations (Flint and Dent). DnaSP Version 4.10 (Rozas et al. 2003) was applied for the analysis. Two estimates of diversity, π and θ, were calculated. π is the average number of nucleotide differences per site between two sequences (Nei 1987), and θ is derived from the total number of segregating sites and corrected for sampling size (Watterson 1975). These estimates were based on single nucleotide polymorphisms only.
To test for neutrality of mutations, Tajima’s D statistic (Tajima 1989), and Fu and Li’s D* and F* statistics (Fu and Li 1993) were applied. These statistics are based on different comparisons of Θ = 4N eμ, where N e equals the effective population size and μ the mutation rate (Watterson 1975). Tajima’s D results from the comparison of Θ based on the number of pair-wise differences and the number of segregating sites between sequences in the sample. Fu and Li’s D* and F* result from comparisons of Θ based on the number of singletons and the number of either segregating sites (D*) or pair-wise differences (F*). The minimum number of recombination events between pairs of non-overlapping SNPs was determined using the four-gamete test (Hudson and Kaplan 1985).
LD between pairs of polymorphic sites (SNPs and insertion/deletion polymorphisms (indels), excluding singletons) in PAL was estimated by the TASSEL software, version 1.9.0 (Thornsberry et al. 2001); http://www.maizegenetics.net/bioinformatics/tasselindex.htm). Various measurements for LD have been developed (reviewed by Gaut and Long 2003) of which squared allele frequency correlations (r 2) (Weir 1996) were chosen for our calculations. The significance of LD between sites was tested by Fisher’s exact test.
For the phylogenetic analysis of allele sequences, the MEGA software version 3 (Kumar et al. 2004) was used with default settings. Bootstrapping, based on 1,000 replications of the dataset, was performed to test phylogenies.
Population structure and association analysis
All lines were genotyped with 101 simple sequence repeat markers (SSRs) providing an even coverage of the maize genome. The employed SSR markers are publicly available (http://www.maizegdb.org/ssr.php). Population structure was inferred from SSR data by using the Structure 2.0 software (Pritchard et al. 2000; Falush et al. 2003). Structure applies a Bayesian clustering approach to identify subpopulations, each modeled by a characteristic set of allele frequencies, in this case based on genotyping data from 101 SSRs. The procedure assigns individuals to these populations, while simultaneously estimating the population allele frequencies. Structure produces a Q matrix that lists the estimated membership coefficients for each individual in each cluster. The Admixture model was applied. A burn-in length of 50.000 followed by 50.000 iterations was used (See the Structure 2.0 documentation at http://www.pritch.bsd.uchicago.edu/).
The estimated Q matrices were used in the subsequent association analysis carried out in TASSEL. This software applies a logistic regression ratio test to calculate, whether the likelihood of the candidate gene distribution (in this case PAL polymorphisms) is associated with either (1) population structure and phenotypic variation or (2) population structure only. The test statistic (Λ), the ratio between these two likelihoods, indicates associations between individual polymorphisms and traits, in this case four quality-related traits. Mean phenotypic values across five environments (Table 1) were applied for the association analysis. In addition, the general linear model (GLM) analysis in TASSEL was employed to identify associations, not considering population structure. All PAL polymorphisms (including singletons) were tested and the P-value for individual polymorphisms was estimated based on 1,000 permutations of the dataset, both for GLM and logistic regression.
Results
Phenotypic data and correlations
Mean phenotypic values across environments were calculated for the overall sample, for within the Flint and Dent pools, and for individual lines (Table 1). Mean phenotypic values for individual lines ranged between 12.29 and 25.81 for WSC, 50.33 and 63.03 for NDF, 67.23 and 77.98 for IVDOM, and 49.59 and 60.99 for DNDF. Overall means were 19.68, 56.06, 73.26, and 56.33 for WSC, NDF, IVDOM, and DNDF, respectively. The phenotypic variation was significantly affected by lines and environments, as well as the interaction between these two (Table 1). Coefficients of correlation between traits and tests of significance are given in Table 2. DNDF was significantly correlated to NDF (−0.32) and IVDOM (0.86). IVDOM was significantly correlated to all other traits, negatively to NDF and positively to WSC and DNDF. The closest positive correlation was observed between DNDF and IVDOM, while the closest negative correlation (−0.89) was observed between NDF and WSC (Table 2).
Sequence alignment and haplotypes
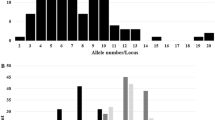
The full PAL alignment spanned 3,594 bp including 453 sites with alignment gaps (indel polymorphisms). In the first exon, two SNPs were identified. In the intron, 23 SNPs and 17 indels of varying size were identified. The largest indel span ∼300 bps in the 3′ end of the intron in a complex manner, not affecting the intron–exon splice site. The two alleles of this indel discriminate the lines into two groups, primarily consisting of Flint and Dent lines, respectively. In the second exon, two 1-bp deletions and 11 SNPs were identified. In total, 39 single nucleotide polymorphic sites (SNPs) were identified (Table 3). Of these, 33 were parsimony informative sites, each allele carried by two or more individuals. The remaining six sites were singletons, i.e., sites in which only one copy of the rare variant was present in the complete sample. No SNPs with more than two variants were identified. While three SNPs in the second exon altered the amino-acid sequence, the remaining SNPs were synonymous mutations, not altering the amino-acid sequence. Of the six singleton sites, five and one were identified in the lines AS31 and AS13, respectively. In total, eight PAL haplotypes were identified based on the 39 SNPs (Table 3). Haplotype 1 comprised the majority (18) of lines, including 15 Flint lines. Haplotypes 6 and 8 comprised four lines each, including seven Dent lines. Haplotype 2 comprised two lines (both Flint), while the remaining haplotypes comprised one line each (Table 4). Considering the intron, exon 2, and 3′ UTR regions, all lines except three (AS09, AS13, and AS31), exhibit one of the two haplotypes, identical to haplotypes 1 and 6 (Table 3) constituted predominantly of Flint and Dent lines, respectively (Table 4).
Nucleotide diversity and selection
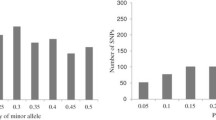
Nucleotide diversity (π) was determined for the Flint and Dent heterotic groups individually and for the combined sample based on the 39 SNPs (Fig. 1 and Table 5). For the combined sample, nucleotide diversity was lowest in the coding regions (π = 0.00248) and highest in the intron (π = 0.00821) and 3′ UTR (π = 0.00751) regions. Overall, nucleotide diversity was π = 0.00424 in the combined sample, and was lower in the Flint lines (π = 0.00166) as compared to the Dent lines (π = 0.00427).
Tajima’s D was not significant when considering either the entire PAL sequence or the non-coding regions of the combined sample (Table 5). However, when considering only the ORF, Tajima’s D was positive and significant. This indicates selection and an excess of alleles with intermediate frequencies at the PAL locus across the 32 lines. Fu and Li’s D* and F* were non-significant in all regions in the combined sample. Within the Flints, Tajima’s D was negative and significant considering the entire region and the ORF. This suggests selection as well as the presence of low-frequency alleles within the Flints. Within the Dents, Tajima’s D was non-significant in all regions while Fu and Li’s D* and F* were both significant considering the ORF. This indicates selection in the ORF and few mutations in more recent generations.
Phylogenetic analysis
Phylogenetic analysis by the neighbor-joining (NJ) method based on the PAL genomic sequence revealed two major clusters, predominantly Flint- and Dent lines, respectively (Fig. 2). In the “Flint” cluster, 17 Flint- and three Dent lines (AS8, AS11, and AS29) grouped together while in the “Dent” cluster 10 Dent and two Flint lines (AS12 and AS13) grouped together.
Linkage disequilibrium (LD) and recombination
LD was estimated between all pairs of polymorphic sites (SNPs and indels) in the PAL genomic sequence (Fig. 3). A plot of r 2 against physical distance for polymorphism pairs indicated that LD persisted (r 2 > 0.2) for the entire length of the PAL locus (Fig. 4). However, as is evident from Fig. 3, LD is not evenly distributed along the locus. Both r 2 values and Fisher’s exact test of LD identified an LD block spanning the 3′ half of the intron, the second exon, and the 3′ UTR. Another LD block was identified spanning the 5′ half of the intron. No LD was detected between these two blocks. Thus, sites in strong LD were predominantly identified in the terminal ∼2.5 kb of the PAL gene. The relative high level of LD is supported by the detection of only two recombination events; one between sites 199 (exon 1) and 948 (intron), and one between sites 2,048 and 3,013 (both exon 2) of the alignment.
LD across the PAL locus. Bp position of polymorphisms in the alignment are given. The vertical bar on the left represents 1st exon-, intron-, 2nd exon-, and 3′UTR regions, respectively. Polymorphisms at bp positions 566, 652, 720, 726, 776, 806, 857, 1,945, 2,086, 2,470, and 3,452 are singletons. Lower left triangle: P-values derived from Fisher’s exact test. Upper right triangle: r 2 values
Population structure and association analysis
Estimation of population structure was performed by Structure based on 101 SSR markers providing an even coverage of the maize genome. Two subpopulations, in agreement with the Flint and Dent pedigree information, were estimated as the most likely subdivision of our plant material (Fig. 5). While most lines exhibited a homogenous genetic background (either Flint or Dent), two Dent lines, AS29, and AS34, harbored ∼5%, and ∼60% of the “Flint genetic background”, respectively (Fig. 5).
Population structure estimates based on 101 SSRs evenly distributed across the maize genome. Vertical bars represent individual maize lines. The area of either green or red illustrates the proportion of either “Flint” or “Dent” genetic background, respectively, harbored in individual lines based on these SSR markers
The population structure estimates was used in TASSEL to test for associations between PAL polymorphisms and WSC, NDF, IVDOM, and DNDF (Table 1). All polymorphisms, including singletons, were considered in the association analysis. By GLM analysis (not considering population structure), all polymorphisms in the 3′ LD block, excluding singletons, and all singletons in the 5′ part of the intron (bp positions 566, 652, 720, 726, 776, 806, and 857), were significantly associated (P<0.05) with NDF (Fig. 6a). No associations were identified for WSC, IVDOM, or DNDF by this approach. By logistic regression analysis (considering population structure), a 1 bp indel at bp position 2,086 was significantly associated (P<0.05) with IVDOM (Fig. 6b). No associations were identified for DNDF, WSC, or NDF, when considering population structure.
Test of associations between PAL polymorphisms and four quality-related traits. Tests were performed excluding (a) and including (b) population structure estimates in the analysis. Individual dots represent the significance (Y-axis) of individual polymorphisms (X-axis). The broken line represents P = 0.05. WSC water soluble carbohydrates, NDF neutral detergent fiber, IVDOM in vitro digestibility of organic matter, DNDF digestibility of neutral detergent fiber
Discussion
Nucleotide diversity and linkage disequilibrium at the PAL locus
In the present study, the genomic sequence of the PAL gene has been obtained for 19 Flint and 13 Dent maize inbred lines. With the exception of lines AS29 and AS34, all lines exhibited a homogenous “Flint” or “Dent” genetic background, defined by SSR markers (Fig. 5). This is in general agreement with pedigree information and with previous studies showing the ability of SSR markers to reliably define heterotic groups in maize (Smith et al. 1997; Senior et al. 1998). While the phylogeny derived from PAL polymorphisms also predicts two main clusters, predominantly Flint and Dent, more interspersions of lines are indicated (Fig. 2). This inconsistency of subpopulations based on multi-locus (SSR)- and single-locus (PAL) data, respectively, is not unexpected. As shown for the tb1 genomic region, single loci polymorphisms produced different phylogenies, depending on the locus in question (Wang et al. 1999; Clark et al. 2004). Strong selection upon a single locus can result in fixation of alleles within (sub)populations, while alleles of other closely linked loci, not under selection, might be randomly distributed across (sub)populations. For the PAL locus, similar to the COMT locus (Zein et al. 2006), most alleles are fixed within the Flint and Dent heterotic groups, indicating selection and/or genetic drift at these loci after the separation of breeding pools.
Population bottlenecks and selection are expected to decrease nucleotide diversity and increase LD at a given locus (Ching et al. 2002; Flint-Garcia et al. 2003). At the PAL locus, selection is indicated in the coding region (Table 5) and nucleotide diversity (π = 0.00424) is within the range of what has been reported for other maize loci (e.g. Thornsberry et al. 2001; Whitt et al. 2002; Palaisa et al. 2003; Clark et al. 2004). However, comparing to loci putatively involved in the biosynthesis of lignin (Guillet-Claude et al. 2004a; b; Zein et al. 2006), the nucleotide diversity at the PAL locus was relatively low. Previously, we have reported an overall nucleotide diversity of π = 0.00834 at the COMT locus among the lines included in this study plus ten additional lines (Zein et al. 2006). Thus, within a similar sample of lines, the overall nucleotide diversity at the COMT locus exceeded that at the PAL locus by two-fold.
Overall, LD persisted (r 2 > 0.2) over the length of the PAL locus (3.7 kb) when considering all polymorphisms, excluding singletons (Fig. 4). Due to population bottlenecks and selection, LD can be expected to be higher among elite breeding lines as compared to among more distantly related genetic resources. In agreement with this, a rapid breakdown of LD (r 2 < 0.1 within few hundreds of bps) has been reported for several loci in diverse sets of maize germplasm (Remington et al. 2001; Tenaillon et al. 2001), while extended LD, up to hundreds of kbs, has been reported in sets of inbred lines (Ching et al. 2002; Jung et al. 2004). However, extended LD was also identified at the sugary1 locus (r 2 > 0.4 over 7 kb) in a set of diverse germplasm (Remington et al. 2001) indicating considerable between-loci variation in LD regardless of the sampled plant material. Different from the PAL locus, LD levels have previously been reported to decline rapidly for several loci involved in lignin biosynthesis (Guillet-Claude et al. 2004a; b). Specifically, we have found r 2 < 0.1 within 2 kb at the COMT locus within an overlapping sample of lines (Zein et al. 2006). This difference in LD-decay could reflect the levels of constrains put on the respective loci by selection.
Two distinct LD blocks were evident at the PAL locus (Fig. 3). While no LD was detected between the 5′ part of the intron and other regions in PAL, extensive LD was detected spanning the 3′ half of the intron, the second exon, and the 3′ UTR (Fig. 3). Though not uncommon (Guillet-Claude et al. 2004a; b), such LD pattern might be due to cloning artifacts or the amplification of segments from different members of a gene family. However, the first of the two amplicons spans the first exon, the intron, and ∼200 bp of the second exon. As the border between the two LD blocks is located in the centre of the intron, this renders cloning artifacts causing the observed LD pattern unlikely. The organization of the two LD blocks in haplotypes (Table 3) and the separation of Flint and Dent lines in haplotypes (Table 4) further argues against artifacts, e.g. a “frame-shift” in the assignment of DNA sequences to individual lines. Extended LD can arise from several processes of population genetics, including population size, population bottlenecks and selection (reviewed by Flint-Garcia et al. 2003). Extended LD might also indicate relatedness between individuals. However, the lines included in the present study originate from several breeding populations of Flint and Dent and are, apart from the isogenic line pair AS20 and AS21, not related. Also, local LD can be a signature of a selective sweep, i.e. a local reduction of genetic variation, caused by the rapid fixation of a beneficial mutation (Kim and Nielsen 2004). Thus, the distinct LD pattern at the PAL locus could reflect genetic drift or selective sweeps within the Flint and Dent pools, fixating different alleles in the respective breeding pools (Tables 3, 4). If caused by a selective sweep, the high LD spanning the 3′ half of the PAL gene (Fig. 3) might indicate causative sites, with regard to phenotype, within this region. In Arabidopsis, PAL mutants were affected in several metabolic pathways, including the monolignol pathway (Rohde et al. 2004). Given a similar function of PAL in maize, functional constrains of the enzyme could restrict mutation- and recombination rates at the gene, resulting in the relatively low nucleotide diversity and high LD observed here.
PAL polymorphisms are associated with forage quality
Previous studies have identified genes involved in the biosynthesis of lignin as promising targets for identification of polymorphic sites associated with forage quality (Guillet-Claude et al. 2004a; b; Lübberstedt et al. 2005). However, the extended LD at the PAL locus has consequences for association analysis. No recombination was detected between bp positions 947 and 1,655 in the intron and between bp positions 2,131 and 2,470 in the second exon (Table 3). Thus, it is not possible to discriminate the effects of individual polymorphisms on phenotypes within these LD blocks in the present maize lines. However, a 1-bp deletion ∼400 bps downstream of the start of the second exon (position 2,086), was associated with IVDOM when considering population structure (Fig. 6b). The deletion introduces a stop codon ∼450 bp into the second exon and could thus affect the functionality of the PAL protein. Consequently, this deletion is a candidate site for deriving a functional marker. However, it is present only in a single line (AS07), and the association needs to be interpreted with caution until validated in more lines. Ultimately, all polymorphisms identified in this study could be evaluated in larger and/or broader collections of germplasm to attempt to enhance both the genetic resolution (i.e. decrease LD) and the power of the association analysis.
Population structure can result in false positive associations, which is controlled by considering population structure in the association analysis (Thornsberry et al. 2001). However, true functional polymorphisms might be confounded with population structure, e.g. between Flint and Dent lines. Consequently, such polymorphisms will not be identified by association analysis when considering population structure. By GLM analysis, not considering population structure, several polymorphisms were associated with NDF, including three non-synonymous SNPs in the second exon (Fig. 6a and Table 3). However, both NDF values and haplotypes were confounded with population structure in the maize lines included in this study (Table 4). Thus, the PAL-NDF associations were not detected when considering population structure. This illustrates a potential problem of considering population structure in the association analysis, specifically within crop plants maintained as separate breeding pools/lines: while the number of false positives can be reduced, “true” causative polymorphisms might be masked, i.e., the number of false negatives might increase. Consequently, the choice of plant material can significantly impact the outcome of the analysis, as illustrated by studies on the Dwarf8 locus in two different sets of maize plant materials (Thornsberry et al. 2001; Andersen et al. 2005).
Deriving functional markers for forage quality traits
Studies in tobacco and Arabidopsis have shown that impaired expression of PAL results in defective lignin formation (Raes et al. 2003; Rohde et al. 2004; Sewalt et al. 1997) and that lignification is crucial for structural integrity of the cell wall and strength of the stem (Chabannes et al. 2001; Jones et al. 2001). Thus, it is conceivable that polymorphisms in PAL could affect forage quality in maize. The three non-synonymous SNPs in the second exon (Table 3) could be considered candidate causative polymorphisms from which functional markers could be derived. Studies of gene expression and enzyme activity could further elucidate the allelic effects of these polymorphisms. However, compared to other genes involved in the monolignol pathway, PAL exhibits low nucleotide diversity and extended LD (Guillet-Claude et al. 2004a; b; Zein et al. 2006), restricting the identification of discrete, causative polymorphisms by association analysis. Consequently, investigations in larger and/or broader sets of maize germplasm are necessary to enhance the genetic resolution at the PAL locus. Alternatively, a series of PAL mutants could be produced by tilling, allowing for comparisons of single polymorphisms, currently in complete LD, in isogenic backgrounds (http://www.genome.purdue.edu/maizetilling/).
PAL is the first enzyme in several phenylpropanoid pathways, catalyzing the production of a number of phenylpropanoids, including monolignols, from phenylalanine (reviewed by Winkel 2004). These phenylpropanoids are functioning as, e.g., structural components (lignins), UV sunscreens, and signaling molecules. In agreement with these diverse functions of phenylpropanoids, it has been shown that PAL mutants in Arabidopsis were affected not only in the monolignol pathway, but that also carbohydrate- and amino acid metabolisms were altered (Rohde et al. 2004). Thus, an allelic shift at a PAL locus could affect (positively or negatively) several traits, restricting selection at the locus. Consequently, genes more downstream and specific to monolignol synthesis, e.g., O-methyltransferase genes (Guillet-Claude et al. 2004a; Lübberstedt et al. 2005), could prove to be more suitable candidates for deriving functional markers for forage quality. However, three unlinked mapping positions in the maize genome (http://www.maizegdb.org) indicate that PAL is organized as a small gene family in maize, as is the case in Arabidopsis (Raes et al. 2003). Thus, alleles at other PAL loci might differentially affect forage quality.
Abbreviations
- IVDOM:
-
In vitro digestibility of organic matter
- WSC:
-
Water soluble carbohydrates
- NDF:
-
Neutral detergent fiber
- DNDF:
-
Digestibility of neutral detergent fiber
- Indel:
-
Insertion–deletion polymorphism
- LD:
-
Linkage disequilibrium
- PAL:
-
Phenylalanine Ammonia-Lyase
- SNP:
-
Single nucleotide polymorphism
References
Andersen JR, Lübberstedt T (2003) Functional markers in plants. Trends Plant Sci 8:554–560
Andersen JR, Schrag T, Melchinger AE, Zein I, Lübberstedt T (2005) Validation of Dwarf8 polymorphisms associated with flowering time in elite European inbred lines of maize (Zea mays L.). Theor Appl Genet 111:206–217
Ballard CS, Thomas ED, Tsang DS, Mandebvu P, Sniffen CJ, Endres MI, Carter MP (2001) Effect of corn silage hybrid on dry matter yield, nutrient composition, in vitro digestion, intake by dairy heifers, and milk production by dairy cows. J Dairy Sci 84:442–452
Barrière Y, Guillet C, Goffner D, Pichon M (2003) Genetic variation and breeding strategies for improved cell wall digestibility in annual forage crops. A review. Anim Res 52:193–228
Barrière Y, Emile JC, Traineau R, Surault F, Briand M, Gallais A (2004) Genetic variation for organic matter and cell wall digestibility in silage maize. Lessons from a 34-year long experiment with sheep in digestibility crates. Maydica 49:115–126
Barrière Y, Alber D, Dolstra O, Lapierre C, Motto M, Ordas A, Van Waes J, Vlasminkel L, Welcker C, Monod JP (2005) Past and prospects of forage maize breeding in Europe.I. The grass cell wall as a basis of genetic variation and future improvements in feeding value. Maydica 50:259–274
Boerjan W, Ralph J, Baucher M (2003) Lignin biosynthesis. Annu Rev Plant Biol 54:519–546
Chabannes M, Ruel K, Yoshinaga A, Chabbert B, Jauneau A, Joseleau JP, Boudet AM (2001) In situ analysis of lignins in transgenic tobacco reveals a differential impact of individual transformations on the spatial patterns of lignin deposition at the cellular and subcellular levels. Plant J 28:271–282
Cherney JH, Cherney DJR, Akin DE, Axtell JD (1991) Potential of brown-midrib, low-lignin mutants for improving forage quality. Adv Agron 46:157–198
Ching A, Caldwell KS, Jung M, Dolan M, Smith OS, Tingey S, Morgante M, Rafalski AJ (2002) SNP frequency, haplotype structure and linkage disequilibrium in elite maize inbred lines. BMC Genet 3:19
Clark RM, Linton E, Messing J, Doebley JF (2004) Pattern of diversity in the genomic region near the maize domestication gene tb1. PNAS 101:700–707
Collazo P, Montoliu L, Puigdomenech P, Rigau J (1992) Structure and expression of the lignin O-methyltransferase gene from Zea mays L. Plant Mol Biol 20:857–867
Falush D, Stephens M, Pritchard JK (2003) Inference of population structure using multilocus genotype data: linked loci and correlated allele frequencies. Genetics 164:1567–1587
Flint-Garcia SA, Thornsberry JM, Buckler ES (2003) Structure of linkage disequlibrium in plants. Annu Rev Plant Biol 54:357–374
Fontaine AS, Barrière Y (2003) Caffeic acid O-methyltransferase allelic polymorphism characterization and analysis in different maize inbred lines. Mol Breed 11:69–75
Fu YX, Li WH (1993) Statistical tests of neutrality of mutations. Genetics 133:693–709
Gaut BS, Long AD (2003) The lowdown on linkage disequilibrium. Plant Cell 15:1502–1506
Grabber JH, Ralph J, Lapierre C, Barrière Y (2004) Genetic and molecular basis of grass cell-wall degradability. I. Lignin-cell wall matrix interactions. CR Biol 327:455–465
Guillet-Claude C, Birolleau-Touchard C, Manicacci D, Fourmann M, Barraud S, Carret V, Martinant JP, Barrière Y (2004a) Genetic diversity associated with variation in silage corn digestibility for three O-methyltransferase genes involved in lignin biosynthesis. Theor Appl Genet 110:126–135
Guillet-Claude C, Birolleau-Touchard C, Manicacci D, Rogowsky P, Rigau J, Murigneux A, Martinant JP, Barrière Y (2004b) Nucleotide diversity of the ZmPox3 maize peroxidase gene: relationships between a MITE insertion in exon 2 and variation in forage maize digestibility. BMC Genet 5:19
Gupta PK, Rustgi S, Kulwal PL (2005) Linkage disequilibrium and association studies in higher plants: present status and future prospects. Plant Mol Biol 57:461–485
Hudson RR, Kaplan NL (1985) Statistical properties of the number of recombination events in the history of a sample of DNA sequences. Genetics 111:147–164
Jones L, Ennos AR, Turner SR (2001) Cloning and characterization of irregular xylem4 (irx4): a severely lignin-deficient mutant of Arabidopsis. Plant J 26:205–216
Jung M, Ching A, Bhattramakki D, Dolan M, Tingey S, Morgante M, Rafalski A (2004) Linkage disequilibrium and sequence diversity in a 500-kbp region around the adh1 locus in elite maize germplasm. Theor Appl Genet 109:681–689
Keith CS, Hoang DO, Barret BM, Feigelman B, Nelson MC, Thai H, Baysdorfer C (1993) Partial sequence analysis of 130 randomly selected maize cDNA clones. Plant Physiol 101:329–332
Kim Y, Nielsen R (2004) Linkage disequilibrium as a signature of selective sweeps. Genetics 167:1513–1524
Kumar S, Tamura K, Nei M (2004) MEGA3: integrated software for molecular evolutionary genetics analysis and sequence aligment. Brief Bioinform 5:150–163
Lübberstedt T, Zein I, Andersen JR, Wenzel G, Krützfeldt B, Eder J, Ouzunova M, Chun S (2005) Development and application of functional markers in maize. Euphytica 146:101–108
Luff G, Schoorl W (1929) Suiker titratie. Chem Weekbl 26:130–134
Morrow SL, Mascia P, Self KA, Altschuler M (1997) Molecular characterization of a brown midrib3 deletion mutation in maize. Mol Breed 3:351–357
Nei M (1987) Molecular evolutionary genetics. Columbia University Press, New York
Palaisa KA, Morgante M, Williams M, Rafalski A (2003) Contrasting effects of selection on sequence diversity and linkage disequilibrium at two phytoene synthase loci. Plant Cell 15:1795–1806
Pedersen JF, Vogel KP, Funnell DL (2005) Impact of reduced lignin on plant fitness. Crop Sci 45:812–819
Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155:945–959
Raes J, Rohde A, Christensen JH, Van de Peer Y, Boerjan W (2003) Genome-wide characterization of the lignification toolbox in Arabidopsis. Plant Physiol 133:1051–1071
Remington DL, Thornsberry JM, Matsuoka Y, Wilson LM, Whitt SR, Doebley J, Kresovich S, Goodman MM, Buckler ES (2001) Structure of linkage disequilibrium and phenotypic associations in the maize genome. PNAS 98:11479–11484
Rohde A, Morreel K, Ralph J, Goeminne G, Hostyn V, De Rycke R, Kushnir S, Van Doorsselaere J, Joseleau JP, Vuylsteke M, Van Driessche G, Van Beeumen J, Messens E, Boerjan W (2004) Molecular phenotyping of the pal1 and pal2 mutants of Arabidopsis thaliana reveals far-reaching consequences on phenylpropanoid, amino acid, and carbohydrate metabolism. Plant Cell 16:2749–2771
Rosler J, Krekel F, Amrhein N, Schmid J (1997) Maize Phenylalanine Ammonia-Lyase has Tyrosine Ammonia-Lyase activity. Plant Physiol 113:175–179
Rozas J, Sanchez-DelBarrio JC, Messeguer X, Rozas R (2003) DnaSP, DNA polymorphism analyses by the coalescent and other methods. Bioinformatics 19:2496–2497
Saghai-Maroof MA, Soliman KM, Jorgensen RA, Allard RW (1984) Ribosomal DNA spacer-length polymorphisms in barley: Mendelian inheritance, chromosomal location, and population dynamics. PNAS 81:8014–8018
Senior ML, Murphy JP, Goodman MM, Stuber CW (1998) Utility of SSRs for determining genetic similarities and relationships in maize using an agarose gel system. Crop Sci 38:1088–1098
Sewalt VJH, Ni W, Blount JW, Jung HG, Masoud SA, Howles PA, Lamb C, Dixon RA (1997) Reduced lignin content and altered lignin composition in transgenic tobacco down-regulated in expression of l-Phenylalanine Ammonia-Lyase or Cinnamate 4-Hydroxylase. Plant Physiol 115:41–50
Smith JSC, Chin ECL, Shu H, Smith OS, Wall SJ, Senior ML, Mitchell SE, Kresovich S, Ziegle J (1997) An evaluation of the utility of SSR loci as molecular markers in maize (Zea mays L.): comparisons with data from RFLPs and pedigree. Theor Appl Genet 95:163–173
Szalma SJ, Buckler ES, Snook ME, McMullen MD (2005) Association analysis of candidate genes for maysin and chlorogenic acid accumulation in maize silks. Theor Appl Genet 110:1324–1333
Tajima F (1989) The effect of change in population size on DNA polymorphism. Genetics 123:597–601
Tenaillon MI, Sawkins MC, Long AD, Gaut RL, Doebley JF, Gaut BS (2001) Patterns of DNA sequence polymorphism along chromosome 1 of maize (Zea mays ssp. mays L.). PNAS 98:9161–9166
Thompson JD, Higgins DG, Gibson TJ (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting,positions-specific gap penalties and weight matrix choice. Nucleic Acids Res 22:4673–4680
Thornsberry JM, Goodman MM, Doebley J, Kresovich S, Nielsen D, Buckler ES (2001) Dwarf8 polymorphisms associate with variation in flowering time. Nat Genet 28:286–289
Tilley JMA, Terry RA (1963) A two stage technique for in vitro digestion of forage crops. J Brit Grassl Soc 18:104–111
VanSoest PJ (1963) Use of detergents in analysis of fibrous feeds. II. A rapid method for determination of fiber and lignin. J Assoc Off Agric Chem 46:829–835
Vignols F, Rigau J, Torres MA, Capellades M, Puigdomenech P (1995) The brown midrib3 (bm3) mutation in maize occurs in the gene encoding Caffeic Acid O-Methyltransferase. Plant Cell 7:407–416
Wang R-L, Stec A, Hey J, Lukens L, Doebley J (1999) The limits of selection during maize domestication. Nature 398:236–239
Watterson GA (1975) On the number of segregating sites in genetical models without recombination. Theor Pop Biol 7:256–276
Weir BS (1996) Genetic data analysis II. Sinauer, Sunderland
Whitt SR, Wilson LM, Tenaillon MI, Gaut BS, Buckler ES (2002) Genetic diversity and selection in the maize starch pathway. PNAS 99:12959–12962
Wilson LM, Whitt SR, Ibanez AM, Rocheford TR, Goodman MM, Buckler ES (2004) Dissection of maize kernel composition and starch production by candidate gene association. Plant Cell 16:2719–2733
Winkel BSJ (2004) Metabolic channeling in plants. Annu Rev Plant Biol 55:85–107
Zein I, Wenzel G, Andersen JR, Lübberstedt T (2006) Nucleotide sequence diversity at the Caffeic acid O-methyltransferase locus in 42 European elite maize inbred lines. Genet Resour Crop Ev (in press)
Acknowledgments
We would like to thank KWS Saat AG (Einbeck) and the German ministry for education and science (BMBF) for financial support of the EUREKA project Cerequal.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by H. H. Geiger.
Jeppe R. Andersen and Imad Zein contributed equally to the manuscript.
Rights and permissions
About this article
Cite this article
Andersen, J.R., Zein, I., Wenzel, G. et al. High levels of linkage disequilibrium and associations with forage quality at a Phenylalanine Ammonia-Lyase locus in European maize (Zea mays L.) inbreds. Theor Appl Genet 114, 307–319 (2007). https://doi.org/10.1007/s00122-006-0434-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00122-006-0434-8