Abstract
Root-knot nematodes Meloidogyne incognita (Kofoid and White) can cause severe yield loss in cotton (Gossypium hirsutum L.). The objectives of this study were to determine the inheritance and genomic location of genes conferring root-knot nematode resistance in M-120 RNR, a highly resistant G. hirsutum line with the Auburn 623 RNR source of resistance. Utilizing two interspecific F2 populations developed from the same M-120 RNR by Gossypium barbadense (cv. Pima S-6) cross, genome-wide scanning with RFLP markers revealed a marker on Chromosome 7 and two on Chromosome 11 showing significant association with the resistant phenotype. The association was confirmed using SSR markers with the detection of a minor and a major dominant QTL on Chromosome 7 and 11, respectively. Combined across the two populations, the major QTL on Chromosome 11 Mi-C11 had a LOD score of 19.21 (9.69 and 9.61 for Pop1 and Pop2, respectively) and accounted for 63.7% (52.6 and 65.56% for Pop1 and Pop2, respectively) of the total phenotypic variation. The minor QTL locus on Chromosome 7 Mi 1 -C07 had a LOD score of 3.48 and accounted for 7.7% of the total phenotypic variation in the combined dataset but was detected in only one population. The allele from the M-120 RNR parent contributed to increased resistance in the Mi-C11 locus, but surprisingly, the Pima S-6 allele contributed to increased resistance in the Mi-C07 locus. The M-120 RNR allele in the Mi-C11 locus, derived from the Auburn 623 RNR, is likely to have originated from the Clevewilt 6 cultivar. Results from this study indicated that the SSR marker CIR316 may replace the laborious greenhouse screening in breeding programs to identify genotypes resistant to M. incognita.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The southern root-knot nematode Meloidogyne incognita Kofoid and White (Chitwood) is the most important endoparasitic pest of cotton (Gossypium hirsutum L.). Found worldwide, root-knot nematode feeding causes root damage and extensive galling, resulting in reduced water and nutrient translocation in the cotton plant (Bridge 1992). Worldwide cotton yield losses due to this pest were estimated to be 10.7% (Sasser and Freckman 1987). In the USA alone, root-knot nematodes cause an estimated loss of 273 million pounds of lint annually (Blasingame and Patel 2005). In addition to direct losses from nematode parasitism, infection by this nematode greatly increases the incidence and severity of Fusarium wilt caused by Fusarium oxysporum f. sp. vasinfectum, which causes an estimated loss of another 54.5 million pounds of lint annually in the USA (Blasingame and Patel 2005). Although nematicides can be effective in controlling this nematode, they are expensive and do not provide season long protection. The development and use of resistant cultivars offers the best management tool for controlling root-knot nematodes.
Three major sources of resistant germplasm are recognized in G. hirsutum, two from the non-Acala Upland (referred herein simply as Upland) and one from the Acala cotton germplasm (Robinson et al. 2001). The first and most important source of resistance in Upland germplasm was from Auburn 623 RNR, which originated from transgressive segregation in a cross between two moderately resistant parents, Clevewilt 6 and Wild Mexican Jack Jones (Shepherd 1974a). Resistance in Auburn 623 RNR was subsequently transferred to several agronomically adapted cultivars through backcrossing, resulting in the release of the M-line series including lines such as ‘M-120 RNR’, ‘M-135 RNR’, and ‘M-155 RNR’, which greatly improved the agronomic qualities while retaining the almost-immune level of resistance of Auburn 623 RNR. Clevewilt 6 was also used to develop the resistant germplasm LA434-RKR. This resistant germplasm provided only a moderate level of protection from root-knot nematodes but was important because it gave rise to two widely planted cultivars in the USA in the early 1990s, Stoneville LA887 and Paymaster H1560, which has moderate root-knot nematode resistance (Robinson et al. 2001). Finally, there is resistance found in a commercial Acala cultivar, Acala NemX (Ogallo et al. 1997). The source of the moderate level of resistance to root-knot nematodes in Acala NemX is uncertain but the donor parent N6072 is thought to have been derived from crossing a Gossypium barbadense L. genotype and the Acala line 1-2302 (Robinson et al. 2001). Like other Acala cotton types, which are only adapted for cotton production in the southwestern USA, Acala NemX is not planted in any other regions of the USA Cotton Belt. A diagrammatic representation of the origin of root-knot nematode resistance in the three germplasm sources can be found in Robinson et al. (2001).
While it is not clear whether these resistant sources have the same genes for resistance, the mode of inheritance of the resistance trait among these sources appears to be different. For example, Shepherd (1974b) reported that the mode of inheritance of the resistance in Auburn 623 RNR, based on crossing with three susceptible cultivars, was incomplete dominance. In working with the M-315 RNR line that was derived from Auburn 623 RNR, McPherson et al. (2004) reported that the resistance in this line was conferred by one dominant and one additive gene. In contrast, Zhou et al. (1999) and Wang et al. (2006a) indicated that a single recessive gene primarily determined resistance in Acala NemX. Thus, the resistance to root-knot nematodes in cotton, simply inherited and governed by one to a few major genes, is consistent with the mode of inheritance in other crop species such as cowpea (Ehlers et al. 2000), soybean (Luzzie et al. 1994), sugar beet (Weiland and Yu 2003), tomato (Messeguer et al. 1991), and peanut (Burow et al. 1996).
Molecular markers can provide a means to address fundamental questions on the genetics of root-knot nematodes resistance in cotton and to assist in breeding for nematode resistant cotton cultivars. For example, they can be used in genetic mapping to uncover the number of genes involved and the effect of each gene in conditioning resistance, thus providing a better understanding of the genetics and perhaps the mechanisms of how the resistance to nematode invasion is conferred by each gene. In addition, the trait-linked marker genotypes can serve as phenotype predictors, thus laborious greenhouse screening for nematode resistance can be delayed until later stages of line development.
DNA markers have been used for mapping root-knot nematode resistance genes in cotton. Bezawada et al. (2003) identified an SSR marker BNL1421, which explained 8% the phenotypic variation in galling index in an F2 population from a cross of the moderately resistant line Clevewilt 6-1 and the susceptible cultivar Stoneville 213. Recently, Wang et al. (2006a) reported the mapping of a major resistance gene in Acala NemX. The resistance gene was mapped at a distance of about 2.1–3.3 cM from the SSR marker CIR316. The resistance genes from these two resistant sources appear to be different from each other because the SSR marker BNL1421 was located on linkage group (LG) A01 (Park et al. 2005), whereas the SSR marker CIR316 was mapped to LG A03 (Wang et al. 2006a). Wang et al. (2006b) has recently assigned LG A01 as Chromosome 13 and LG A03 as Chromosome 11.
Genetic mapping of the root-knot resistance gene(s) in the Auburn 623 RNR source would allow us to determine if the resistance genes are the same or different from those mapped in other resistant sources. Further, the identification of markers linked to the resistance genes would facilitate its deployment in breeding for cultivars with improved nematode resistance and assist in combining resistance genes from other resistant sources to obtain more durable or higher levels of resistance. The objectives of this study were to determine the inheritance and genomic location of gene(s) conferring resistance to root-knot nematodes in M-120 RNR, a highly resistant G. hirsutum line that was derived from the Auburn 623 RNR germplasm source.
Materials and methods
Mapping population and resistance screening
Two genetically similar F2 populations were developed by self-pollinating different F1 plants from a cross between a highly resistant Upland cotton line M-120 RNR (Shepherd et al. 1996) and the susceptible G. barbadense cv. Pima S-6. The two F2 populations (referred herein as Pop1 and Pop2) consisted of 138 (Pop1) and 107 (Pop2) progenies. Each population along with three plants of each parent and F1 (except two F1 plants in Pop1) was planted in a completely randomized design in separate sections of the greenhouse. At 3 weeks after seedling emergence, both populations were inoculated with approximately, 8,000 eggs of M. incognita race 3 (approximately, 450 eggs 150 cm−3 soil) (Wang et al. 2006a). Nematode eggs were collected from tomato roots (Lycopersicon esculentum L.) by agitating roots in 0.5% sodium hypochlorite solution for 2 min (Hussey and Barker 1973) at 1 h before inoculation. Inoculum was distributed into 2 holes about 2.5 cm deep and covered with soil. Pots were watered immediately following inoculation. The soil temperatures varied between 17 and 28°C.
Plants were evaluated for their ability to host M. incognita reproduction at 56 and 58 days after inoculation for Pop1 and Pop2, respectively. The phenotypic data commonly used to determine resistance reaction to nematodes were collected; they include root galling index, total number of eggs extracted, and the mean number of eggs/gram of fresh root. Roots were washed free of soil, evaluated for galling, weighed, cut into 5 cm pieces, and agitated in a 1% sodium hypochlorite solution in a 1 l flask for 4 min (Hussey and Baker 1973). Eggs were collected and rinsed with tap water on nested 150- over 25-mm-pore sieves. Root galling was evaluated using the 0–10 scale where, 0 = no galling, 1 = 1–10% of the root system galled, 2 = 11–20% of the roots system galled, etc., with 10 = 91–100% of the root system galled (Bridge and Page 1980). Roots of each plant were harvested and weighed immediately prior to extraction of eggs. Eggs were extracted from the whole root system and counted. The mean number of eggs/gram of fresh root (eggs/g of root) was evaluated in order to standardize egg counts.
RFLP and SSR analysis
Laboratory techniques for DNA extraction, restriction digestion, gel electrophoresis, southern blotting, probe labeling, southern hybridization, and autoradiography were as described in Reinisch et al. (1994). Four restriction enzymes (EcoRI, EcoRV, HindIII, and XbaI) were used to reveal polymorphisms between the two parents. Probes were selected at approximately 20 cM intervals for all 13 linkage groups based on a cotton consensus map (Paterson AH, unpublished data). The origins of all the genomic or cDNA libraries from which RFLP probes were drawn from is described by Rong et al. (2004).
SSR primer sequences were drawn from the following sources: BNL primers from the Research Genetics Co. (Huntsville, AL, USA, http://www.resgen.com); JESPR primers from Reddy et al. (2001); TM primers from Dr. John Yu, USDA-ARS, Crops Germplasm Research Unit, TX, USA; CIR primers from Nguyen et al. (2004); NAU primers from Han et al. (2004, 2006). PCR amplifications were performed as described in Zhang et al. (2002), using a Peltier Thermal Cycler (MJ Research, Waltham, MA, USA) and PCR products were electrophoresed on a 10% non-denatured polyacrylamide gel using a Bio-Rad Protean II xi electrophoresis apparatus (Bio-Rad, Hercules, CA, USA). For primer CIR069, PCR product of the two parents was no polymorphic on a non-denatured polyacrylamide gel, but polymorphism was revealed when electrophoresed on a denatured polyacrylamide gel using the protocol as described by Nguyen et al. (2004).
Three CIR primers that produced weak amplification were assumed to have short sequences (15–18 bases), which resulted in an unstable annealing of primer and genomic DNA. These CIR primers were redesigned with a longer sequence based on their corresponding genomic DNA sequences in Genbank using the Primer Premier 5.0 program. The redesigned primer sequence for CIR316 was 5′-CCG TCC TTA CAG GCA CTA CCA CC-3′ and 5′-TTC TGG CGA CTT CAC CAC ATT-3′, for CIR196 was 5′-CGA TTG GTG AAG GGA GGA AAG-3′ and 5′-GAC AGA GGG AAT GCT CAA A-3′, and for CIR069 was 5′-GTC ACT GCT ATA CAC TTT CCA T-3′ and 5′-AGT ATT GGG CTT TGA TTT GA-3′.
Mapping and statistical analysis
A subset of 16 highly resistant and 16 highly susceptible F2 individuals (8 individuals from each population) was initially selected based on the egg count phenotype to screen for putative genomic regions associated with root-knot nematode resistance. The highly resistant plants have total egg counts from 0 to 600, with a mean of 300, while the highly susceptible plants have egg counts from 42,000 to 446,400, with a mean of 106,912. These 32 individuals were individually screened with a total of 188 RFLP markers selected from the cotton genetic map of Rong et al. (2004), which provides an average of 20 cM interval coverage for the entire cotton genome. A single-factor analysis was tested on the RFLP marker genotype and the resistance phenotype using the QGene 3.6 program (Nelson 1997). RFLP markers that showed significant association with the resistance phenotype were further tested using the entire population to confirm genetic linkage. Five plants in each population had a root weight that was less than 2 g and were excluded from the data analysis; therefore, data analysis was performed with only 133 individuals in Pop1 and 102 individuals in Pop2. For genomic regions on Chromosome 7 and 11, in which significant associations were detected by RFLP markers, a total of 51 SSR markers that targeted either of these chromosomes or their corresponding homoeologous counterparts on Chromosome 16 and 21 were selected based on the genetic linkage map of Han et al. (2004, 2006), and Nguyen et al. (2004). The SSR markers were tested on all F2 individuals and a localized linkage map for each of the two chromosomes was constructed.
Genetic mapping and QTL analyses were performed on each population separately and combined across populations. Linkage maps were constructed using the MAPMAKER/Exp Version 3.0b Software (Lander 1987). A LOD score of 4.0 and a 50 cM maximum distance were used. Marker order was confirmed with the “ripple” command. Recombination frequencies were converted into map distances (cM) using the Kosambi mapping function (Kosambi 1944). QTLs were identified by composite interval mapping (Zeng 1994) using Windows QTL Cartographer 2.5 (Basten et al. 2001). QTLs were declared significant if the corresponding LR score were greater than 11.5 (equal to a LOD score of 2.5). The percent of phenotypic variance (PV) explained by a QTL (R2) was estimated at highest probability peaks.
QTL Cartographer assumes that the quantitative data under consideration is normally distributed. However, as with most disease resistance data, none of the data sets for root-knot nematode resistance in this study fit this assumption. A common method to normalize the data used in QTL-mapping is to conduct log transformation. However, transforming to normalize the data could misrepresent the differences among individuals for the trait by pulling the skewed tails of the distribution toward the center, thus reducing one’s ability to detect QTLs (Mutschler et al. 1996). Therefore, in view of this shortfall, both the non-transformed and log10 (x + 1) transformed data were used in QTL mapping.
Results
Classification and segregation of resistance
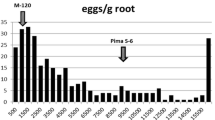
In the screening for both populations, resistant parent M-120 RNR was associated with a lower galling index, total eggs count, and eggs/g of root than the susceptible parent Pima S-6 (Table 1). Further, the F1 plants showed high levels of resistance, with the galling index, total egg count, and eggs/g of root not significantly different from the resistant parent M-120 RNR (Table 1). These data indicated that resistance was mainly controlled by dominant genes (Table 1). The phenotypic distributions of Pop1 and Pop2 for root galling index, total egg count, and the eggs/g of root are presented in Fig. 1. All three phenotypes displayed a non-normal distribution (skewness exceeding 1.0), with individuals skewed towards the resistant parent (Table 1). This result further supports the F1 data in that the resistance trait is governed by dominant genes. The coefficients of correlation among the three traits were significant (Table 2), suggesting that they measure the same genetic factor, albeit with different precision. Galling index was correlated (P < 0.01) with eggs/g of root at a value of r = 0.53 and 0.67 and with total egg count at r = 0.44 and 0.74 in Pop1 and Pop2, respectively, whereas eggs/g of root was correlated with total egg count at r = 0.89 and 0.92 in Pop1 and Pop2, respectively.
Preliminary marker by trait association analysis
Preliminary association analysis using 188 RFLP markers on 16 highly resistant and 16 highly susceptible F2 progenies identified seven markers (A1593, pAR1–56a, G1112b, G1158b, pAR188, pAR111 and pGH243) significantly associated with nematode resistance (P ≤ 0.05) based on the galling index data. These markers were further tested using all the retained progenies in Pop1 and Pop2 with a threshold for declaring significant association set at P ≤ 0.001 to reduce type I errors. The association was confirmed for markers pAR111, pGH243 and G1158b, while the remaining markers failed to reach the significance threshold. Both pAR111 and pGH243 were mapped to the distal end of Chromosome 11 (previously LG A03) at about 10 cM from each other, and G1158b was mapped on Chromosome 7 (Rong et al. 2004). These three markers were also tested for association with the total egg count and eggs/g of root phenotypes, only pAR111 showed significant (P < 0.0001) association with both phenotypes. Based on the galling data, the coefficients of determination (R2) calculated for pAR111, pGH243 and G1158b were 16.2, 8.1 and 5.3%, respectively, in the combined dataset. These results suggest that a resistance gene may be present near these markers, but the linkage is not sufficiently tight to explain a large amount of the phenotypic variation. The two chromosome regions (on Chromosome 7 and 11) were investigated in more detail by surveying SSR markers in order to find markers more closely linked to the resistance genes.
In addition to RFLP markers, we also tested the SSR marker BNL1421 on Chromosome 13, which was reported by Bezawada et al. (2003) to be significantly associated with galling index in an F2 population from a cross of the moderately resistant line Clevewilt 6–1 and the susceptible cultivar Stoneville 213. A single-factor analysis for both populations showed no association between this marker and galling index, total egg count, or eggs/g of root phenotypes.
Construction of localized linkage map and QTL mapping
A total of 51 SSR markers, mapped to Chromosome 7 and 11 or to their corresponding homoeologous chromosomes on Chromosome 16 and 21, were selected based on the cotton genetic linkage maps of Han et al. (2004, 2006) and Nguyen et al. (2004), and tested for polymorphism between M-120 RNR and Pima S-6. Thirty-four markers are from Chromosome 11, 11 from Chromosome 7, 4 from Chromosome 21 and 2 from Chromosome 16. Thirty-nine primers amplified to DNA fragments that were polymorphic between the two parents and were tested on both F2 populations. The SSR data were combined with the RFLP data to develop a localized LG for Chromosome 7 and Chromosome 11. In total, 17 markers were mapped on Chromosome 11, which spanned 196.1 cM, and 7 markers were mapped on Chromosome 7, which covered 79.7 cM. The marker order for both linkage groups and distances between markers were in close agreement with the public genetic maps (Rong et al. 2004; Han et al. 2004, 2006; Nguyen et al. 2004).
QTL mapping for nematode resistance on Chromosome 11
On Chromosome 11, a major QTL for galling index was detected in the interval flanked by two co-dominant SSR markers CIR069 and CIR316 in both Pop1 and Pop2. In Pop1, this locus was located only 2 cM from CIR316; using non-transformed data and log-transformed data, the LOD score was 9.69 and 9.68, respectively, and the PV explained by this locus was 52.56 and 34.44%, respectively (Table 3). In Pop2, this locus was located only 4 cM from CIR316; using non-transformed data and log-transformed data, the LOD score was 9.61 and 7.21, respectively, and the PV explained by this locus was 65.65 and 31.67%, respectively (Table 3). In both populations, this QTL interval was also significantly associated with total eggs and eggs/g of root phenotypes for both log-transformed and non-transformed data, however the PVs explained were lower than those found from the galling index phenotype.
For the combined dataset, the locus near CIR316 had a LOD score of 19.21 and accounted for 63.7% of the variation in galling index with non-transformed data (Fig. 2; Table 3). With log-transformed data, the LOD score was 8.51 and the locus accounted for only 35.84% of PV, but the composite interval mapping results confirmed that this QTL has a large genetic effect. The outcomes of the association analyses with total eggs and eggs/g of root were also highly significant, but the PVs explained by this locus were much lower than from the galling index phenotype. For total eggs, the PV for the non-transformed and log transformed data were 8.91 and 14.39%, respectively, and for eggs/g of root, the PV for the non-transformed and log transformed data were 16.09 and 15.0%, respectively. The allele from the resistant parent M-120 RNR (identified by a 340 bp DNA fragment amplified using the primer CIR316) increased nematode resistance for all of the measured traits; using non-transformed data, it reduced galling index by 2.68, reduced total eggs by 13,540, and reduced the eggs/g of root by 1,859. This QTL, which showed a completely dominant gene action, was given the name Mi-C11 for resistance to M incognita on Chromosome 11 (see Discussion).
QTL mapping for nematode resistance on Chromosome 7
On Chromosome 7, a QTL for galling index, total egg count and eggs/g of root was detected in the interval flanked by the SSR marker NAU474b and the RFLP marker G1158b in Pop2 only. This locus was located only 6 cM from G1185b; using non-transformed data and log-transformed data, the LOD score was 5.31 and 5.02, respectively, and the PV explained by this locus was 32.50 and 33.33%, respectively (Table 4).
For the combined dataset, the locus near G1158b (Fig. 3; Table 4) had a LOD score of 3.48 and accounted for 7.7% of the variation in galling index using either the non-transformed or log-transformed data. The interval NAU474b-G1158b was also significantly associated with the total eggs and eggs/g of root. The association with the total eggs phenotype accounted for 5.35 and 9.4% of the PV with non-transformed data and log-transformed data, respectively, while for eggs/g of root phenotype accounted for only 7.42% of the PV with log-transformed data. The association for the non-transformed eggs/g of root phenotype was below the significance threshold for declaring a QTL. Interestingly, the allele that confers resistance for all phenotypes in this QTL originated from the susceptible parent, Pima S-6 (identified by a 235 bp fragment amplified using the primer NAU474b). The low PV and d/a > 1 indicated that this was a minor QTL (Table 4) with an over dominance gene action. This locus was given the name Mi-C07 for resistance to M. incognita on Chromosome 7.
Origin of the major root-knot nematode resistance gene in the M-120 RNR line
To determine the origin of the Mi-C11 locus, we tested the SSR marker CIR316 on the parental lines; Auburn 623 RNR, Auburn 634 RNR, Clevewilt 6, Wild Mexican Jack Jones, and the cultivar Coker 201 which was used as recurrent parent in the backcrossing program to develop the M-120 RNR line (Fig. 4). In addition, we have also tested seven other Auburn 634 RNR-derived root-knot nematode resistant M-series lines, M-92 RNR, M-240 RNR, M-249 RNR, M-272 RNR, M-315 RNR, M-331 RNR, M-725 RNR, as well as the nematode resistant cultivars Acala NemX and Stoneville LA887. A 340 bp DNA fragment amplified using primer CIR316 (Fig. 5, see arrow), which showed tight linkage to the root-knot nematode resistant locus, was present in M-120 RNR, Auburn 623 RNR, Auburn 634 RNR, and Clevewilt 6, but was absent in Wild Mexican Jack Jones and Coker 201. Further, this DNA fragment was present in all other resistant M-lines derived from the Auburn 623 RNR resistance source. These results strongly suggest that the Mi-C11 resistant locus in M-120 RNR originated from the resistant parent, Clevewilt 6. Interestingly, this DNA fragment also was present in the cultivars LA887 and Acala NemX, suggesting that the two cultivars were either carrying the same resistance gene from the Auburn 623 RNR source or had the same amplicon site by chance due to a low level of genetic diversity in the G. hirsutum gene pool (Van Becelaere et al. 2005).
Diagrammatic pedigree of nematode resistant germplasm M-120 RNR. RC recurrent parent (refer to Robinson et al. 2001)
Origin of root-knot nematode resistance locus on Chromosome 11 amplified by SSR primer CIR316. Lane: 1 Marker, 2 Pima S-6, 3 M-120 RNR, 4 Auburn 634 RNR, 5 Auburn 623 RNR, 6 Clevewilt 6, 7 Wild Mexican Jack Jones, 8 Coker 201, 9 M-92 RNR, 10 M-240 RNR, 11 M-249 RNR, 12 M-272 RNR, 13 M-315 RNR, 14 M-331 RNR, 15 M-725 RNR, 16 Acala NemX, 17 LA887
Discussion
Previous inheritance studies (McPherson et al. 1995, 2004; Zhou et al. 1999) showed that the root-knot nematode resistance in M-lines derived from the Auburn 623 RNR source was simply inherited and controlled by one to a few major genes. In this study, QTL analysis showed that the resistance in M-120 RNR was due to one major dominant QTL, which explained as much as 52.6–65.6% of the PV in two genetically similar F2 population. The genomic location of this QTL was determined using QTL Cartographer to be on Chromosome 11. McPherson et al. (2004) reported that the inheritance of the resistance trait in the M-315 RNR line was conferred by one dominant and one additive gene, which they named Mi 1 and Mi 2 , respectively. Since the near immunity level of resistance in both M-120 RNR and M-315 RNR was backcrossed from the same Auburn 623 RNR donor parent, many believed that these 2 lines, as well as others derived from Auburn 623 RNR, carry the same major genes for resistance (Shepherd et al. 1989; Robinson et al. 2001; McPherson et al. 1995, 2004). If this hypothesis holds true, then the major dominant QTL on Chromosome 11 reported in this study is likely the same as the Mi 1 locus reported by McPherson et al. (2004). We named this locus Mi-C11 by following the conventional nomenclature for QTLs where acronym of the scientific name of the pathogen causing the disease in lowercase following by the chromosome name.
In addition to the Mi-C11 locus, a minor QTL was also detected on Chromosome 7. This Mi-C07 locus, which is likely to be codominant and explained only 7.7% of PV, originated from the susceptible parent Pima S-6. A possible explanation for this observation is that the breeding for a higher yield potential during cultivar development could fortuitously select for genotypes with greater tolerance, albeit still susceptible to nematodes parasitism. Cultivars and germplasm lines with different levels of tolerance to root-knot nematodes are recognized (Davis and May 2003; Robinson et al. 1999), some of which could carry minor QTLs conferring resistance. This result supports the notion that QTLs with positive effects may be recovered from an inferior parent (Xiao et al. 1996; Chee et al. 2005). Alternatively, this minor QTL could be an analytical artifact. A major QTL for root weight was identified in Pop1 and in the combined dataset at the interval NAU845-NAU1048, about 25 cM from the Mi-C07 locus (Fig. 3). This root weight QTL for the combined dataset had a LOD score of 5.04 and accounted for 28.8% PV with the Pima S-6 allele conferring a 3.52 g increased in root weight (Table 4). Gall ratings of all but the most susceptible plants indicated that nematode reproduction was well below the carrying capacity of the root systems, so the amount of root material should not have limited reproduction, and plants with equal levels of resistance should have an equal number of galls. Therefore, a plant with a larger root system would appear to be more resistant based on galling because the rating scale is based on a percentage of the system covered with galls. The QTL for resistance at the Mi-CO7 locus also was identified based on the number of eggs produced, which should not be significantly affected by the size of the root system until carrying capacity begins to limit population levels. Further study is needed to verify whether the resistance locus on Chromosome 7 is authentic.
Pedigree accounts indicated that the root-knot nematode resistance in M-120 RNR could be traced back to Auburn 623 RNR, which was derived from the cross Clevewilt 6 and Wild Mexican Jack Jones (Fig. 4). Consequently, it has long been debated which parental line contributed the genes for the near-immunity in Auburn 623 RNR (Shepherd 1974b; McPherson et al. 2004). Our result, from tracking the 340 bp DNA fragment amplified by primer CIR316 which showed tight linkage to the root-knot nematode resistance locus, showed that this DNA fragment was present in M-120 RNR, Auburn 623 RNR, Auburn 634 RNR, Clevewilt 6, but was absent in Wild Mexican Jack Jones and Coker 201. Based on this observation, we speculate that the dominant Mi-C11 locus in M-120 RNR originated from the Clevewilt 6 parent. Further, the presence of this DNA fragment in all the tested M-series lines support this result and suggest that resistance in these lines is mainly conferred by this locus.
Interestingly, this DNA fragment also was present in the cultivars LA887 and Acala NemX. The resistance in LA887 can be traced back to the cultivar Bayou, which was derived from a cross between Clevewilt 6 and Deltapine 15 (Robinson et al. 2001). Therefore, it is conceivable that the gene for resistance in LA887 was allelic to the Mi-C11 locus of M-120 RNR. However, Acala NemX shares no known common parentage with Auburn 623 RNR or LA887 (Robinson et al. 2001). Thus, we are uncertain if Acala NemX carries the same resistance gene as Auburn 623 RNR. A recent genetic mapping experiment suggests that the gene conferring resistance in Acala NemX is located on Chromosome 11 and is linked to the same SSR marker (CIR316) as Mi-C11 (Wang et al. 2006a).
Although there is considerable coincidence in the location of the resistance loci detected in this study compared with loci detected in Acala NemX, there are two lines of evidence suggesting that the two genes may not be allelic. First, the resistant gene in Acala NemX was located 2.6 cM from CIR316 in the region of CIR316 and BNL1066 (Wang et al. 2006a), whereas in our study the resistance gene was located 2–4 cM above CIR316 but in the region of CIR316 and CIR069. Second, the two genes are dissimilar in their inheritance pattern: the segregation pattern in this study showed that the resistant gene in M-120 RNR was clearly dominant while that in Acala NemX was recessive (Wang et al. 2006a; Zhou et al. 1999). It has been well documented that recessive and dominant R genes are unrelated mechanistically (Grube et al. 2000). Further allelism studies are required to resolve this question.
The highly resistant germplasm line Auburn 623 RNR has been in existence for more than 3 decades and the resistance in this line has been transferred to lines with acceptable agronomic qualities. Yet, none of the currently grown commercial cotton cultivars expresses a similar high level of resistant to root-knot nematodes. The slow progress in developing resistant cultivars is believed to be due to the current screening process which is tedious, time consuming and destructive for identifying resistance genotypes. Using the SSR primer CIR316 as a selection marker, post hoc analysis of the combined dataset indicated that if we select only the progeny carrying the homozygous M-120 RNR allele, 80% (33/41) of the progeny would have a galling index of 3 or lower, and would be resistant to root-knot nematodes. This result indicates that the easily handled co-dominant SSR marker could be effective in screening for nematode resistant progenies in a breeding population. The use of this marker could save considerable time and effort by precluding the necessity of using greenhouse screening for this subterranean phenotype. Further, it could be used to ensure that the gene was not lost during cultivar development.
References
Basten CJ, Weir BS, Zeng ZB (2001) QTL Cartographer, Version 1.15. Department of Statistics, North Carolina State University, Raleigh
Bezawada C, Saha S, Jenkins JN, Creech RG, McCarty JC (2003) SSR marker(s) associated with root knot nematode resistance gene(s) in cotton. J Cotton Sci 7:179–184
Blasingame D, Patel MV (2005) Cotton disease loss estimate committee report. In: Proceedings of Beltwide Cotton Conference, New Orleans, LA. Nat Cotton Counc Am, Memphis, pp 259–262
Bridge J (1992) Nematodes. In: Hollocks RJ (ed) Cotton diseases. CAB Int, Wallingford, pp 33–353
Bridge J, Page SLR (1980) Estimation of root-knot nematode infestation levels on roots using a rating chart. Trop Pest Manage 26:296–298
Burow MD, Simpson CE, Paterson AH, Starr JL (1996) Identification of peanut (Arachis hypogaea L.) RAPD markers diagnostic of root-knot nematode (Meloidogyne arenaria (Neal) Chitwood) resistance. Mol Breed 2:369–379
Chee P, Draye X, Jiang C-X, Decanini L, Delmonte TA, Bredhauer R, Smith CW, Paterson AH (2005) Molecular dissection of phenotypic variation between Gossypium hirsutum and G. barbadense (cotton) by a backcross-self approach: I. Fiber Elongation. Theor Appl Genet 111:757–763
Davis RF, May OL (2003) Relationships between tolerance and resistance to Meloidogyne incognita in cotton. J Nematol 35:411–416
Ehlers JS, Matthews WC, Hall AE, Roberts PA (2000) Inheritance of a broad-based form of root-knot nematode resistance in cowpea. Crop Sci 40:611–618
Grube RC, Radwanski ER, Jahn M (2000) Comparative genetics of disease resistance within the Solanaceae. Genetics 155:873–887
Han ZG, Guo WZ, Song XL, Zhang TZ (2004) Genetic mapping of EST-derived microsallites from the diploid Gossypium arboretum in allotetraploid cotton. Mol Gen Genomics 272:308–327
Han ZG, Wang CB, Song XL, Guo WZ, Gou JY, Li CH, Chen XY, Zhang TZ (2006) Characteristics development and mapping of Gossypium hirsutum derived EST-SSRs in allotetraploid cotton. Theor Appl Genet 112:430–439
Hussey RS, Barker KR (1973) A comparison of methods of collecting inocula of Meloidogyne spp. including a new technique. Plant Dis Rep 57:1025–1028
Kosambi DD (1944) The estimation of map distances from recombination values. Ann Eugen 12:172–175
Lander ES, Green P, Abrahamson J, Barlow A, Daly MJ, Lincoln SE, Newburg L (1987) MAPMAKER: an interactive computer package for constructing primary genetic linkage maps of experimental and natural populations. Genomics 1:174–181
Luzzie BM, Boerma HR, Hussey RS (1994) A gene for resistance to southern root-knot nematode in soybean. J Hered 85:484–486
McPherson MG, Jenkins JN, McCarty JC, Watson CE (1995) Combining ability analysis of root-knot nematode resistance in cotton. Crop Sci 35:373–375
McPherson MG, Jenkins JN, Watson CE, McCarty JC (2004) Inheritance of root-knot nematode resistance in M-315 RNR and M78-RNR cotton. J Cotton Sci 8:154–161
Messeguer R, Ganal M, Devicente MC, Young ND, Bolkan H, Tanksley SD (1991) High-resolution RFLP map around the root-knot nematode resistance gene (MI) in tomato. Theor Appl Genet 82:529–536
Mutschler MA, Doerge RW, Liu SC Kuai JP, Liedl BE, Shapiro JA (1996) Analysis of pest resistance in the wild tomato Lycopersicon pennellii: QTLs controlling acylsugar level and composition. Theor Appl Genet 92:709–718
Nelson JC (1997) QGENE: Software for marker-based genomic analysis and breeding. Mol Breed 3:239–245
Nguyen TB, Giband M, Brottier P, Risterucci AM, Lacape JM (2004) Wide coverage of the tetraploid cotton genome using newly developed microsatellite markers. Theor Appl Genet 109:167–175
Ogallo JL, Goodell PB, Eckert J, Roberts PA (1997) Evaluation of NemX, a new cultivar of cotton with high resistance to Meloidogyne incognita. J Nematol 29:531–537
Park YH, Alabady MS, Sickler B, Wilkins TA, Yu J, Stelly DM, Kohel RJ, El-Shihy OM, Cantrell RG, Ulloa M (2005) Genetic mapping of new cotton fiber loci using EST-derived microsatellites in an interspecific recombinant inbred line (RIL) cotton population. Mol Gen Genomics 274:428–441
Reddy OUK, Pepper AE, Ibrokhim A, Saha S, Jenkins JN, Brooks T, Bolek Y, El-Zik KM (2001) New dinucleotide and trinucleotide microsatellite marker resources for cotton genome research. J Cotton Sci 5:103–113
Reinisch AJ, Dong JM, Brubaker C, Stelly D, Wendel JF, Paterson AP (1994) A detailed RFLP map of cotton (Gossypium hirsutum x Gossypium barbadense): chromosome organization and evolution in a disomic polyploid genome. Genetics 138:829–847
Robinson AF, Cook CG, Percival AE (1999) Resistance to Rotylenchulus reniformis and Meloidogyne incognity race 3 in the major cotton cultivars planted since 1950. Crop Sci 39:850–858
Robinson AF, Bowman DT, Cook CG, Jenkins JN, Jones JE, May LO, Oakley SR, Oliver MJ, Roberts PA, Robinson M, Smith CW, Starr JL, Stewart JM (2001) Nematode Resistance In: Kirkpatrick TL Rothrock CS (ed) Compendium of cotton diseases. The American Phytopathological Society, St. Paul, pp 68–72
Rong JK, Abbey C, Bowers JE, Brubaker CL Chang C, Chee PW, Delmonte TA, Ding X, Garza JJ, Marler BS, Park C, Pierce GJ, Rainey KM, Rastogi VK, Schulze SR, Tronlinde NL, Wendel JF, Wilkins TA, Wing RA, Wright RJ, Zhao X, Zhu L, Paterson AH (2004) A 3347-locus genetic recombination map of sequence-tagged sites reveals features of genome organization, transmission and evolution of cotton (Gossypium). Genetics 166:389–417
Sasser JN, Freckman DW (1987) A world perspective on nematology: The role of the society. In: Veech JA, Dickson DW (eds) Vistas on nematology: a commemoration of the twenty-fifth anniversary of the Society of Nematologists. Society of Nematologists, Hyattsville, pp 7–14
Shepherd RL (1974a) Registration of Auburn 623 RNR cotton germplasm. Crop Sci 14:911
Shepherd RL (1974b) Transgressive segregation for root-knot nematode resistance in cotton. Crop Sci 14:872–875
Shepherd RL, Parrott WL, McCarty JC, Jenkins JN (1989) Notice of release of nine root-knot nematode resistance germplasm lines of upland cotton Gossypium hirsutum L. USDA/Mississippi Agric For Exp Stn Memo
Shepherd RL, McCarty JC, Jenkins JN (1996) Registration of nine cotton germplasm lines resistance to root-knot nematode. Crop Sci 36:820
Van Becelaere G, Lubbers EL, Paterson AH, Chee PW (2005) Pedigree vs. RFLP based genetic similarity estimates in cotton. Crop Sci 45:2281–2287
Wang C, Ulloa M, Roberts P (2006a) Identification and mapping of microsatellite markers linked to a root-knot nematode resistance gene (rkn1) in Acala NemX cotton. Theor Appl Genet 112:770–777
Wang K, Song X, Han Z, Guo W, Yu JZ, Sun J, Pan J, Kohel RJ, Zhang T (2006b) Complete assignment of the chromosomes of Gossypium hirsutum L. by translocation and fluorescence in situ hybridization mapping. Theor Appl Genet 113:73–80
Weiland JJ, Yu MH (2003) A cleaved amplified polymorphic sequence (CAPs) marker associated with root-knot nematode resistance in sugarbeet. Crop Sci 43:1814–1818
Xiao J, Grandillo S, Ahn S, Yuan L, Tanksley SD, McCouch SR (1996) Genes from wild rice improve yield. Nature 384:223–224
Zeng ZB (1994) Precision mapping of quantitative trait loci. Genetics 136:1457–1468
Zhang J, Guo WZ, Zhang TZ (2002) Molecular linkage map of allotetraploid cotton (Gossypium hirsutum L. × Gossypium barbadense L.) with a haploid population. Theor Appl Genet 105:1166–1174
Zhou E, Star JL, Smith CW (1999) Inheritance of resistance to Meloidogyne incognita in the cotton cultivar Acala NemX. J Nematol 31:584–585
Acknowledgments
We thank the Paterson lab for sharing the RFLP probes, the Zhang lab for SSR primer sequences and acknowledge the financial support from the Georgia Cotton Commission and the Cotton Incorporated.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by B. Friebe.
Rights and permissions
About this article
Cite this article
Shen, X., Van Becelaere, G., Kumar, P. et al. QTL mapping for resistance to root-knot nematodes in the M-120 RNR Upland cotton line (Gossypium hirsutum L.) of the Auburn 623 RNR source. Theor Appl Genet 113, 1539–1549 (2006). https://doi.org/10.1007/s00122-006-0401-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00122-006-0401-4