Abstract
Leaf rust caused by the fungus Hemileia vastatrix is the most devastating disease of arabica coffee (Coffea arabica). Therefore, developing leaf rust-resistant varieties has been a breeding objective of the highest priority in many countries. The purpose of the present work was to gain insight into the mechanism of introgression into C. arabica of a leaf rust resistance gene from C. liberica (i.e. SH3 resistance factor) and to identify associated molecular markers. An F2 progeny (i.e. 101 individuals) derived from a cross between Matari, an arabica accession and liberica-introgressed line S.288, was evaluated for resistance against three different races of H. vastatrix. The progeny segregated for the SH3 gene in a 3:1 ratio, as expected for a single dominant gene. Amplified fragment length polymorphism analysis of a population subset using 80 different primer combinations revealed that at least half of the total polymorphism observed in the population is associated with introgression of C. liberica chromosome fragments. Furthermore, 15 primer combinations generating candidate marker bands associated with the SH3 resistance gene were used to analyse the whole F2 population. A total of 34 marker bands originating from S.288 and attributable to introgression were scored. None exhibited segregation distortion. Linkage analysis revealed only three distinct introgressed fragments corresponding to a total length of 52.8 cM. Twenty-one markers were strongly associated (LOD score >14) with the SH3 gene and were grouped together in a single linkage group of 6.3 cM. The results are discussed in relation to the efficient use of genetic resources in arabica breeding.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Coffee leaf rust, popularly known as orange rust, is caused by the obligate parasitic fungus Hemileia vastatrix and is a major disease which greatly limits arabica coffee (Coffea arabica) production in almost all growing countries around the world. In Brazil, the leading producer of arabica coffee, the crop losses due to this disease are estimated to be around 30% if control measures are not taken (Kushalappa and Eskes 1989). In India, leaf rust is a great threat to arabica cultivation as the climatic factors in coffee tracts are favourable to a high disease build-up, leading to crop losses of up to 70% in susceptible cultivars, if proper control measures are not adopted (Anonymous 2000). Although it is rather difficult to estimate precisely the global impact of this rust, the economic damage to world arabica coffee production has been estimated to be between $(US)1 billion and 2 billion per year (Van der Vossen 2001) due to crop losses of 20–25%. In view of the economics and to minimise the chemical input for disease management, the development and cultivation of tolerant cultivars is the most effective and viable option. Therefore, the development of coffee varieties resistant to coffee leaf rust has been a breeding objective of the highest priority in many countries and in India since the 1920s.
The allotetraploid species C. arabica is characterised by low genetic diversity. In addition, most cultivars are derived from the few trees that survived various efforts to spread arabica growing world-wide (Anthony et al. 2002). The transfer of resistance genes from either spontaneous accessions collected in the primary centre of diversity or diploid wild relative Coffea species has therefore been a constant priority of coffee breeding programmes (Bettencourt and Rodrigues 1988). Nine major dominant resistance factors to H. vastatrix have been so far inferred on the basis of the gene-for-gene concept. The resistance to leaf rust of coffee plants therefore appears to be conditioned by at least nine resistance genes designated as SH1–SH9, either singly or in combination, while the corresponding virulences have been indicated as V1–V9 (Bettencourt and Rodrigues 1988). Of these resistance factors, SH1, SH2, SH4 and SH5 have been found in C. arabica. The other genes, SH6, SH7, SH8 and SH9, have been introgressed from the diploid species C. canephora, while SH3 probably originates from another diploid species, C. liberica (Wagner and Bettencourt 1965; Vishveshwara 1974; Bettencourt and Rodrigues 1988).
Efforts to obtain durable resistance to coffee leaf rust have had a long history of initial successes followed by disappointments because of the appearance of new virulent races of rust fungus (Van der Vossen 2001). To date as many as 40 physiological races are known to be infecting different coffee genotypes in various coffee-growing countries (Rodrigues et al. 1993; Sreenivasan et al. 1994). The resistance genes identified in C. arabica, used either singly or in combination, have not provided durable resistance to most of the races of rust fungus. In contrast, the SH3 gene from C. liberica as well as certain genes from C. canephora have provided long-lived protection under field conditions (Srinivasan and Narasimhaswamy 1975; Ramachandran and Srinivasan 1979; Sreenivasan et al. 1994). Consequently, the combined used of resistance genes introgressed from diploid coffee species, either by accumulating them into one genotype (pyramid varieties) or using them in composite varieties, is expected to provide durable resistance (Kushalappa and Eskes 1989; Moreno and Alvarado 2000). However, the combined introgression into outstanding cultivars of various genes of resistance appears to be a very difficult task in an acceptable time-frame through traditional breeding approaches. An availability of molecular markers linked to SH genes and other genes conditioning resistance to coffee leaf rust would be extremely useful, especially in the context of the gene pyramiding approach (Santaram and Sreenath 2000; Lashermes et al. 2000a; Van der Vossen 2001).
The aim of the present study was to identify molecular markers closely linked to the SH3 gene for leaf rust resistance and to gain insight into the mechanism of introgression of a leaf rust resistance gene into C. arabica from C. liberica. F2 plants derived from a cross between a liberica-introgressed line carrying the SH3 gene and an accession of C. arabica were evaluated for leaf rust resistance and analysed using molecular makers. The amplified fragment length polymorphism (AFLP) approach was mainly selected as this technique allows simultaneous analysis of large number of marker loci throughout the genome. Identification of genetic markers linked to resistance would be a first step towards marker-facilitated breeding and map-based positional cloning strategies in coffee.
Materials and methods
Plant material
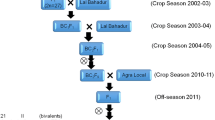
The plant material analysed consisted of an F2 population of 101 individuals derived from a cross between two Coffea arabica inbred lines, Matari and S.288. Matari is a C. arabica accession and shows high susceptibility to rust due to the absence of major rust resistance genes, while S.288 is a C. arabica line introgressed with a C. liberica genome (Prakash et al. 2002) and reported to carry the SH3 and SH5 rust resistance genes (Rodrigues et al. 1975). In order to produce the F2 individuals, two F1 trees (H.536/17 and H.536/12) from the Matari × S.288 cross were self-pollinated under controlled conditions at the Centro de Investigacao das Ferrugens do Cafeeiro (CIFC, Oeiras, Portugal). In addition to the above F2 population and the two parents, six selected accessions of C. arabica representing the different diversity groups (Anthony et al. 2002) and two accessions of C. liberica were also included for marker introgression analysis.
Evaluation of rust resistance
Evaluation of the entire F2 population for leaf rust resistance was carried out at CIFC (Oeiras, Portugal) as detailed by Bettencourt and Rodrigues (1988). Rust resistance patterns were assessed based on artificial inoculations under greenhouse/controlled conditions using selected rust races. The rust races screened, their virulent gene combinations along with the rust reaction group are presented in Table 1. The races of Hemileia vastatrix were selected to allow a segregation analysis of the SH3 resistance gene. The rust uredospores (1 mg/pair of leaves) were inoculated onto the lower side of the fully expanded tender leaves, which were then gently spread with a camel-hair brush. The inoculated leaf surfaces were sprayed with distilled water and the plants were kept in moist chambers (18–24°C) or enveloped in wet plastic bags for 48 h under dark/dim light conditions. The reaction type was evaluated 12–15 days after inoculation using a descriptive scale (D’Oliveira 1957; Eskes and Toma-Braghini 1982).
DNA isolation and AFLP assay
DNA was extracted from fresh leaf samples and the AFLP protocol described by Vos et al. (1995) was basically followed with minor modifications to suit coffee DNA as reported by Lashermes et al. (2000b). For each sample, 500 ng of genomic DNA was digested using restriction enzymes EcoRI and MseI. Restricted DNA fragments were ligated with EcoRI and MseI adapters using T4 DNA ligase (Gibco BRL, Gaithersburg, Md.). In pre-selective amplification, 5 μl of a tenfold-diluted ligation mixture was amplified using primers complementary to the adapters with one additional selective 3′ nucleotide—E+ (A or C) combined with M+ (A or C)—where E and M were the EcoRI and MseI primers, respectively, while A and C represent selective nucleotides at their 5′ ends. All of the primers of the E group (EcoRI end) and M group (MseI end) include the sequence 5′-GACTGCGTACCAATTC and 5′-GATGAGTCCTGAGTAA, respectively. The reaction mixture was diluted 1/30, and 10 μl of the solution was used for amplification using two sets of primers each with three selective nucleotides. The EcoRI primers were end-labelled with γ-[33P]-ATP using T4 polynucleotide kinase. PCR amplifications were carried out in an MJ Research Thermal controller (MJ Research, Waltham, Mass.). The amplification cycle consisted of an initial cycle of 94°C for 30 s, 65°C for 30 s, 72°C for 60 s, followed by a continuation of the same cycle 12 times but with a lowering of the annealing temperature by 0.7°C per cycle. This was followed by 32 cycles, each of 94°C for 30 s, 56°C for 30 s, 72°C for 60 s. Amplification products were electrophoresed on a 6% denaturing poly-acrylamide gel with 8 M urea and 1×TBE. The dried gels were exposed to Kodak Biomax-MS film.
Data analysis
The polymorphic AFLP bands were designated in order of decreasing fragment size and according to the primer combinations used. AFLP markers were manually scored as binary data with presence indicated by a ‘1’ and absence by ‘0’. The segregation pattern of each marker, both associated and non-associated with rust resistance, in the F2 population was subjected to χ2 analysis to test the goodness-of-fit to the expected Mendelian segregation ratio. Linkage analysis and map construction were performed using the computer programme mapmaker ver. 3.0b (Lander et al. 1987). Multipoint analysis was used to designate the most likely order of the markers. Recombination frequencies were converted into map distances or centiMorgan (cM) values by using the Kosambi (1944) function.
Results
Rust resistance pattern in the F2 population
The F2 individuals derived from a cross between the susceptible line Matari and the line S.288 carrying the SH3 and SH5 rust resistance genes were evaluated for resistance against three different races of H. vastatrix showing different virulence factor combinations (Table 1). As anticipated from the gene-for-gene concept, three groups of plants, each with different resistance patterns, were identified. These groups were interpreted as indicating the presence of the SH3 gene in association or not with the SH5 gene (group A), the presence of the SH5 gene alone (group B) or the absence of both SH3 and SH5 genes (group C). Among the 101 F2 plants analysed in this study, the SH3 resistant genes appeared to be present in 79 individuals (78%) and absent in 22 individuals (22%), a segregation pattern that fits the 3:1 ratio expected for a single dominant gene (χ2=0.558, P=0.455). Furthermore, the identification of 19 plants that could be classified in group B and three plants classified in group C is consistent (χ2=1.73, P=0.187) with an independent segregation of the SH3 and SH5 genes in the F2 population (making this assumption, the expected ratios are 1:15 and 3:13 for groups B and C, respectively).
AFLP polymorphism
A total of 80 primer combinations were screened on a subset of DNA samples that included both parents (i.e. Matari and S.288), an F1 plant and two F2 individuals as well as six accessions of C. arabica representing the different diversity groups and two accessions of C. liberica. Between 20 and 50 clearly amplified products per sample were detected, the number depending on the genotype and primer combination used for the AFLP reaction. A total of 260 polymorphic bands were scored among Matari, S.288 and their progeny. Of these 260 polymorphic bands, 154 were found to originate from S.288, and 81% of these were also seen in at least one of the C. liberica accessions included in the analysis; these latter marker bands were never seen in any of the other arabica accessions analysed and were therefore considered to be markers introgressed from C. liberica. The remaining 106 polymorphic bands originated from Matari. The data indicate that at least half of the total polymorphism observed in the F2 (Matari × S.288) population is associated with the introgression of C. liberica chromosome fragments.
Of the initially screened primer combinations, 60 that gave parental polymorphism were selected for further analysis. These 60 primers were used to screen a set of 15 plants, including the two parents, F1 and 12 F2 (Matari × S.288) plants (six plants each of group A containing SH3 gene and group B without SH3). Twenty-one marker bands derived from 15 different primer combinations were identified (Fig. 1) as candidate markers associated with the SH3 resistance gene (i.e. markers exhibited by the six plants classified in group A and absent in the six plants classified in group B).
Segregation and linkage analysis
The 15 primer combinations generating the candidate markers (Table 2) were used to analyse the whole F2 (Matari × S.288) population (101 plants). A total of 57 polymorphic bands were scored in the population, of which 35 marker bands were found to originate from S.288, while the remaining 22 polymorphic bands derived from Matari. All marker bands from S.288, with only one exception, displayed an introgressed origin. The segregation ratios in the F2 population of all the 35 marker bands derived from S.288 corresponded well with the 3:1 inheritance pattern expected for a dominant marker (Table 2). Linkage analysis in the F2 population of those marker bands using an LOD threshold of 5.0 resulted in three linkage groups (namely 1, 2 and 3) of 21, 6 and 7 markers, respectively (Fig. 2). Only the marker not attributed to introgression remained unlinked. When the LOD score was decreased to 2.5, no modification in linkage association was detected.
Genetic linkage map of the three introgressed fragments identified in the S.288 line as deduced from the AFLP analysis of the F2 (Matari × S.288) population. Molecular marker names are indicated on the right side, while the estimated map distances in centiMorgans are shown on the left. The SH3 gene for resistance to rust in coffee is located in group 1
All 21 candidate markers associated with the SH3 resistance gene were grouped together in a single linkage group (group 1) of 6.3 cM. The two other linkage groups ( groups 2 and 3) covered 21.4 and 25.1 cM, respectively. The linkage between the candidate markers belonging to group 1 and the SH3 resistance gene was confirmed when following analysis of the whole F2 population (LOD score >14).). The size of the marker bands ranged from 60 bp to 390 bp (Table 2). Among the markers, marker M8 co-segregated perfectly with the SH3 gene and exactly positioned with the SH3 locus at one end of linkage group, while all other markers flanked on the other side (Fig. 2). Hence, it could be inferred that the SH3 resistance gene was located close to marker M8.
Reliability of the markers
In order to validate the reliability of the identified markers linked to resistance, AFLP marker data of the eight markers (M2, M7, M6, M1, M4, M3, M5 and M8) were compared with the AFLP data generated earlier for C. liberica introgressed lines evolved in India (Prakash et al. 2002). This material, not yet evaluated for the presence of the SH3 resistance gene, included eight F2 individuals (S.795) and nine F4 plants (S.1934) derived from a cross between the same introgressed parent, S.288 and Kent, another arabica accession. All eight markers were found to be present in these progenies, and segregation data of the markers confirmed the order of markers on the map deduced in the present study (Table 3). These findings validated the position of markers on the localised map and confirmed the recombination as well as the transferability of the identified markers.
Discussion
Efficient use of genetic variation available in the wild relatives of crop species depends on the genetic determinism of the desirable trait and the ability to introgress desirable DNA segments from one species into the genome of another.
The segregation data obtained in the present study confirm the hypothesis of a single dominant gene for the SH3 resistance factor. Rodrigues et al. (1975) reported that SH genes are dominant and condition total susceptibility to compatible races and specific resistance to incompatible races. Accordingly, coffee plants with the SH3 resistance gene are expected to manifest susceptibility to the corresponding virulence factor (V3). Further, on studying the segregation pattern of the SH3 in F2 hybrids, Kukhang et al. (1993) reported a variable rust reaction to a virulent rust isolate and suggested that SH3 resistance was more complex than could be explained by a single dominant gene and attributed the same to the relative durability of this resistance under field conditions in India. However, the rust screening data of the present population provide convincing evidence for the simple inheritance of the SH3 gene for rust resistance, as observed in earlier screening studies (Wagner and Bettencourt 1965). Nevertheless, this interpretation did not exclude the possibility that the SH3 locus corresponds to a complex cluster of several resistance genes in tandem array, as often reported in plants (Goff et al. 2002; Richly et al. 2002).
The AFLP technique appears to be highly efficient in generating DNA markers in the coffee plant material analysed in this study. In accordance with previous reports (Lashermes et al. 2000b; Prakash et al. 2002), a large part of the detected polymorphism is related to the introgression of foreign genetic material in C. arabica. With respect to the S.288 line carrying the SH3 gene, almost all identified marker bands appeared to originate from introgressions of C. liberica chromosome fragments. The line S.288 is the selfed offspring of S.26, a natural hybrid between C. arabica and C. liberica, which was used as the main source for rust resistance in India. Based on the analysis of the number of introgressed markers, Prakash et al. (2002) suggested that the introgression in S.288 was restricted to a few chromosome segments. In the present study, only three distinct introgressed fragments were identified, corresponding to a total length of 52.8 cM. The chromosomes of coffee species are rather uniform in shape and size (Lashermes et al. 1999). If we assume a mean length of 100 cM per chromosome in C. arabica (Lashermes et al. 2000a) and the lack of significant recombination restriction in the region of introgressed fragments, as reported for C. canephora chromosome segments (Herrera et al. 2002), one can estimate that the introgression of C. liberica chromosome fragments in S.288 represents approximately one-half the chromosome equivalent of C. liberica distributed over two or three chromosomes.
Furthermore, it was interesting to note that all the markers corresponding to the three C. liberica introgressed fragments exhibited no segregation distortion and that recombination events occurred between markers. These findings support the earlier reported view that the introgression of genes into C. arabica from diploid species like C. canephora appears not to be limited by differences in either sequence homology or chromosomal structure (Herrera et al. 2002; Noir et al. 2003).
A probable consequence of the introgressed origin of the SH3 resistance gene was that the AFLP technique proved to be a powerful tool for identifying linked DNA markers. A total of 21 markers tightly linked to the SH3 rust resistance gene were identified. Similarly, Noir et al. (2003) demonstrated the efficiency of AFLP in identifying markers linked to an introgressed major gene (Mex-1) conferring resistance to root-knot nematode (Meloidogyne exigua) in C. arabica. In both cases, introgressed materials involving two different diploid species (C. canephora and C. liberica) were used as the source of resistance and almost all genetic polymorphism in such materials, as revealed by molecular markers, was attributed to the introgression process (Lashermes et al. 2000b; Prakash et al. 2002).
It is apparent that the usefulness of the identified AFLP markers linked to the SH3 gene in marker-assisted selection (MAS) will depend on the distance and orientation of the markers with respect to the gene (Frisch and Melchinger 2001; Hospital 2001). High efficiencies require small distances between the markers and the genes, thereby minimising the chances of recombination between the marker and the gene of interest. In our study, all of the markers linked to the SH3 gene were distributed at a distance of 6.3 cM, and the recombination within the introgressed region was also evident as six recombinants were identified in the population. However, no recombinant individuals were detected between the SH3 and marker M8. In fact, this marker was found to co-segregate perfectly with SH3, which determines the high selection efficiency of this marker for resistance genotypes.
The identification of molecular markers linked to rust resistance genes is an important starting point towards improving the selection efficiency in coffee breeding programmes. This is the first report on DNA markers linked to a gene conferring rust resistance in coffee. As the resistance in coffee is known to be governed by the gene-for-gene concept, there is a potential for high similarities among the SH resistance genes, and this molecular information generated on SH3 affords insight and scope to approach other SH genes. Further, the identified markers may also be used for pyramiding of the different resistance sources (Duvick 1996; Liu et al. 2000). As breeding for rust resistance is an important part of coffee breeding programmes, efforts are being made to convert the identified AFLP markers into sequence-specific PCR-based markers (Koebner et al. 1998; Arru et al. 2003). Thus, the findings of the present study are the first step towards exploitation of MAS and map-based gene cloning strategies in coffee.
References
Anonymous (2000) Coffee guide. Coffee Board, Bangalore
Anthony F, Combes MC, Astorga C, Bertrand B, Graziosi G, Lashermes P (2002) The origin of cultivated Coffea arabica L. varieties revealed by AFLP and SSR markers. Theor Appl Genet 104:894–900
Arru L, Faccini N, Govoni C, Cattivelli L, Pecchioni N, Delogu G, Stanca AM, Vale G (2003) The PCR-based marker MWG2018 linked to the RDG2A leaf stripe resistance gene is a useful tool for assessing barley resistance in breeding programs. Crop Sci 43:1036–1042
Bettencourt AJ, Rodrigues CJ Jr (1988) Principles and practice of coffee breeding for resistance to rust and other disease. In: Clarke RJ, Macrae R (eds) Coffee, vol 4. Agronomy. Elsevier, London, pp 199–234
D’Oliveira B (1957) As ferrugens do cafeeiro. Rev Café Port 4:5–15
Duvick DN (1996) Plant breeding, an evolutionary concept. Crop Sci 36:539–548
Eskes AB, Toma-Braghini M (1982) Assessment methods for resistance to coffee leaf rust (Hemileia vastatrix Berk. and Br.). Plant Prot Bull 29:56–66
Frisch M, Melchinger AE (2001) The length of the intact donor chromosome segment around a target gene in marker-assisted backcrossing. Genetics 157:1343–1356
Goff SA, Ricke D, Lan TH, Presting G, Wang R, Dunn M, Glazebrook J, Sessions A, Oeller P, Varma H, Hadley D, Hutchison D, Martin C, Katagiri F, Lange BM, Moughamer T, Xia Y, Budworth P, Zhong J, Miguel T, Paszkowski U, Zhang S, Colbert M, Sun WL, Chen L, Cooper B, Park S, Wood TC, Mao L, Quail P, Wing R, Dean R, Yu Y, Zharkikh A, Shen R, Sahasrabudhe S, Thomas A, Cannings R, Gutin A, Pruss D, Reid J, Tavtigian S, Mitchell J, Eldredge G, Scholl T, Miller RM, Bhatnagar S, Adey N, Rubano T, Tusneem N, Robinson R, Feldhaus J, Macalma T, Oliphant A, Briggs S (2002) A draft sequence of the rice genome (Oryza sativa L. ssp. japonica). Science 296:92–100
Herrera JC, Combes MC, Anthony F, Charrier A, Lashermes P (2002) Introgression into the allotetraploid coffee (Coffea arabica L.): segregation and recombination of the C. canephora genome in the tetraploid interspecific hybrid (C. arabica × C. canephora). Theor Appl Genet 104:661–668
Hospital F (2001) Size of donor chromosome segments around introgressed loci and reduction of linkage drag in marker-assisted backcross programs. Genetics 158:1363–1379
Koebner RMD, Kirsch F, Torpe C, Prins R (1998) AFLPs as a source of STS markers in alien introgression. In: Slinkard AE (ed) Proc 9th Int Wheat Genet Symp, vol 1. University Extension Press, Saskatoon, pp 118–122
Kosambi DD (1944) The estimates of map distances from recombination values. Ann Eugen 12:172–175
Kukhang TD, Mawardi S, Eskes AB (1993) Studies on the inheritance of the SH3 resistance factor to coffee leaf rust. In: ASIC (ed) Proc 15th Assoc Sci Int Café (ASIC) Colloq. ASIC, Paris, pp 776–778
Kushalappa AC, Eskes AB (1989) Advances in coffee rust research. Annu Rev Phytopathol 27:503–531
Lander ES, Green P, Abrahamson J, Barlow A, Daly MJ, Lincoln SE, Newburg L (1987) mapmaker: an interactive computer package for constructing primary genetic linkage maps of experimental and natural populations. Genomics 1:174–181
Lashermes P, Combes MC, Robert J, Trouslot P, D’Hont A, Anthony F, Charrier A (1999) Molecular characterisation and origin of the Coffea arabica L. genome. Mol Gen Genet 261:259–266
Lashermes P, Combes MC, Topart P, Graziosi G, Bertrand B, Anthony F (2000a) Molecular breeding in coffee (Coffea arabica L.) In: Sera R, Soccol S, Pandey A, Roussos AR (eds) Coffee biotechnology and quality. Kluwer, Dordrecht, pp 101–112
Lashermes P, Andrzejewski S, Bertrand B, Combes MC, Dussert S, Graziosi G, Trouslot P, Anthony F (2000b) Molecular analysis of introgressive breeding in coffee (Coffea arabica L.). Theor Appl Genet 100:139–146
Liu J, Liu D, Tao W, Li W, Wang S, Chen P, Cheng S, Gao D (2000) Molecular marker-facilitated pyramiding of different genes for powdery mildew resistance in wheat. Plant Breed 119:21–24
Moreno G, Alvarado G (2000) La variedad Colombia: veinte anos de adopcion y comportalmiento frente a nuevas razas de la roya del cafeto. Cenicafe Tech Bull 22:32
Noir S, Anthony F, Bertrand B, Combes MC, Lashermes P (2003) Identification of a major gene (Mex-1) from Coffea canephora conferring resistance to Meloidogyne exigua in Coffea arabica. Plant Pathol 52:97–103
Prakash NS, Combes MC, Naveen KS, Lashermes P (2002) AFLP analysis of introgression in coffee cultivars (Coffea arabica L.) derived from a natural interspecific hybrid. Euphytica 124:265–271
Ramachandran M, Srinivasan CS (1979) Four generations of selection for resistance to race I of leaf rust in arabica cv. S.288 × ‘Kents’. Indian Coffee 43:159–161
Richly E, Kurth J, Leister D (2002) Mode of amplification and reorganization of resistance genes during recent Arabidopsis thaliana evolution. Mol Biol Evol 19:76–84
Rodrigues CJ Jr, Bettencourt AJ, Rijo L (1975) Races of the pathogen and resistance to coffee rust. Annu Rev Phytopathol 13:49–70
Rodrigues CJ Jr, Varzea VM, Godinho IL, Palma S, Rato RC (1993) New physiological races of Hemileia vastatrix. In: Proc 15th ASIC Colloquium (Montpellier). ASIC, Paris, pp 318–321
Santaram A, Sreenath HL (2000) Genetic fingerprinting of coffee genotypes with varying resistance to rust. In: Muraleedharan N, Rajkumar R (eds) Recent advances in plantation crops research. pp 57–62
Sreenivasan MS, Ram AS, Prakash NS (1994) Search for new sources of resistance to coffee rust. In: Central Coffee Research Institute (ed) Project report on “Pathology and improvement of coffee for the main diseases”, a collaborative project of CRRC, Portugal and CCRI, India. CRRC/CCRI, Portugal/India, p 24
Srinivasan KH, Narasimhaswamy RL (1975) A review of coffee breeding work done at the Government coffee experiment station, Balehonnur. Indian Coffee 34:311–321
Van der Vossen HAM (2001) Coffee breeding practices. In: Clarke RJ, Vitzthum OG (eds) Coffee recent developments. Agronomy, vol 1. Blackwell, London, pp 184–201
Vishveshwara S (1974) Periodicity of Hemileia in arabica selection. Indian Coffee 38:49–51
Vos P, Hogers R, Bleeker M, Reijans M, Van der Lee T, Hornes M, Frijters A, Pot J, Peleman J, Kuiper M, Zabeau M (1995) AFLP: a new technique for DNA fingerprinting. Nucleic Acids Res 23:4407–4414
Wagner M, Bettencourt AJ (1965) Inheritance of reaction to Hemileia vastatrix Berk & Br. in Coffea arabica L. Progress report 1960–1965, Coffee Rusts Research Center, Oeira
Acknowledgements
This work is part of the Agropolis programme (2001–2003)—Advanced research platform for genomics and biotechnology. We wish to thank Drs. R. Naidu, C.S. Srinivasan, H.L. Sreenath, and A. Santaram of the Coffee Board, India for their encouraging support extended during the period of the investigation. N.S. Prakash is indebted to the Coffee Board, Government of India for financing of the study abroad .
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by C. Möllers
Rights and permissions
About this article
Cite this article
Prakash, N.S., Marques, D.V., Varzea, V.M.P. et al. Introgression molecular analysis of a leaf rust resistance gene from Coffea liberica into C. arabica L.. Theor Appl Genet 109, 1311–1317 (2004). https://doi.org/10.1007/s00122-004-1748-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00122-004-1748-z