Abstract
Gray leaf spot (GLS) is a serious fungal disease caused by Magnaporthe grisea, recently reported on perennial ryegrass (Lolium perenne L.), an important turfgrass and forage species. This fungus also causes rice blast and many other grass diseases. Rice blast is usually controlled by host resistance, but durability of resistance is a problem. Little GLS resistance has been reported in perennial ryegrass. However, greenhouse inoculations in our lab using one ryegrass isolate and one rice-infecting lab strain suggest presence of partial resistance. A high density linkage map of a three generation Italian × perennial ryegrass mapping population was used to identify quantitative trait loci (QTL) for GLS resistance. Potential QTL of varying effect were detected on four linkage groups, and resistance to the ryegrass isolate and the lab strain appeared to be controlled by different QTL. Of three potential QTL detected using the ryegrass isolate, the one with strongest effect for resistance was located on linkage group 3 of the MFB parent, explaining between 20% and 37% of the phenotypic variance depending on experiment. Another QTL was detected on linkage group 6 of the MFA parent, explaining between 5% and 10% of the phenotypic variance. The two QTL with strongest effect for resistance to the lab strain were located on linkage groups MFA 2 and MFB 4, each explaining about 10% of the phenotypic variance. Further, the QTL on linkage groups 3 and 4 appear syntenic to blast resistance loci in rice. This work will likely benefit users and growers of perennial ryegrass, by setting the stage for improvement of GLS resistance in perennial ryegrass through marker-assisted selection.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Perennial ryegrass (Lolium perenne L.) is a valuable cool-season turf and forage grass, extensively used on golf courses, athletic fields, and lawns. It is a diploid (2n=2x=14), outcrossing, self-incompatible species. Its positive attributes include excellent forage quality that makes it one of the most important pasture grass species in temperate regions (Jones et al. 2002). In addition, its fast establishment and versatility as a turfgrass (Hannaway et al. 1999), as well as its color, upright growth habit, tolerance of low mowing height and soil compaction, and absence of thatch, contribute to its wide use on golf course fairways (Uddin et al. 2003a). As a member of the Festuceae tribe of the Pooideae subfamily of the Poaceae family (Yaneshita et al. 1993), it is taxonomically related to oat (Avena sativa L.), barley (Hordeum vulgare L.), wheat (Triticum aestivum L. em Thell), and rice (Oryza sativa L.) (Kellogg 2000).
Gray leaf spot has become a serious problem on perennial ryegrass (Viji et al. 2001; Williams et al. 2001) since the time it was first reported on golf course fairways in Pennsylvania in 1992 (Landschoot and Hoyland 1992). The causal agent, the ascomycete fungus Magnaporthe grisea, also causes rice blast disease on rice, as well as foliar diseases on many grasses, such as blast on wheat (Viji et al. 2001), barley (Sato et al. 2001), and gray leaf spot on other turf and forage grasses such as tall fescue (Festuca arundinacea Schreb.), St. Augustinegrass [Stenotaphrum secundatum (Walt.) Kuntze] and Italian ryegrass (Lolium multiflorum Lam.) (Viji et al. 2001; Williams et al. 2001).
Under warm, humid conditions, gray leaf spot can completely destroy mature ryegrass plants in a matter of days (Landschoot and Hoyland 1992). Fungicide use is important in managing this disease on turf (Vincelli and Dixon 2002). However, strains of M. grisea resistant to one of the most effective classes of fungicides, the strobilurins, have been reported (Vincelli and Dixon 2002). Due to the high genetic variability of the pathogen, possible development of resistance to the remaining effective fungicides, thiophanate-methyl and certain demethylation inhibitors, is also a concern (Vincelli and Dixon 2002).
Host resistance is an attractive, environmentally sound control strategy, which has been well-studied and tested in other hosts of M. grisea. For example, in rice the rice blast disease is largely controlled by host resistance. Many major, race-specific genes conferring complete blast resistance have been found (Hittalmani et al. 2000; Chauhan et al. 2002; Wang et al. 1994; Tabien et al. 2000), as well as quantitative trait loci (QTL) that control minor gene or partial resistance in rice (Tabien et al. 2002; Fukuoka and Okuno 2001; Wang et al. 1994) and in barley (Sato et al. 2001).
While major gene blast resistance is easier to manipulate genetically, it can often break down (Chauhan et al. 2002). On the other hand, partial resistance may be more durable though generally more difficult to manipulate. For example, rice cultivars such as ‘Moroberekan’ and ‘IR36’, which have been found to contain genes for both major and partial resistance (Wang et al. 1994), remained resistant under years of disease pressure and inoculation with many M. grisea isolates (Tabien et al. 2000; Wang et al. 1994). Similar resistance to gray leaf spot in perennial ryegrass would be valuable in improving the utility of this species, as well as in reducing the level of dependence on environmentally harmful pesticides.
For perennial ryegrass, little resistance has been reported in commercially available cultivars (Williams et al. 2001; Bonos et al. 2004), while some perennial ryegrass plant introductions from outside the US seem to show some level of resistance, particularly from eastern European sources (Hoffmann and Hamblin 2001; Bonos et al. 2004). Some materials from the US were also found to be resistant (Bonos et al. 2004). However, it is not known how well these materials will retain their resistance in a field setting, especially if planted for a number of years, or how well the introduced materials will adapt to conditions in the US. More detailed knowledge of the number and degree of effect of the genes involved would indicate the amount of breeding work required to transfer the resistance into perennial ryegrass cultivars.
A valuable tool to study the genetics of both complete and partial disease resistance is the genetic linkage map, which has been widely used to study resistance to M. grisea in rice (Chen et al. 2003; Fukuoka and Okuno 2001; Hittalmani et al. 2000; Tabien et al. 2000, 2002; Wang et al. 1994) and barley (Sato et al. 2001). QTL analysis is important in finding molecular markers linked to gray leaf spot resistance genes. These markers will be effective for pyramiding of resistance genes from different sources into a single cultivar, and for selecting for resistance to multiple diseases over several environments (Melchinger 1990).
In perennial ryegrass, several genetic linkage maps have been reported. One of the first (Bert et al. 1999) was based on dominant AFLP markers. More recent maps employ codominant markers such as SSRs and RFLPs (Jones et al. 2002; Warnke et al. 2004; Sim et al. 2005), which have more power in QTL mapping, as they can distinguish up to four allelic classes in segregating progeny from two heterozygous parents. RFLP-based maps have an additional advantage, in that markers common between ryegrass and well-studied grasses such as rice, wheat, and barley, can be used to align ryegrass linkage groups to published grass maps, and utilize synteny between ryegrass and other grasses. The map used in this study (Sim et al. 2005) includes a high density of heterologous cereal RFLPs, facilitating comparisons of QTL locations between ryegrass and rice.
The objectives of this study were to map QTL using two M. grisea strains against a ryegrass mapping population, and then use the results to determine the source of resistance in the population and compare the map locations of resistance loci between ryegrass and rice.
Materials and methods
Plant materials, fungal strains, and inoculation
The ryegrass genetic mapping population originally developed by Dr. R. Barker (USDA-ARS, Corvallis, OR, USA) consisted of 156 progeny individuals derived from the cross between two heterozygous ryegrass clones, MFA and MFB (Warnke et al. 2004). Both of these parental clones are Italian × perennial ryegrass hybrids, derived from separate crosses between two different grandparental clones of the perennial ryegrass cultivar ‘Manhattan’, termed Manh-1 and Manh-3, and two different grandparental clones of the Italian ryegrass cultivar ‘Floregon’, which were not available for this study due to the annual nature of this species. Further details on population development are given in Warnke et al. (2004).
Two perennial ryegrass clonal genotypes, 4LB-2 and L4B-5 were derived from a cross among clones of the commercial perennial ryegrass cultivars ‘Linn’ and ‘SR4400’. Additionally, five clonal genotypes of Floregon Italian ryegrass, Flor-1 through Flor-5, were established from seed of the same seedlot that generated the Italian ryegrass grandparents of the mapping population. These genotypes were tested for their gray leaf spot reaction along with the parental and grandparental clonal genotypes.
Ryegrass clonal genotypes were prepared for inoculation by asexual division and transplanting into flats of conical pots (RLC-7 Super “Stubby” Cell, Stuewe and Sons Inc., Corvallis, OR, USA) filled with soilless potting media (Scott’s Metro-Mix 366-P). These cone pots were arranged in a randomized complete block design with four replicates per genotype. Fertilizer (Peter’s 20-18-20) was applied once weekly until plants had reached 5–10 cm in height, usually after 3– weeks. Plants were always inoculated when leaf blades were still young.
Five isolates were tested including one lab strain 6082, which was provided by Dr. S. Leong at the University of Wisconsin and is capable of infecting both rice and ryegrass, as well as four field isolates collected from diseased perennial ryegrass golf course fairways. These were GG9 and GG12, collected by Dr. M. Farman at the University of Kentucky, and Lin00 and BL00, collected by Dr. A. Hamblin at the University of Illinois. All isolates used in this study were stored as frozen stocks on filter paper at −20°C, and then cultured for spore production on oatmeal agar plates at room temperature under continuous lighting for 2–3 weeks. These inoculations were carried out by misting conidial suspensions in 0.2% gelatin solution onto the plants, with spore concentrations adjusted to 1–3×105 spores/ml using a hemocytometer. The mist chamber method described below was followed for the initial test.
Isolates GG9 and 6082 were chosen based on sporulation capacity in culture and virulence, and then used to inoculate the complete mapping population, to generate phenotypic data for QTL mapping. For all mapping population experiments, 152 of the original 156 progeny were inoculated. In mapping population experiment 1 using GG9, the spore suspension was atomized onto the flats of plants in conical pots and allowed to dry for about 1 h. The plants were then kept in a mist chamber, which had the same temperature (approximately 20–25°C) and photoperiod (approximately 12 h) as the surrounding greenhouse, for 3 days to allow symptom development. In mapping population experiments 2 and 3 using GG9, and mapping population experiment 4 using 6082, the flats of plants in conical pots were placed into plastic boxes with moist soil, sprayed with the spore suspension, and the boxes were individually covered with plastic bags tied closed at the end to retain leaf wetness. These sealed boxes were then placed in a growth chamber maintained at 28C±3°C, with 12-h photoperiod, for 30–36 h. The growth chamber temperature of 28°C was chosen since this is the optimum temperature for infection (Moss and Trevathan 1987; Uddin et al. 2003b). The growth chamber inoculation method appeared to be preferable as lesion types produced were much more consistent than with the mist chamber method, which had shown pronounced mixing of lesion types on the same plant. However, results of the mist chamber experiment were included to determine whether use of different assay methods influenced QTL detection.
At the end of the respective incubation times, the plants were moved to a greenhouse, where lesions were scored 5–7 days after inoculation. Lesions were scored following the scale and descriptions in Table 1. Where mixing of lesion types occurred on the same plant, they tended to be more severe on younger leaves, thus the lesion type on the youngest leaves of each plant was used, as recommended for rice blast race testing (Atkins et al. 1967). In each of the four experiments the mean lesion type over the four replicates was used for subsequent QTL analyses.
Statistical analysis of gray leaf spot reaction in the progeny, including ANOVA testing for a significant genotype effect, correlation analysis between the four experiments, and estimating genetic and error variance components, was conducted using JMP software (SAS Institute, Cary, NC). Broad sense heritability was calculated using the formula \( {\text{H}}_b^2 = \sigma _g^2 /\left( {\sigma _g^2 + \sigma _e^2 /4} \right), \) where \( \sigma _g^2 \) is the genetic variance and \( \sigma _e^2 \) is the error variance divided by 4, corresponding to the number of clonal replicates of each genotype, as in Calenge et al. (2004).
QTL mapping and analysis
Two linkage maps were constructed separately for each parent, MFA and MFB, using the ‘DH’ or doubled haploid population type option in Joinmap® (Van Ooijen and Voorrips 2001) and the same marker data used in Sim et al. (2005), which had used an integrated map representing both parental maps. In brief, the two parental maps were constructed using the RAPD, RFLP, AFLP, SSR, isozyme, and morphological marker data described in Warnke et al. (2004), and then supplemented with additional RFLP data from Sim et al. (2005). AFLP markers are the same as those used in the aforementioned maps, except that the names have been converted to the KeyGene format while retaining the A and P notation, which indicates inheritance from Italian or perennial ryegrass (Warnke et al. 2004). Naming of RAPD and other markers is also described in Warnke et al. (2004).
Analyses of QTL were performed using the two parental maps and marker data sets separately, again using the DH population type option available in MapQTL® software (Van Ooijen et al. 2002). The maps were analyzed separately mainly due to the unequal rates of recombination observed in the MFA and MFB parents (Warnke et al. 2004), but also to enable analysis of QTL present in each parent map completely independently of the other parent.
Quantitative trait locus analyses were conducted for all four experiments, using each parental map separately, for a total of eight analyses. The phenotypic variable was the mean lesion score over the four replicates in each of the four experiments. Interval mapping (Lander and Botstein 1989; Jung et al. 1996) was used to estimate the map location, LOD score, and phenotypic effect of potential QTL, in terms of the percentage of phenotypic variance explained. An additional test of QTL detected by interval mapping was performed using the nonparametric, single marker-based Kruskal-Wallis analysis. This was to check whether markers linked to QTL detected by interval mapping were significant when examined individually. For this analysis, a genome-wide P-value of less than 0.00017 was chosen based on a comparison-wise P-value of 0.05 divided by 300, the approximate number of markers in each parental map. Additionally, phenotypic means of the two marker allele classes, ‘a’ and ‘b’, present in each of the two parental maps were determined using this method, since it provides an arithmetic mean rather than a mean calculated by maximum likelihood methods.
After potential QTL were detected by interval mapping and Kruskal-Wallis analysis, markers closely linked to the QTL were selected as cofactors, and tested using the automatic cofactor selection procedure in MapQTL, with the P-value cutoff for elimination of a cofactor set at P=0.02, the default value in MapQTL. Using the set of cofactors, multiple QTL mapping was conducted using the MQM option of MapQTL. Similar to the interval mapping analyses, the map location, LOD score, and percentage of phenotypic variance explained were estimated. LOD significance thresholds were determined using the permutation test of MapQTL with 1,000 iterations. Specifically, for each of the eight combinations of trait and parental map, the 5% genome-wide threshold was determined. In summary, a QTL was considered as significant only if it exceeded genome-wide 5% threshold, and/or had a P-value of less than 0.00017 in the Kruskal-Wallis analysis. However, markers that were consistently significant at P=0.01 via Kruskal-Wallis analysis over experiments were presented for illustrative purposes in Table 4, despite having LOD scores lower than the genome-wide threshold.
Map locations of QTL were compared with the approximate locations of previously reported rice blast resistance loci and QTL (Sallaud et al. 2003, Chen et al. 1999, 2003, Causse et al. 1994; Chauhan et al. 2002, Zhu et al. 2004, and Zenyabashi et al. 2002), using syntenic relationships between rice and ryegrass (Sim et al. 2005).
Results
Ryegrass isolates GG9, GG12, BL00, and Lin00 produced similar patterns of disease reaction on the mapping parents, grandparents, and check clones. The four perennial ryegrass clones, grandparents Manh-1 and Manh-3 and check clones 4LB-2 and L4B-5, showed large, expanding type 4 lesions when inoculated with ryegrass isolates, causing the typical bending, twisting, or blighting of leaves associated with severe gray leaf spot (Table 2). Parental clones MFA and MFB showed reduced symptoms compared to the perennial genotypes, ranging from type 2 to type 3 lesions over the ryegrass isolates (Table 2). The reaction of these clones was sensitive to plant condition. When these clones grow vigorously, they would show full-sized type 3 lesions, as shown for isolates BL00 and Lin00 (Table 2).
However, the rice-infecting strain 6082 produced a differential reaction between the mapping parents MFA and MFB (Table 2), which was not observed with the ryegrass isolates. These two clones showed mostly type 2 and type 4 lesions, respectively (Table 2). For the perennial ryegrass grandparent clones, Manh-1 showed a resistant reaction, while Manh-3 grandparent showed an intermediate, type 3 reaction (Table 2). Further, strain 6082 produced approximately the same number of lesions as the ryegrass isolates tested, but the sizes of the susceptible lesions were slightly smaller.
These results suggested that the Italian ryegrass cv. ‘Floregon’ is the source of resistance in this population (Table 2). Of the five clones tested (Flor-1 through Flor-5), three were resistant to GG9 (score approximately 2, similar to resistant progenies), one was intermediate with type 3 lesions, and one was susceptible. When two of the clones were tested against strain 6082, one clone showed resistance while the other was susceptible (Table 2), indicating that these genotypes may possess resistance to diverse M. grisea strains. Based on these results, isolates GG9 and 6082 were chosen for use in QTL mapping.
Phenotypic analysis of gray leaf spot reaction in the mapping population
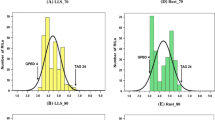
Frequency distributions of gray leaf spot reaction of the entire mapping population in four different experiments are presented in Fig. 1. Score 0 was not observed on any genotype in the four experiments, and score 1 did not occur separately from score 2. Using ryegrass isolate GG9, transgressive segregation was observed (Fig. 1b, c), suggesting that the MFA and MFB parents carry resistance against isolate GG9. The graphs from the three GG9 inoculations appeared skewed towards susceptibility, and with more plants tending towards resistant reactions than intermediate (type 3) reactions. The 6082 inoculation showed a different pattern (Fig. 1d) with more plants having intermediate, type 3 reactions than either resistant or susceptible reactions. The reaction of the mapping parents was much closer to the extremes of the frequency distribution, suggesting a lack of transgressive segregation for resistance to this strain.
Frequency distributions of GLS phenotypic data from four greenhouse inoculations, based on the mean score over four clonal replicates of each progeny genotype. For each experiment, the mean GLS reaction for each of the two mapping parents, MFA and MFB, is given. a GG9, mist chamber experiment. b GG9, first growth chamber experiment. c GG9, second growth chamber experiment. d 6082, growth chamber experiment
The two growth chamber experiments produced a coefficient of 0.92, while the mist chamber experiment showed correlations of approximately 0.79 with the two growth chamber experiments (all three P-values less than 0.0001). These results provide evidence that the phenotypic assay is reproducible, particularly in the growth chamber. The results of the 6082 inoculation were only very weakly correlated with the three GG9 inoculations, with coefficients ranging from 0.15 to 0.22 and P-values ranging from 0.07 to 0.008. This provides further evidence of the different segregation pattern between the two isolates.
ANOVA tests performed for all four mapping population experiments showed a highly significant genotype effect (Table 3). All three GG9 experiments showed similar mean gray leaf spot reaction scores, as well as high broad-sense heritabilities, while the 6082 experiment mean was somewhat lower (Table 3). When heritability was calculated using all three GG9 experiments together, the value was 0.895 with σ2 g=0.443 and σ2 e=0.206, while for the two growth chamber experiments, the value was 0.932 with σ2 g=0.562 and σ2 e=0.164.
Gray leaf spot QTL mapping
Results of QTL mapping using Kruskal-Wallis, interval mapping, and MQM analysis are presented in Table 4, the linkage maps used for the analysis are presented in Fig. 2, and LOD plots derived from MQM analysis are presented in Fig. 3. Multiple QTL were detected in each experiment, with three potential genomic regions in each of the three GG9 experiments, and two regions in the 6082 experiment. Results from interval mapping were similar to those from MQM analysis (Table 4), although the LOD plots showed much wider intervals, as compared to MQM (data not shown).
Linkage maps of the MFA×MFB population used for QTL analysis in this study, constructed separately using the RFLP, RAPD, AFLP, SSR, isozyme, and morphological marker data from Warnke et al. (2004) and Sim et al. (2005). AFLP markers are the same as those in the above maps, with the names expressed in KeyGene format. Linkage groups for both maps are numbered according to Warnke et al. (2004). a MFA parent map. b MFB parent map
LOD score plots for QTLs for GLS resistance obtained from four greenhouse inoculations, based on the mean over four clonal replicates of each progeny genotype, using MQM analysis. The different experiments are indicated by different shapes within each diagram. For QTLs relating to isolate GG9, three experiments are presented: the mist chamber experiment (shown with open triangles), the first growth chamber experiment (shown with filled squares), and the second growth chamber experiment (shown with filled circles). For strain 6082, the growth chamber experiment is presented (shown with filled triangles). Positions of significant markers associated with the respective traits, as mentioned in Table 4, are indicated below each linkage group. a MFB linkage group 3. b MFA linkage group 6. c MFA linkage group 2. d MFB linkage group 4
For the three GG9 experiments, two genomic regions with strong effects on resistance were noted, along with one region of weaker effect. The QTL with highest LOD and percentage of phenotypic variance explained was located on linkage group 3 of the MFB parent map (Table 4, Fig. 3a). Over the three experiments, the LOD score ranged from 6.8 to 14.7 with a percentage of phenotypic variation explained of 20–37%, derived from both interval mapping and MQM analysis (Table 4). RFLP marker CDO460 was closest to this QTL, and the ‘a’ allele appears to be associated with reduced lesion score. This allele was derived from the Italian ryegrass parent of MFB, thus supporting Floregon as the source of this resistance QTL. In all three GG9 experiments, the LOD score for this QTL exceeded the genome-wide 5% significance threshold as established by permutation analysis.
The next most significant QTL detected using GG9 was located on linkage group 6 of the MFA parent map (Fig. 3b). The LOD score ranged from 3.36 to 3.45 in the first and second growth chamber experiments, but was only 1.70 to 1.82 in the mist chamber experiment (Table 4). The percentage of phenotypic variation explained was approximately 10% in the first and second growth chamber experiments, but was only about 6% in the mist chamber experiment. For this QTL, the ‘b’ allele of RAPD marker C19.390, corresponding to absence of the band, was associated with reduced lesion score. RAPD screening of Manh-1 parent of MFA showed that the band, that is, the susceptible allele, was inherited from Manh-1 parent. Thus, the resistance allele of this QTL is also derived from Italian ryegrass. This QTL had peak LOD scores exceeding the 5% genome-wide significance threshold in both growth chamber experiments, but not in the mist chamber experiment (Table 4).
One potential minor QTL was noted on MFA linkage group 2, with LOD scores ranging from 1.57 to 2.56 and percentages of phenotypic variation ranging from 4.9% to 7.8% based on MQM analysis (Table 4). The AFLP marker A-E33M62109 was most closely linked to this minor QTL. It did not exceed the genome-wide significance threshold, but was included because it was detected in all three GG9 experiments at P=0.01 via Kruskal-Wallis analysis.
For the inoculation experiment using the lab strain 6082, two potential QTL were detected (Table 4). These QTL were located on MFA linkage group 2 and MFB linkage group 4 (Fig. 3c, respectively). The QTL on MFB linkage group 4 was closely linked to the RAPD marker E3.650 (Table 4, Fig. 3d), while the QTL on MFA linkage group 2 was closely linked to the RAPD marker E8.575 (Table 4, Fig. 3c). Both QTL had LOD scores exceeding the 5% genome-wide significance threshold according to interval mapping, while only the QTL on MFA linkage group 2 exceeded the threshold according to MQM analysis (Table 4).
Discussion
Interpretation of QTL for resistance to GLS
In this study, a QTL mapping approach was utilized to uncover four genomic regions potentially associated with GLS resistance in ryegrass. Two of these were uniquely associated with resistance to a perennial ryegrass field isolate, GG9, while one was uniquely associated with resistance to a lab strain, 6082. One QTL, on MFA linkage group 2, was detected using both isolates. Thus, it may be effective against widely divergent pathogen genotypes, such as ryegrass field isolates like GG9 and strains with similar genetics to 6082. Results suggest that the population is segregating for at least two different sets of resistance loci, which are differentially effective against the two isolates, or that there is only one set of loci against which different isolates can cause differing amounts of disease.
For resistance to GG9, three QTL were located, on MFB linkage group 3, MFA linkage group 6, and MFA linkage group 2, which in total explained from 32.3 to 53.0% of the phenotypic variance over the three experiments (Table 4). However, the broad-sense heritability of resistance to isolate GG9 was rather high, ranging from 0.895 to 0.932.
One possible reason why the total variance explained was not as high as might be expected from the heritabilities is segregation distortion. Warnke et al. (2004) have reported that a region of MFB linkage group 3, including CDO460, and a region of MFA linkage group 6, which is closely linked to but does not include C19.390, show distorted segregation. Since these two regions include the two most significant QTL for resistance to GG9, the estimates of effect on phenotype may have been changed. Also, it is possible that additional low effect QTL, such as the one on MFA linkage group 2, are segregating in the population yet remain undetected.
Nature of GLS resistance and role of environmental variables
The resistance in this population appears to be partial resistance, involving differences in lesion size rather than presence or absence of lesions. Resistant plants often show few small type 3 lesions, with a reduced amount of pathogen reproduction compared to that of large, expanding type 4 lesions on susceptible progenies. Also, the disease reaction was sensitive to vigor of the youngest leaves, as might be modified by nitrogen fertilization, optimum light intensity, and temperature and soil conditions. In rice, blast disease severity has been shown to be increased by nitrogen fertilizer application (Bonman 1992). Partial resistance to M. grisea has been reported in other species, such as tall fescue (Tredway et al. 2003), rice (Seebold et al. 2001; Zenyabashi et al. 2002), and barley (Sato et al. 2001).
Comparative mapping of GLS resistance QTL between ryegrass and rice
Based on the extensive synteny between rice and ryegrass (Sim et al. 2005), positions of QTL can be compared across these species. For the QTL on MFB linkage group 3 against ryegrass isolate GG9, there are several syntenous blast resistance genes and QTL in rice. Major blast resistance genes Pi-t and Pi-24(t) (Sallaud et al. 2003), Pi-sh, and Pi-27(t) (Zhu et al. 2004) have been mapped to rice chromosome 1 (Fig. 4a), which is syntenic with perennial ryegrass linkage group 3 (Jones et al. 2002; Sim et al. 2005). Rice blast resistance QTLs mapping to rice chromosome 1 have been detected by Wang et al. (1994) as well as Chen et al. (2003). Although several blast resistance genes (Pi-b, Pi-25(t), and Pi-tq5) map to rice chromosome 2 (Sallaud et al. 2003), none of these are syntenous with the QTL detected on MFA linkage group 6 (Fig. 4b).
Comparative relationships between rice and ryegrass highlighting locations of blast resistance loci and QTL. Approximate QTL location from this study is indicated by a diamond symbol in each of the three ryegrass linkage groups. Deduced approximate positions of the blast resistance genes or QTLs are indicated by vertical lines at the right of the rice chromosomes. Blast resistance genes are italicized and indicated by symbols beginning with “Pi”. Ryegrass mapping information is derived from Sim et al. (2005), which used a consensus map produced by integrating the MFA and MFB maps. RFLP markers generated by the same probes and mapped on both species are connected by lines. The two genomic regions indicated by vertical lines at the right of the ryegrass linkage group 4 are syntenous with rice chromosome 3. a Ryegrass linkage group 3. b Ryegrass linkage group 6. c Ryegrass linkage group 4
Resistance to strain 6082 in rice mapped to chromosome 11, conferring a hypersensitive, type 1 lesion (Chauhan et al. 2002). This is different from the partial resistance observed in ryegrass. Markers CDO520, CDO844, and E3.650 map to ryegrass linkage group 4 (Fig. 2b) and are significantly associated with a QTL for resistance to isolate 6082. CDO520 also maps to rice chromosome 11 (Chen et al. 1999), near several blast resistance genes, such as Pi-44(t) (Chen et al. 1999), Pi-1 and Pi-m (Sallaud et al. 2003), Pi-lm2 (Tabien et al. 2000), as well as a single dominant QTL conferring partial blast resistance (Fig. 4c) (Zenyabashi et al. 2002). However, these loci map approximately 40–50 cM from the Pi-CO39(t) locus (Chauhan et al. 2002), suggesting that Pi-CO39(t) is not syntenous with the QTL for resistance to isolate 6082 on ryegrass linkage group 4.
As blast resistance genes occur in clusters (Zhu et al. 2004), there are practical breeding applications of these results. Since the rice genome has been sequenced, the chromosomal regions syntenous with the GLS QTLs can be searched for sequences matching resistance gene motifs, in a synteny-based candidate gene approach (Armstead et al. 2004). Once identified, the candidate resistance genes from rice can be used to construct PCR-based markers for ryegrass, so that actual resistance genes underlying the QTL from this study can be utilized for marker-assisted selection for GLS resistance, rather than a linked anonymous cDNA clone.
Implications for breeding for improved GLS resistance
Currently, perennial ryegrass cultivars show limited resistance to GLS (Bonos et al. 2004), although some plant introductions of perennial ryegrass show variation in GLS reaction under greenhouse and field conditions (Bonos et al. 2004, Hoffmann and Hamblin 2001). Bonos et al. (2004), using open-pollinated single plant progeny evaluations, have revealed some partially resistant perennial ryegrass selections. Further, they reported that the resistance is under strong genetic control, with evidence for a single major gene (Bonos et al. 2004), in contrast with the current study which suggests multiple genes are important.
Similarly, Italian ryegrass has shown variation in reaction to GLS in field and greenhouse tests as well (Trevathan et al. 1994), with the most resistant plant introductions originating from Europe (Trevathan et al. 1994). Further, the cultivar Floregon was subjected to unintentional selection for resistance due to a natural gray leaf spot outbreak in Florida during a breeding cycle (R. Barker, personal communication). Results of this study also provide evidence for GLS resistance in Italian ryegrass, as well as its potential as a genetic resource in this respect.
The GLS resistance in our population may represent a different source of resistance to that found by Bonos et al. (2004), which would allow for pyramiding of different resistances into a perennial ryegrass cultivar. Most importantly, the resistance segregating in our population has been characterized in terms of the potential number of genes, and of the nature of resistance. These characteristics are important for estimating how easily resistance can be transferred to new cultivars. Markers tightly linked to the resistance loci in this population will allow for marker-assisted selection. This will become even more important as more sources of gray leaf spot resistance are incorporated into breeding programs (Bonos et al. 2004), as the use of markers linked to each unique resistance gene will enable the tracking of each one through the cultivar development process.
References
Armstead IP, Turner LB, Farrell M, Skøt L, Gomez P, Montoya T, Donnison, IS, King IP, Humphreys MO (2004) Synteny between a major heading-date QTL in perennial ryegrass (Lolium perenne L.) and the Hd3 heading-date locus in rice. Theor Appl Genet 108:822–828
Atkins JG, Robert AL, Adair CR, Goto K, Kozaka K, Yanagida R, Yamada M, Matsumoto S (1967) An international set of rice varieties for differentiating races of Piricularia oryzae. Phytopathology 57:297–301
Bert PF, Charmet G, Sourdille P, Hayward MD, Balfourier F (1999) A high-density molecular map for ryegrass (Lolium perenne L.) using AFLP markers. Theor Appl Genet 99:445–452
Bonman JM (1992) Blast. In: Webster RK, Gunnell PS (eds) Compendium of rice diseases. APS Press, St. Paul, pp 14–17
Bonos SA, Kubik C, Clarke BB, Meyer WA (2004) Breeding perennial ryegrass for resistance to gray leaf spot. Crop Sci 44:575–580
Calenge F, Faure A, Goerre M, Gebhardt C, Van de Weg WE, Parisi L, Durel C-E (2004) Quantitative trait loci (QTL) analysis reveals both broad-spectrum and isolate-specific QTL for scab resistance in an apple progeny challenged with eight isolates of Venturia inaequalis. Phytopathology 94:370–379
Causse M, Fulton T, Cho Y, Ahn S (1994) Saturated molecular map of the rice genome based on an interspecific backcross population. Genetics 138:1251–1274
Chauhan R, Farman ML, Zhang H-B, Leong SA (2002) Genetic and physical mapping of a rice blast resistance locus, Pi-CO39(t), that corresponds to the avirulence gene AVR1-CO39 of Magnaporthe grisea. Mol Gen Genomics 267:603–612
Chen D-H, dela Viña M, Inukai T, Mackill DJ, Ronald PC, Nelson RJ (1999) Molecular mapping of the blast resistance gene, Pi44(t), in a line derived from a durably resistant rice cultivar. Theor Appl Genet 98:1046–1053
Chen H, Wang S, Xing Y, Xu C, Hayes PM, Zhang Q (2003) Comparative analyses of genomic locations and race specificities of loci for quantitative resistance to Pyricularia grisea in rice and barley. Proc Natl Acad Sci USA 100:2544–2549
Fukuoka S, Okuno K (2001) QTL analysis and mapping of pi21, a recessive gene for field resistance to rice blast in Japanese upland rice. Theor Appl Genet 103:185–190
Hannaway D, Fransen S, Cropper J, Teel M, Chaney M, Griggs T, Halse R, Hart J, Cheeke P, Hansen D, Klinger R, Lane W (1999) Perennial ryegrass (Lolium perenne L.). Oregon State University Extension Publication PNW503
Hittalmani S, Parco A, Mew TV, Zeigler RS, Huang N (2000) Fine mapping and DNA marker-assisted pyramiding of the three major genes for blast resistance in rice. Theor Appl Genet 100:1121–1128
Hoffman N, Hamblin A (2001) Reaction of perennial ryegrass to gray leaf spot following inoculation in the field, 2000. Biol Cult Tests 16:T56
Jones ES, Mahoney NL, Hayward MD, Armstead HI, Jones JG, Humphreys MO, King IP, Kishida T, Yamada T, Balfourier F, Charmet G, Forster JW (2002) An enhanced molecular marker based genetic map of perennial ryegrass (Lolium perenne) reveals comparative relationships with other Poaceae genomes. Genome 45:282–295
Jung G, Coyne D, Skroch P, Nienhuis J, Arnaud-Santana E, Bokosi J, Ariyarathne H, Steadman J, Beaver J, Kaeppler S (1996) Molecular markers associated with plant architecture and resistance to common blight, web blight and rust in common beans (Phaseolus vulgaris L.). J Am Soc Hort Sci 121(5):794–803
Kellogg E (2000) The grasses: a case study in macroevolution. Ann Rev Ecol Syst 31:217–238
Lander E, Botstein D (1989) Mapping Mendelian factors underlying quantitative traits using RFLP linkage maps. Genetics 121:185–199
Landschoot P, Hoyland B (1992) Gray leaf spot of perennial ryegrass turf in Pennsylvania. Plant Dis 76:1280–1282
Melchinger AE (1990) Use of molecular markers in breeding for oligogenic disease resistance. Plant Breeding 104:1–19
Moss MA, Trevathan LE (1987) Environmental conditions conducive to infection of ryegrass by Pyricularia grisea. Phytopathology 77:863–866
Sallaud C, Lorieux M, Roumen E, Tharreau D, Berruyer R, Svestasrani P, Garsmeur O, Ghesquire A, Notteghem J-L (2003) Identification of five new blast resistance genes in the highly blast-resistant rice variety IR64 using a QTL mapping strategy. Theor Appl Genet 106:794–803
Sato K, Inukai T, Hayes PM (2001) QTL analysis of resistance to the rice blast pathogen in barley (Hordeum vulgare). Theor Appl Genet 102:916–920
Seebold KW, Kucharek TA, Datnoff LE, Correa-Victoria FJ, Marchetti MA (2001) The influence of silicon on components of resistance to blast in susceptible, partially resistant, and resistant cultivars of rice. Phytopathology 91:63–69
Sim S, Chang T, Curley J, Warnke S, Barker R, Jung G (2005) Chromosomal rearrangements differentiating the ryegrass genome from the Triticeae, oat, and rice genomes using common heterologous RFLP probes. Theor Appl Genet 110:1011–1019
Tabien RE, Li Z, Paterson AH, Marchetti MA, Stansel JW, Pinson SRM (2000) Mapping of four major rice blast resistance genes from ‘Lemont’ and ‘Teqing’ and evaluation of their combinatorial effect for field resistance. Theor Appl Genet 101:1215–1225
Tabien R, Li Z, Paterson AH, Marchetti MA, Stansel JW, Pinson SRM (2002) Mapping QTLs for field resistance to the rice blast pathogen and evaluating their individual and combined utility in improved varieties. Theor Appl Genet 105:313–324
Tredway LP, Stevenson KL, Burpee LL (2003) Components of resistance to Magnaporthe grisea in ‘Coyote’ and ‘Coronado’ tall fescue. Plant Dis 87:906–912
Trevathan LE, Moss MA, Blasingame D (1994) Ryegrass blast. Plant Dis 78:113–117
Uddin W, Viji G, Vincelli P (2003a) Gray leaf spot (blast) of perennial ryegrass turf-an emerging problem for the turfgrass industry. Plant Dis 87:880–889
Uddin W, Serlemitsos K, Viji G (2003b) A temperature and leaf wetness duration-based model for prediction of gray leaf spot of perennial ryegrass turf. Phytopathology 93:336–343
Van Ooijen JW, Voorrips RE (2001) JoinMap® 3.0: software for the calculation of genetic linkage maps. Plant Research International, Wageningen
Van Ooijen JW, Boer MP, Jansen RC, Maliepaard C (2002) MapQTL® version 4.0: software for the calculation of QTL positions on genetic maps. Plant Research International, Wageningen
Viji G, Wu B, Kang S, Uddin W, Huff DR (2001) Pyricularia grisea causing gray leaf spot of perennial ryegrass turf: population structure and host specificity. Plant Dis 85:817–826
Vincelli P, Dixon E (2002) Resistance to QoI (Strobilurin-like) fungicides in isolates of Pyricularia grisea from perennial ryegrass. Plant Dis 86:235–240
Wang G-L, Mackill DJ, Bonman JM, McCouch SR, Champoux MC, Nelson RJ (1994) RFLP mapping of genes conferring complete and partial resistance to blast in a durably resistant rice cultivar. Genetics 136:1421–1434
Warnke SE, Barker RE, Jung G, Sim S, Rouf Mian MA, Saha MC, Brilman LA, Dupal MP, Forster JW (2004) Genetic linkage mapping of an annual x perennial ryegrass population. Theor Appl Genet 109:294–304
Williams DW, Burrus PB, Vincelli P (2001) Severity of gray leaf spot in perennial ryegrass as influenced by mowing height and nitrogen level. Crop Sci 41:1207–1211
Yaneshita M, Ohmura T, Sasakua T, Ogihara Y (1993) Phylogenetic relationships of turfgrasses as revealed by restriction fragment analysis of chloroplast DNA. Theor Appl Genet 87:129–135
Zenbayashi K, Ashizawa T, Tani T, Koizumi S (2002) Mapping of the QTL (quantitative trait locus) conferring partial resistance to leaf blast in rice cultivar Chubu 32. Theor Appl Genet 104:547–552
Zhu M, Wang L, Pan Q (2004) Identification and characterization of a new blast resistance gene located on rice chromosome 1 through linkage and differential analyses. Phytopathology 94:515–519
Acknowledgements
We thank Drs. Mark Farman and Andy Hamblin for providing us the perennial ryegrass isolates, and Drs. Michael Havey and Michael Casler for critical reviews for the manuscript. We thank Jung lab members for their excellent technical assistance. Funding for this project was provided by the United States Golf Association and partially by the Wisconsin Turfgrass Association.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by T. Lübberstedt
Rights and permissions
About this article
Cite this article
Curley, J., Sim, S.C., Warnke, S. et al. QTL mapping of resistance to gray leaf spot in ryegrass. Theor Appl Genet 111, 1107–1117 (2005). https://doi.org/10.1007/s00122-005-0036-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00122-005-0036-x