Abstract
The restoration of the C-type cytoplasmic male sterility (Cms) has been a common agriculture practice in the production of hybrid seed for many years. In this study, a series of crosses between select sterile and restorer lines, as well as a backcross population of (Cms-C77 × 6233) × 6233, were used to investigate the restoration of C-type Cms. Our results demonstrated that there was an inhibitor of the Rf5 restorer gene. This inhibitor gene, Rf-I, maps to chromosome 7 and is tightly linked with SSR markers, umc2326 and umc2327, at a genetic distance 4.7 and 3.4 cM, respectively. After analyzing our data combined with previous studies, we propose that the restoration of C-type Cms has two dominant genes, Rf4 and Rf5. Rf4 has the ability to restore all genotypes of Cms-C lines; however, there exists an inhibitor for the other restorer gene, Rf5; thus, it can restore only those genotypes of Cms-C lines lacking the Rf-I inhibitor.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Cytoplasmic male sterility (Cms) has important economic and logistic advantages in maize for the production of hybrid seed. More than 40 sources of Cms have been found and classified into three major groups; these are designated the Cms-T, Cms-S, and Cms-C groups (Beckett 1971). Compared to T- and S-Cms, the fertility restoration of Cms-C has been found to be very complex in previous analyses. Out of them, Duvick (1972) reported that restoration of Cms-C fertility was controlled by a dominant gene, Rf4. Khey-Pour et al. (1981) also found this gene to be sufficient for Cms-C restoration. However, Josephson and Morgan (1978) proposed that fertility restoration in Cms-C was conditioned by the complementary action of the dominant allele of two genes, Rf4 and Rf5, which have since been mapped to chromosomes 8 and 5, respectively (Sisco 1991; Tang et al. 2001a, b). Meanwhile, Chen et al. (1979) considered that two dominant restorer genes in Cms-C had duplicate action. Further complicating the system, Vidakovic (1988) demonstrated the existence of three dominant and complementary genes for full restoration of fertility in Cms-C, adding the gene, Rf6. Vidakovic et al. (1997a, b) later reported these complementary genes, Rf4, Rf5, and Rf6, were indeed not the sole genetic system for fertility restoration in Cms-C of maize. Thus, the fertility restoration mechanisms of Cms-C remain unresolved, and, in practice, it is difficult to select restorer lines for some genotypic sterile lines. The objective of this study was to resolve the mechanism of Cms-C restoration and map the gene of an inhibitor of the restorer gene, Rf5, with SSR markers.
Materials and methods
Plant materials and phenotype evaluation
The inbred line 6233 with C-type male cytoplasm was selected from restorer individuals of a F2 population derived from a cross between Cms-C Mo17 and Fengke1, and self-pollinated to F7, which the restorer line, Fengke1, had two dominant duplicate restorer genes, Rf4 and Rf5 (Chen et al. 1979; Tang et al. 2001b). The inbred line 6233 was tested for the restorer gene Rf5 using tightly linked molecular markers and an allelism test. The fertility of some F1 progeny derived from crosses between the sterile lines Cms-CMo17, Cms-C77, Cms-Es87-1, Cms-Es478, Cms-C237, and Cms-CZong3 and the restorer lines 6233, A619, Guang10-2, Dan958, and Fengke1, as well as from a backcross (Cms-C77 × 6233) × 6233 were evaluated in Zhengzhou and Sayan, China.
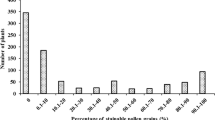
In the field, the crosses between select sterile lines and restore lines were sown in one or three plots respectively; each plot was 4 m in length and contained 15 plants, seeds of the BC1 populations were sown using the single-seed planting method in nine plots. The percentage restoration was evaluated by monitoring anthesis of each plant daily. The criterion of fertility was then rated on a scale of 0–5 based on the percentage of exserted anthers: (0) no anthers exserted; (1) > 0–5% anthers exserted; (2) 5–25% anthers exserted; (3) 25–50% anthers exserted; (4) 51–75% anthers exserted; (5) more than 75% anthers exserted. At the same time, the dehiscence of anthers was graded on a scale of I to III: (I) no dehiscent anthers; (II) few dehiscent anthers; (III) many normal, dehiscent anthers. To identify the fertility of each plant, a combined score of 4/III or 5/III was regarded as restored, while combined scores of 0/I and 1/I were recorded as sterile (Duvick et al. 1961; Li et al. 1963).
Linkage analysis
Genomic DNA was extracted from young leaves using the cetyltrimethylammonium bromide (CTAB) method (Murrsay and Thompson 1980). Bulked segregant analysis (BSA) was used to detect the linkage of the Rf-I gene. Fertile bulk and sterile bulk were made by mixing equal amounts of DNA from ten fertile plants and ten sterile plants, respectively (Michelmore et al. 1991).
In accordance with their bin location, a total of 521 SSR markers were chosen from the maize genome database (http://www.maizegdb.org) to detect polymorphism of the two parents and bulks. The products amplified using SSR primers were separated by electrophoresis on 6% polyacrylamide gels and visualized by silver-staining. Based on the SSR data and the phenotype of the segregating individuals, linkage analysis was performed with JOINMAP version 3.0 (Stam 1993).
Results
Identification the restorer gene Rf5 in inbred line 6233
The SSR markers, bnlg1711 and bnlg1346, which are tightly linked with the Rf5 gene, were used to identify the genotype of the restorer lines 6233 (Tang et al. 2001b), A619 (Rf4Rf4rf5rf5; Sisco 1991), Fengke1 (Rf4Rf4Rf5Rf5; Chen et al. 1979; Tang et al. 2001b), Guang10-2 (Rf4Rf4rf5rf5; Tang et al. 2001b), and Dan958, as well as the sterile lines, Cms-C Mo17 and Cms-ES 87-1, which were identified simultaneously as a control. The results from PCR amplification demonstrated that the inbred line, 6233, had one restorer gene, Rf5 (The figures are not shown).
For further genotypic identification of the inbred line 6233, the F2 population, including 213 derived from the cross, Cms-C 6233 × A619, were evaluated in the summer of 2004. There were 189 fertile plants and 16 sterile plants in this population, and the segregating ratio of male fertile to male sterile plants fell to the theoretical ratio 15:1 (χ2 = 0.85). This result also indicated that the inbred line 6233 had a single restorer gene, Rf5.
Identification the Rf5 restorer inhibitor gene, Rf-I
The F1 progeny of crosses between five sterile lines, and the restorer line 6233 were evaluated in the summer of 2003 in Zhengzhou (Table 1). The F1 progeny of the sterile lines Cms-CMo17, Cms-Es87-1, Cms-Es 478, Cms-C237, and Cms-CZong3 and the restorer line 6233 presented full restoration; however, the F1 progeny of Cms-C77 and 6233 were sterile. They also exhibited sterility in the winter of 2003 and 2004 in Sanya, as well as in the summer of 2004 at Zhengzhou (Table 2). It was very interesting that only the F1 progeny between Cms-C77 and 6233 presented sterility.
Samples from the backcross population of (Cms-C77 × 6233) × 6233 were also evaluated, finding 59 sterile plants and 56 fertile plants out of 123 individuals, a ratio of restored to sterile plants following the theoretical ratio 1:1 by the chi-squared test (Table 2). These results demonstrate that the sterile line, Cms-C77, has a dominant inhibitor gene, which is named Rf-I that can inhibit expression of the restorer gene, Rf5. At the same time, the younger leaves of this backcross population were harvested for DNA extraction and linkage analysis.
To further analyze the genetic effect of this inhibitor gene, a set of F1 progeny from the sterile line Cms-C77, crossed to the restorer lines A619, Guang10-2, Fengke1, Dan958, and 6233, were evaluated in the summer of 2004 in Zhengzhou (Table 1). All of the F1 progeny, except those from Cms-C77 × 6233, exhibited restoration of fertility. This indicated that the inhibitor gene, Rf-I, has no effect on the Rf4 restorer gene, in contrast to the effect on Rf5.
Mapping the dominant inhibitor gene, Rf-I
Out of the 521 SSR markers used in this study, the primers umc1978, bnlg1904, umc2326, umc2327, umc1666, bnlg2203, and umc2325 amplified polymorphisms between the two parents and two bulks; all of these primers reside on the chromosomal bin 7.02. To determine the chromosomal location of the Rf-I gene, DNA from 115 individuals of the backcross population of (Cms-C77 × 6233) × 6233 was amplified with these SSR primers. The patterns of amplification for some individuals using the SSR marker, umc2327, are shown in Fig. 1 as examples. The linkage map of the SSR markers and Rf-I gene were calculated with JOINMAP, and showed in Fig. 2, the genetic distances between the Rf-I and the SSR primers umc2326 as well as umc2327 were 4.7 and 3.4 cM, respectively.
Discussion
Vidakovic et al. (1997b) reported that C-group Cms had perhaps two different restoration mechanisms, one restoration system was the three dominant complementary genes (Rf4, Rf5, Rf6), and the second one was a duplication or a parallel and independent genetic device for fertility restoration in Cms-C. In this study, our results revealed that the restorer gene, Rf5, has a corresponding inhibitor gene, Rf-I, but this inhibitor has no effect on the Rf4 restorer gene. Our results are similar to those of Qin et al. (1990), who used C-type and Y-type Cms to investigate the fertility restoration of two Cms, they demonstrated that the Y-type Cms had an inhibitor gene called Rf7 that most likely inhibits expression of the Rf5 gene. Moreover, the restorer genes, Rf4 and Rf5, were dominant to both C-group and Y-type Cms, and there is little difference between these two types of Cms (Qin et al. 2001). It is possible the inhibitor genes Rf-I and Rf7 are indeed the same gene, but this remains to be seen. Combining the previous studies, we propose that the restoration of C-Cms has two dominant genes (Chen et al. 1979; Sisco 1991; Tang et al. 2001b), one is the major restorer gene, Rf4 (Duvick 1972; Khey-Pour et al. 1981; Sisco 1991), which has restorer activity over all types of C-Cms; the other restorer gene, Rf5, has an inhibitor, so it can restore only some genotypes of C-Cms—those without the inhibitor gene, Rf-I.
In the utilization of C-Cms, many maize breeders have faced the difficulty that some C-group sterile lines are not restored. The reason for this problem is perhaps that the sterile line harbors the inhibitor gene Rf-I, and the restorer line being used has the Rf5 gene. To overcome this problem, restorer lines with Rf4, such as A619 (Sisco 1991; Tang et al. 2001a) and Guang10-2 (Tang et al. 2001b), should be used as the donor for selecting new restorer lines, because the Rf4 restorer is effective in any genotype of C-Cms.
References
Beckett JB (1971) Classification of male sterile cytoplasms in maize (Zea mays L.). Crop Sci 11:724–726
Chen WC, Luo FH, Ji LY (1979) Some genetic aspects of the C-type cytoplasmic male-sterility in maize and its use in breeding. Acta Agronom Sin 5(4):21–28
Duvick DN (1972) Potential usefulness of new cytoplasmic male sterile and sterility system. In: Proceedings of the 27th annual corn and sorghum research conference, pp 197–201
Duvick DN, Snyder RJ, Anderson EC (1961) The chromosomal location of Rf1, a restorer gene for cytoplasmic pollen sterile maize. Genetics 46:1245–1252
Josephson LM, Morgan TE (1978) Genetics and inheritance of fertility restoration of male sterile cytoplasms in corn. In: Proceedings of the 33rd corn and sorghum research conference 7:13
Khey-Pour A, Grace VE, Everett HL (1981) Genetics of fertility restorations in the C-group of cytoplasmic male sterility in maize. Genetics 98:379–388
Li JX, Dai JR, Xu QF (1963) Study on the inheritance of cytoplasmic male-sterile and fertility restoration in maize. Acta Agronom Sin 2:339–362
Michelmore RW, Paran I, Kesseli V (1991) Identification of markers linked to disease resistance genes by bulked segregant analysis: a rapid method to detect markers in specific genomic region is by using segregating population. Pro Natl Aca Sci USA 88:9828–9832
Murray MG, Thompson WF (1980) Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res 8:4321–4325
Qin TC, Xu ML, Deng DX (1990) Cytoplasmic male-sterility: identification of the number of restorer genes. Maize Genet Coop News Lett 64:124
Qin TC, Xu ML, Deng DX, Chen RL, Bian YL (2001) Study on male-sterility in Zea mays VIII. RFLP analysis of mtDNA of YII-1 type maize male-sterile cytoplasm. Acta Agronom Sin 27:185–189
Sisco PH (1991) Duplication complicates genetic mapping of Rf4 a restoring gene for cms-C cytoplasmic male sterility in corn. Crop Sci 31:1263–1266
Stam P (1993) Construction of integrated genetic linkage maps by means of a new computer package: JoinMap. Plant J 3:739–744
Tang JH, Ji HQ, Huang ZW, Ji LY, Wang CH (2001a) Mapping of the main restorer gene (Rf4) of C-cytoplasmic male sterility in maize with RFLP. J Henan Agric Univ 35:99–102
Tang JH, Liu ZH, Chen WC, Hu YM, Ji LY (2001b) Mapping the main restorer genes of C-cytoplasmic male sterility using SSR method. Acta Agronom Sin 34:592–596
Vidakovic M (1988) Genetics of fertility restoration in cytoplasmic male sterility of the C-type (Cms-c) in maize. Maydica 33:51–64
Vidakovic M, Vancetovic J, Vidakovic M (1997a) Complementary genes Rf4, Rf5 and Rf6 are not the unique genetic system for fertility restoration in cmsC of maize (Zea mays L.). Maize Genet Coop News Lett 71:10
Vidakovic M, Vancetovic J, Vidakovic M (1997b) The existence of a duplicated or parallel genetic system for fertility restoration in cmsC of maize (Zea mays L.). Maydica 42:313–316
Acknowledgements
This work was supported by the National High-technology Project of China and National Natural Science Foundation of China.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by R. Hagemann
Rights and permissions
About this article
Cite this article
Hu, Y.M., Tang, J.H., Yang, H. et al. Identification and mapping of Rf-I an inhibitor of the Rf5 restorer gene for Cms-C in maize (Zea mays L.). Theor Appl Genet 113, 357–360 (2006). https://doi.org/10.1007/s00122-006-0302-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00122-006-0302-6