Abstract
Cytoplasmic male sterility (CMS) of rice (Oryza sativa L.) was first reported using the cytoplasm of a Chinese wild rice, Oryza rufipogon Griff. strain W1. However, it was not possible to characterize this ms-CW-type CMS in more detail until a restorer line had been developed due to the lack of restorer genes among cultivars thus far tested. The breeding of a restorer line (W1-R) was eventually achieved by transferring the restorer gene(s) of W1 to a cultivar. We report here the characterization of the ms-CW pollen grains and mapping of the restorer gene for ms-CW-type CMS. Pollen grains of the male-sterile plants appeared to be normal and viable based on the fluorochromatic reaction test, but they did not germinate on normal stigmas. The 1:1 segregation of fertile and sterile plants in a BC1F1 population from a cross between W1-R and a maintainer line demonstrated that fertility restoration is controlled by a single gene. The fertile seed set of all the F2 plants examined indicated that the fertility restoration functions gametophytically. We designated the fertility restorer gene Rfcw. Using cleaved amplified polymorphic sequence (CAPS) and simple sequence repeat (SSR) markers, we localized Rfcw to chromosome 4 with a genetic distance of 0.6 cM from the nearest SSR marker.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Cytoplasmic male sterility (CMS) is a maternally inherited trait that results in the inability to produce fertile pollen and is a common occurrence in higher plants. A CMS cytoplasm of rice was first reported by Katsuo and Mizushima (1958) in the W1 strain of a Chinese wild rice species [Oryza rufipogon Griff. (= O. sativa L. f. spontanea in the original literature)]. An ms-CW-type CMS rice was obtained from backcrossing the Japonica cultivar Fujisaka 5 (Oryza sativa L.) to strain W1. The gene symbol of the cytoplasm was designated [ ms-CW], namely, cytoplasm from Chinese wild rice (Kinoshita 1984). Since then, various types of CMS, such as the ms-bo-type originating from Chinsurah boro II (Shinjyo 1969), ms-ld-type from Lead rice (Watanabe 1971), ms-WA-type from a wild abortive line (Lin and Yuan 1980) and ms-HL-type from red awned wild rice (Rao 1988), have been identified in rice (for a review, see Kinoshita 1984).
In some CMS lines, pollen fertility can be recovered by a nuclear-encoded gene known as a fertility restorer gene (Rf). The most studied CMS/Rf system in rice is the ms-bo-type cytoplasm and a fertility restorer gene, Rf1. Rf1 has been shown to be located on chromosome 10 (Shinjyo 1975), and molecular cloning of Rf1 has been recently achieved (Kazama and Toriyama 2003; Akagi et al. 2004; Komori et al. 2004). Rf2 for ms-ld-type CMS has been shown to reside on chromosome 2 (Shinjyo and Sato 1994), while Rf3 and Rf4 for ms-WA-type CMS have been mapped on chromosomes 1 and 10, respectively (Yao et al. 1997; Zhang et al. 1997). Two fertility restorer genes, Rf5 and Rf6(t) for ms-HL-type CMS, have been shown to be located on chromosome 10 (Liu et al. 2004) as has the Rf-D1(t) restorer gene for ms-D1-type CMS (Tan et al. 2004). These fertility restorer genes, except for the fertility restorer genes for ms-WA-type CMS, are known to function gametophytically with respect to fertility restoration. On the contrary, the fertility restorer genes Rf3 and Rf4 for ms-WA-type CMS function in a sporophytic manner (for review, see Kinoshita 1984).
In spite of the progress in elucidating these CMS/Rf systems, characterization and mapping of a restorer gene(s) for ms-CW-type CMS has not yet been carried out. This is due to the lack of any restorer genes among cultivars thus far tested (Ishimine and Shinjyo 1978). Breeding of a restorer line (W1-R) has been achieved by transferring the restorer genes of W1 to an ms-CW-type CMS line through the process of anther culture (Toriyama and Hinata 1987).
We report here the characterization of pollen grains of an ms-CW-type CMS line and molecular mapping of the restorer gene. Pollen viability and pollen-tube germination were observed using the fluorochromatic reaction (FCR) test and aniline blue staining, respectively. The fertility restorer gene, which was designated Rfcw, was mapped on rice chromosomes using BC1F1 and the F2 populations derived from the cross between the restorer line W1-R and a maintainer line.
Materials and methods
Plant materials
An ms-CW-type CMS line (W1-A) was obtained from a backcross between a Chinese wild rice, strain W1 (Oryza rufipogon Griff.), and a Japonica cultivar Reimei (O. sativa L.). The Japonica cultivar Taichung 65 was used as the maintainer line. A fertility restorer line (W1-R) was derived from strain W1 through a process of anther culture and backcrossing (Toriyama and Hinata 1987). BC1F1 and F2 populations used for genetic analysis and linkage analysis were derived from a cross between W1-R (female) and Taichung 65 (male). Cytoplasmic background and seed set traits of the plant materials used in this study are listed in Table 1.
Observations of pollen viability and pollen germination
Mature anthers of W1-R and W1-A were harvested and the pollen grains stained with a 1% iodine-potassium iodide solution for the observation of starch accumulation (Toriyama and Hinata 1987). Stained pollen grains were observed under an optical microscope. For the FCR test, mature pollen was stained with 2 mg/ml fluorescein diacetate solution and then observed under a fluorescent microscope (Heslop-Harrison et al. 1984). For the pollen-tube germination test, stigmas of W1-R and W1-A were fixed in 3:1 (v/v) ethanol/acetic acid solution 6 h after flowering and incubated with 1 N KOH solution at 50°C for 60 min. The stigmas were subsequently incubated with 0.1% aniline blue solution at 50°C for 60 min and observed under a fluorescent microscope (Sato et al. 2004).
Genetic inheritance analysis of the fertility restorer gene(s)
Ninety-six plants of the BC1F1 generation and 96 F2 plants were planted in a paddy field at Tohoku University in Sendai, Japan in 2003. Individuals were categorized into two groups, completely sterile and fertile.
Genetic linkage analysis
Genomic DNA was extracted from the fresh leaves of 96 BC1F1 plants and 608 F2 plants with extraction buffer (200 mM Tris-HCl, pH 7.5, 250 mM NaCl, 25 mM EDTA, 0.5% sodium dodecyl sulfate) and then centrifuged at 15,000 rpm for 5 min. The supernatant was collected, and a 1.0 volume of isopropanol was added. Following a centrifugation at 15,000 rpm for 5 min, the pellet was dissolved in 1× TE. Genotypes were determined using 24 simple sequence repeat (SSR) markers (RM1306, RM1350, RM1359, RM1388, RM1817, RM1973, RM2197, RM2634, RM3042, RM3120, RM3335, RM3625, RM3866, RM4108, RM4355, RM4691, RM4771, RM5314, RM5352, RM5631, RM6704, RM7049, RM8068, and RM8144; McCouch et al. 2002), 11 cleaved amplified polymorphic sequence (CAPS) markers (C213, C50, C727A, C777, R1427, R1862, R2171, R2232, R2349, S10091, S1461-2; http://rgp.dna.affrc.go.jp/publicdata/caps/index.html) and one sequence-characterized amplified region (SCAR) marker (KT04) developed by Shirasawa et al. (2004). The PCR profile for SSR markers was performed as described by McCouch et al. (2002); that for CAPS markers was performed as described at http://rgp.dna.affrc.go.jp/publicdata/caps/index.html. The amplification products were electrophoresed on a 2.0% (w/v) agarose gel in 1× TAE buffer and, if necessary, the amplification products were separated by electrophoresis on 8% denaturing polyacrylamide gel. The PCR product was stained with ethidium bromide and visualized by UV illumination. An 8% denaturing polyacrylamide gel was visualized by applying the silver staining procedure. Recombination frequencies and genetic distances were determined with mapmaker/exp ver. 3.0 (Lincoln et al. 1992).
Additional SSR markers
Four additional markers were designed on chromosome 4 to determine the genotypes of 608 F2 plants. PCR primers flanking these SSRs were as follows:
AT15-1, 5′-GTGTGGCAAGTGTTGCTTCA-3′ and 5′-AGGTCATGTCTCCTCTCATC-3′; AT14-1, 5′-AGTATTCTCTGTCTGGTGGC-3′ and 5′-TATGGAAGCCAGTAGCGACA-3′; AT12-1, 5′-GAAGCCTGATAGGTCGATGT-3′ and 5′-TAGCCAGCACTGCAATTTGC-3′; AT10.5-1; 5′-AGATAGCATCCCCGTTAGCT-3′ and 5′-TTCCAGTCTAGTCCACCATC-3′.
Results and discussion
Comparison of pollen viability and pollen germination between W1-R and W1-A
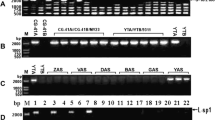
Pollen grain morphology and pollen germination of the restorer line W1-R and the CMS line W1-A were compared. No morphological differences were visible in the mature pollen grains under light microscopy (Fig. 1a, b). Staining with a I2-KI solution indicated that the pollen grains of both W1-A and W1-R had normal starch accumulation (Fig. 1c, d). Pollen viability was assessed using the FCR test. Mature pollen grains of W1-A and W1-R showed FCR positive fluorescence (Fig. 1e, f), but did not show such fluorescence following fixation (Fig. 1g, h), indicating that pollen grains of both lines have normal plasma membrane permeability and esterase activity. Aniline blue staining of a stigma 6 h after flowering revealed pollen-tube germination in W1-R but no germination in W1-A (Fig. 1g, h), indicating that the pollen of W1-A does not possess the ability to germinate on a stigma. Pollen grains of a maintainer line, Taichung 65, showed staining and FCR identical to those of W1-R (data not shown).
Pollen grains of ms-CW-type CMS rice (W1-A, left panels) are indistinguishable from those of a restorer line (W1-R, right panels), but the former do not germinate on stigmas. a, b Mature pollen grains, c, d pollen grains stained with I2-KI solution, e, f FCR test of pollen grains—the green fluorescence indicates FCR-positive signals, g, h FCR test following fixation of the pollen grains, showing negative FCR, I, j aniline blue staining of pollen tubes on stigma 6 h after flowering. Bars: (a–f) 10 μm; (I–j) 100 μm
Characterization of an ms-CW-type CMS strain has been limited to a few reports in the Japanese literature. Sasahara and Katsuo (1965) reported that they were unable to observe localization of starch grains to the polar site of the germ pore of ms-CW-type pollen and that these pollen grains did not germinate on artificial culture media. The morphologically normal appearance of ms-CW-type pollen grains is quite different from the features of pollen grains of other CMS in rice. Pollen abortion is evident at the late bicellular pollen stage in ms-bo-type CMS (Shinjyo 1969) and at the early bicellular pollen stage in ms-HL-type CMS (Rao 1988). Pollen grains of both types of CMS were not stained with I2-KI solution. An ms-WA -type CMS plant produces shrunken pollen grains which are almost empty (Lin and Yuan 1980). Pollen abortion of ms-CW-type CMS is likely to start at the latest developmental stage among the CMS rice lines characterized to date.
Genetic inheritance of Rfcw
Using 96 plants of the BC1F1 population of (W1-R × Taichung 65) × Taichung 65, we analyzed the genetic inheritance of the fertility restoration to determine whether it is controlled by a single locus or by multiple loci. The appearance of 42 fertile plants and 54 sterile plants fit a 1:1 segregation (χ2=1.50, 0.25>P>0.01), indicating the Mendelian inheritance of a single dominant gene. The F1 plant (W1-R × Taichung 65) was considered to segregate pollen with a restorer gene and pollen without a restorer gene at a ratio of 1:1. We also used 96 F2 plants to determine whether the fertility restoration behaves in a gametophytic manner or a sporophytic manner. If the fertility restoration behaves in a sporophytic manner, the pollen fertility of the F1 plant is determined by the genotype of the sporophyte, meaning that all the pollen—with or without a restorer gene—of the F1 plant is fertile and participates in fertilization. Thus, 25% of the F2 plants would be sterile. If fertility restoration behaves in a gametophytic manner, the genotype of the individual pollen grain determines its own fertility. Therefore, pollen without a restorer gene in the F1 plant is unable to germinate on a stigma, and all the F2 plants would be fertile. In our analysis, all of the F2 plants were found to show a fertile seed-set phenotype, indicating that the fertility restoration is controlled in a gametophytic manner. The F2 population was considered to contain two groups of plants, one carrying the restorer gene homozygously and the other carrying it heterozygously. We were unable, however, to distinguish between the two groups because individual pollen grains with a restorer gene and those without one were indistinguishable based on I2-KI staining and the FCR test as shown in Fig. 1. In conclusion, the fertility restoration of ms-CW-type CMS is gametophytically controlled by a single gene. We designated this gene Rfcw.
Genetic linkage analysis of Rfcw
Using 24 SSR, 11 CAPS and one SCAR marker in 96 BC1F1 plants, we carried out a genetic linkage analysis of Rfcw. Five SSR markers (RM1359, RM3042, RM3866, RM1388, RM3335) and one CAPS marker (R1427) on chromosome 4 showed a linkage to Rfcw (Fig. 2). Rfcw was flanked by RM3866 and RM1388. The map positions of the SSR markers on the restriction fragment length polymorphism (RFLP) linkage map are 70.9 cM and 77.9 cM, respectively. To further specify the location of Rfcw, we designed four additional SSR markers, AT10.5-1, AT12-1, AT14-1 and AT15-1, on the region between RM3866 and RM1388 (Fig. 2) and screened for recombinant individuals. Of the 608 F2 plants, six recombinant plants were identified using RM3866, and two were identified using AT10.5-1. The genetic distance between RM3866 and Rfcw was calculated to be 2.0 cM, and that between Rfcw and AT10.5-1 to be 0.6 cM (Fig. 2). Various fertility restorer genes have been identified and mapped on rice chromosomes (Kinoshita 1984). However, Rfcw is the first fertility restorer gene to be identified on chromosome 4.
Rfcw is mapped between two SSR markers on chromosome 4. Linkage distances between each marker are calculated based on the analysis of 96 BC1F1 plants and shown to the left of the chromosome. The map position of Rfcw between RM3866 and AT10.5-1 is based on the analysis of 608 F2 plants and is shown to the right of the chromosome. The arrowhead indicates the centromere
The fertility restorer gene for ms-bo-type CMS, Rf1, has recently been cloned (Kazama and Toriyama 2003; Akagi et al. 2004; Komori et al. 2004). Rf1, like the fertility restorer genes of petunia and radish (Bentolila et al. 2002; Brown et al. 2003; Koizuka et al. 2003), encodes a pentatricopeptide repeat (PPR) containing protein. The PPR motif is characterized by the presence of tandem arrays of a degenerate 35-amino-acid repeat (Small and Peeters 2000; Lurin et al. 2004). A member of the PPR gene family is a possible candidate for Rfcw. It is also possible that Rfcw does not encode a PPR protein and that the restoration mechanism is distinct, as in the case of the maize fertility restorer rf2, which encodes aldehyde dehydrogenase (Cui et al. 1996). Based on the genomic sequence of Nipponbare, it is predicted that the physical distance between two SSR markers, RM3866 and AT10.5-1, is approximately 900 kb, although Nipponbare does not have an Rfcw gene. We are currently working on fine-mapping and genomic walking of the corresponding region of W1-R in order to carry out molecular cloning of Rfcw.
References
Akagi H, Nakamura A, Yokozeki-Misono Y, Inagaki A, Takahashi H, Mori K, Fujimura T (2004) Positional cloning of the rice Rf-1 gene, a restorer of BT-type cytoplasmic male sterility that encodes a mitochondria-targeting PPR protein. Theor Appl Genet 108:1449–1457
Bentolila S, Alfonso AA, Hanson MR (2002) A pentatricopeptide repeat-containing gene restores fertility to cytoplasmic male-sterile plants. Proc Natl Acad Sci USA 99:10887–10892
Brown G, Formanova N, Jin H, Wargachuk R, Dendy C, Patil P, Laforest M, Cheung WY, Landry BS (2003) The radish Rfo restorer gene of Ogura cytoplasmic male sterility encodes a protein with multiple pentatricopeptide repeats. Plant J 35:262–272
Cui X, Wise RP, Schnable PS (1996) The rf2 nuclear restorer gene of male-sterile T-cytoplasm maize. Science 272:1334–1336
Heslop-Harrison J, Heslop-Harrison Y, Shivanna KR (1984) The evaluation of pollen quality, and a further appraisal of the fluorochromatic (FCR) test procedure. Theor Appl Genet 67:367–375
Ishimine Y, Shinjyo C (1978) Distribution of fertility restoring genes in commercial lowland-rice varieties cultivated in Japan for two male sterile cytoplasms of rice. Bull Coll Agr Univ Ryukyus 25:665–672
Katsuo K, Mizushima U (1958) Studies on the cytoplasmic difference among rice varieties, Oryza sativa L. 1. On the fertility of hybrids obtained reciprocally between cultivated and wild varieties. Jpn J Breed 8:1–5
Kazama T, Toriyama K (2003) A pentatricopeptide repeat-containing gene that promotes the processing of aberrant atp6 RNA of cytoplasmic male-sterile rice. FEBS Lett 544:99–102
Kinoshita T (1984) Gene analysis and linkage map. In: Tsunoda S, Takahashi N (eds) Biology of rice. Japan Sci Soc Press, Tokyo, pp 184–274
Koizuka N, Imai R, Fujimoto H, Hayakawa T, Kimura Y, Kohno-Murase J, Sakai T, Kawasaki S, Imamura J (2003) Genetic characterization of pentatricopeptide repeat protein gene, orf687, that restores fertility in the cytoplasmic male-sterile Kosena radish. Plant J 34:407–415
Komori T, Yamamoto T, Takemori N, Kashihara M, Matsushima H, Nitta N (2004) Map-based cloning of a fertility restorer gene, Rf-1, in rice (Oryza sativa L.). Plant J 37:315–325
Lin SC, Yuan LP (1980) Hybrid rice breeding in China. In: IRRI (eds) Innovative approaches to rice breeding. IRRI, Manila, Philippines, pp 35–51
Lincoln S, Daly M, Lander E (1992) Constructing genetic maps with mapmaker/exp. 3.0. Whitehead Institute Technical Report 3rd edn. Whitehead Institute for Biochemical Research, MA
Liu X-Q, Xu X, Tan Y-P, Li S-Q, Hu J, Huang J-Y, Yang D-C, Li Y-S, Zhu Y-G (2004) Inheritance and molecular mapping of two fertility-restoring loci for Honglian gametophytic cytoplasmic male sterility in rice (Oryza sativa L.). Mol Genet Genomics 276:586–594
Lurin C, Andres C, Aubourg S, Bellaoui M, Bitton F, Bruyere C, Caboche M, Debast C, Gualberto J, Hoffman B, Lecharny A, Le Ret M, Martin-Magniette M-L, Mireau H, Peeters N, Renou J-P, Szurek B, Taconnat L, Small I (2004) Genome-wide analysis of Arabidopsis pentatricopeptide repeat proteins reveals their essential role in organelle biogenesis. Plant Cell 16:2089–2103
McCouch SR, Teytelman L, Xu Y, Lobos KB, Clare K, Walton M, Fu B, Maghirang R, Li Z, Xing Y, Zhang Q, Kono I, Yano M, Fjellstrom R, Declerck G, Schneider D, Cartinhour S, Ware D, Stein L (2002) Development and mapping of 2240 new SSR markers for rice (Oryza sativa L.). DNA Res 9:199–207
Rao YS (1988) Cytohistology of cytoplasmic male sterile lines in hybrid rice. In: Smith WH, Bostian LR, Cervantes (eds) Hybrid rice. IRRI, Manila, Philippines, pp 115–128
Sasahara T, Katsuo K (1965) Studies on the cytoplasmic difference among rice varieties, Oryza sativa L. III. On the abortive pollen of Fujisaka No. 5-type plants with the cytoplasm of Chinese wild variety, Oryza sativa f. spontanea. (with English summary, p 196) Jpn J Breed 15:43–48
Sato Y, Okamoto S, Nishio T (2004) Diversification and alteration of recognition specificity of the pollen ligand SP11/SCR in self-incompatibility of Brassica and Raphanus. Plant Cell 16:3230–3241
Shinjyo C (1969) Cytoplasmic-genetic male sterility in cultivated rice, Oryza sativa L. II. The inheritance of male sterility. Jpn J Genet 44:149–156
Shinjyo C (1975) Genetical studies of cytoplasmic male sterility and fertility restoration in rice, Oryza sativa L. Sci Bull Coll Agr Univ Ryukyus 22:1–57
Shinjyo C, Sato S (1994) Chromosomal location of fertility-restoring gene Rf2. Rice Genet Newsl 11:93–95
Shirasawa K, Kishitani S, Nishio T (2004) Conversion of AFLP markers to sequence-specific markers for closely related lines in rice by use of the rice genome sequence. Mol Breed 14:283–292
Small ID, Peeters N (2000) The PPR motif—a TPR-related motif prevalent in plant organellar proteins. Trends Biochem Sci 25:46–47
Tan XL, Tan YL, Zhao TH, Zhang XM, Hong RK, Jin SL, Liu XR, Huang DJ (2004) Identification of the Rf gene conferring fertility restoration of the CMS Dian-type 1 in rice by using simple sequence repeat markers and advanced inbred lines of restorer and maintainer. Plant Breed 123:338–341
Toriyama K, Hinata K (1987) Anther culture application to breeding of a restorer for a male-sterile cytoplasm of wild rice (ms-CW). Jpn J Breed 37:469–473
Watanabe Y (1971) Establishment of cytoplasmic and genetic male-sterile lines by means of Indica-Japonica cross. Oryza Cuttack 8[Suppl 2]:9–16
Yao FY, Xu CG, Yu SB, Li JX, Gao YJ, Li XH, Zhang Q (1997) Mapping and genetic analysis of two fertility restorer loci in wild-abortive cytoplasmic male sterility system of rice. Euphytica 98:183–187
Zhang G, Bharaji TS, Lu Y, Virmani SS, Huang N (1997) Mapping of the Rf-3 nuclear fertility-restoring gene for WA cytoplasmic male sterility in rice using RAPD and RFLP markers. Theor Appl Genet 94:27–33
Acknowledgements
This study was partially supported by a grant for “Functional analysis of genes relevant to agriculturally important traits in rice genome” from the Ministry of Agriculture, Forestry and Fisheries, Japan, and by a Grant-in-Aid from the Ministry of Education, Science and Culture, Japan.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by T. Sasaki
Rights and permissions
About this article
Cite this article
Fujii, S., Toriyama, K. Molecular mapping of the fertility restorer gene for ms-CW-type cytoplasmic male sterility of rice. Theor Appl Genet 111, 696–701 (2005). https://doi.org/10.1007/s00122-005-2054-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00122-005-2054-0