Abstract
One hundred and fifty F2–F3 families from a cross between two inbred sunflower lines FU and PAZ2 were used to map quantitative trait loci (QTL) for resistance to white rot (Sclerotinia sclerotiorum) attacks of terminal buds and capitula, and black stem (Phoma macdonaldii). A genetic linkage map of 18 linkage groups with 216 molecular markers spanning 1,937 cM was constructed. Disease resistances were measured in field experiments for S. sclerotiorum and under controlled conditions for P. macdonaldii. For resistance to S. sclerotiorum terminal bud attack, seven QTL were identified, each explaining less than 10% of phenotypic variance. For capitulum attack by this parasite, there were four QTL (each explaining up to 20% of variation) and for P. macdonaldii resistance, four QTL were identified, each having effects of up to 16%. The S. sclerotiorum capitulum resistance QTL were compared with those reported previously and it was concluded that resistance to this disease is governed by a considerable number of QTL, located on almost all the sunflower linkage groups.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
White rot and black stem, caused respectively by Sclerotinia sclerotiorum (Lib.) de Bary and Leptosphaeria lindquistii Frezzi (Phoma macdonaldii Boerema L.), are important pathogens of sunflower (Helianthus annuus L.). They can significantly restrict the productivity of sunflower when this oil crop is grown in humid and temperate environments.
Although S. sclerotiorum infections on capitula probably produce the greatest loss, destruction of the terminal buds by this fungus may be economically serious under favourable conditions (Peres et al. 1989). Resistance to S. sclerotiorum is under polygenic control (Vear et al. 1992; Castaño et al. 1993; Bert et al. 2002) and to date, no complete resistance is available in cultivated sunflower, but differences in susceptibility exist (Tourvieille et al. 1996).
Phoma black stem is caused by the soil-borne fungus P. macdonaldii Boerema. The fungus overwinters as pycnidia and possibly mycelium in infected residue. Conidia are released from pycnidia and spread to neighbouring plants by means of rain splashing or insects. Dark black lesions start on the stem at the base of the leaf petioles and spread along the stems. When Phoma girdles the stem base, symptoms of premature ripening and early drying may occur resulting in small heads and seeds that are empty or not completely filled, reducing seed and oil yields. The stem may be rotted completely, rendering the infected plants weak and susceptible to lodging. Yield losses attributable to Phoma have been variable, but may often be significant (Penaud and Peres 1994). Black stem can occur at any time during the growing season, but is most common after flowering and the disease is most severe when abundant moisture is available during and after flowering. No control measures are totally effective (Roustaee et al. 2000). Some cultivars are more tolerant than others, but no hybrids have been identified as being immune to the disease.
Most agronomic characters, including resistance to pathogens, show continuous variation and are strongly influenced by environmental conditions. It is well accepted that quantitative inheritance most often results from the segregation of multiple genetic factors. Identification of these is an important goal in genetics and breeding, now helped by the use of molecular markers. During the past two decades, DNA molecular markers have been used successfully to develop genetic maps in numerous plant species and to detect loci controlling quantitative trait variations. Several F2 or RIL genetic maps have been developed for the sunflower, based on RFLP (Gentzbittel et al. 1995), AFLP (Flores Berrios et al. 2000a, b), DALP (Langar et al. 2003) and microsatellites (Tang et al. 2002). Many of the maps are applicable to only one population, but the linkage groups defined in the first map to be published (Gentzbittel et al. 1995) have since been used for a wide range of populations (e.g. Mestries 1996; Gentzbittel et al. 1998, 1999; Lu et al. 1999; Bert et al. 2002, 2003; Radwan et al. 2002).
Such genetic maps have allowed the mapping of quantitative trait loci (QTL) for a wide range of characteristics such as somatic embryogenesis and in vitro organogenesis (Flores Berrios et al. 2000a, b), photosynthesis and water status (Hervé et al. 2001), growing degree days to flowering and photoperiod response (Leon et al. 2001), seed oil content, developmental and agronomic traits (Bert et al. 2003; Mokrani et al. 2002). Mapping of QTL associated with disease resistance in sunflowers has concerned resistance to S. sclerotiorum, P. macdonaldii, phomopsis stem canker (Diaporthe helianthi), and downy mildew (Plasmopara halstedii). Concerning S. sclerotiorum resistance, Mestries (1996), Mestries et al. (1998) and Genzbittel et al. (1998 and unpublished) studied F2/F3 populations from four crosses involving five inbred sunflower lines with measurements of resistance to leaf and capitulum attacks. Bert et al. (2002) used a different F2/F3 population to study the same S. sclerotiorum resistances, together with phomopsis resistance. The QTL reported in these studies were based on the linkage groups defined by Gentzbittel et al. (1995, 1999) so they can be compared. In contrast, the only QTL for resistance to P. macdonaldii reported previously (Rachid Al-Chaarani et al. 2002) used a different map, although in an RIL population of which one parent was common with the F2/F3 studied by Mestries (1996).
This paper reports studies of QTL for S. sclerotiorum and P. macdonaldii resistance in a further F2/F3 population from a cross between two inbred lines of different genetic origins from those previously published. This material permitted studies of resistance to S. sclerotiorum terminal bud attack in addition to capitulum attack, as well as for P. macdonaldii resistance, and thus allowed comparisons between the locations of all these QTL. More generally, the objective was to compare the QTL detected for capitulum resistance to S. sclerotiorum in all the different genotypes so far reported by our group; their number, frequency and co-localisations, in order to obtain a better overall understanding of resistance to this disease in cultivated sunflower.
Materials and methods
Sunflower genotypes
The two parental inbred lines of the cross were bred by the INRA (Institut National de la Recherche Agronomique, France). FU, an unbranched maintainer selected from genotypes of Rumanian and Russian origin, shows a high level of resistance to S. sclerotiorum capitulum attack. PAZ2, a male fertility restorer line, bred from a cross between Yugoslav and French restorers and a population from Zambia, exhibits the apical branching gene (b1) phenotype, a high level of capitulum resistance to S. sclerotiorum but some susceptibility to terminal bud attack by this parasite. This line had been shown to have a higher level of resistance to black stem than FU. The male fertile forms of the two lines were crossed. The F1 plants were selfed by covering the capitula with grease-proof paper bags a few days before flowering to obtain the 150 F2 plants, which were in turn selfed to obtain the 150 F3 families.
Experimental plan and resistance tests
S. sclerotiorum field trials were conducted in 2000 on the 150 F3 families in a randomised block design with two replications of 25 plants. At the star bud stage (1 month before flowering) S. sclerotiorum attack on terminal buds was observed. Plants were counted as infected if the young leaves around the developing capitulum showed the characteristic pale brownish-pink colour, or in rare cases, if the rot had reached the main stem. The percentage of infected plants was calculated. The ascospore test used was that described by Tourvieille and Vear (1984). Capitula were infected with an ascospore suspension at the beginning of flowering. The test gave measurements of percentage attack and a latency index, which was the number of days between infection and the appearance of symptoms compared with the mean delay for two control inbreds infected on the same day.
The P. macdonaldii resistance tests were carried out on seedlings in a growth chamber, in 2001. A monopycniospore isolate of the parasite (MP6) from naturally infected plants in the south-west of France was used in this study, designed as a randomised complete block with two replications of 26–44 seedlings for each of the 150 genotypes. Twelve-day-old seedlings were inoculated at the junction of the cotyledon petiole and hypocotyl with 20 μl of a pycniospore suspension (106 pycniospore per ml of water) as described by Rachid Al-Chaarani et al. (2002). Seven days after inoculation, the two cotyledon petioles of the seedling were scored according to the proportion of the petiole area exhibiting disease symptoms (necrosis), giving a figure between 1 (no symptoms) and 9 (cotyledons completely necrotic).
Molecular analysis and map construction
RFLP probes (available upon request to I. Jouan) were chosen among those used for the sunflower CARTISOL linkage map (Gentzbittel et al. 1995, 1999), both for their regular distribution over all linkage groups and their polymorphism between FU and PAZ2, using two restriction enzymes for DNA digestion, EcoRI and HindIII. DNA extraction, digestion by restriction enzymes and Southern hybridisation were carried out as described previously (Gentzbittel et al. 1995). RFLP markers were dominantly and codominantly scored.
The AFLP protocol developed by Vos et al. (1995) was followed with minor modifications as described in Bert et al. (1999) using the silver nitrate staining method. Sixty-four AFLP primer combinations were tested in order to compare fingerprint patterns and determine those which produced clearly detectable bands and revealed high levels of polymorphism. Among the 64 primer pairs tested, ten were selected (E32/M48, E32/M49, E32/M50, E32/M59, E32/M60, E32/M61, E32/M61, E33/M48, E33/M59 and E33/M61) for mapping. AFLP markers were named according to their primer pair combination and their estimated molecular size, and were scored dominantly.
For data scoring, segregation distortion was checked and highly significantly (P<0.001) skewed markers were removed from further analyses in order to avoid false linkages in map construction. The software package JOINMAP 2.0 (Stam 1993) was used to estimate segregation distortion and determine linkage groups and the software Mapmaker 3.0b (Lander et al. 1987) was used to order loci and construct the map. Analyses were performed with a LOD score threshold of 4.0 and a maximum of recombination value θ=0.40 for grouping and ordering markers. Kosambi’s mapping function was applied for map distance calculation.
Statistical analyses and QTL detection
Before analysis, the normality of the different traits were assessed according to the Shapiro and Wilk test (PROC UNIVARIATE of SAS; SAS Institute Inc.). When necessary, data were converted using the arc-sinus-square root (for percentage capitulum attack), and the square root (for percentage terminal bud attack) to fit a normal distribution.
Broad-sense heritabilities were calculated as followed: h 2=σ2 G /σ2 G +σ2 e . Phenotypic correlation analyses among traits were also performed using Statgraphics/Plus to detect associations between resistances tests to the two pathogens.
Data sets were analysed by generalised linear modelling (GLM) using the SAS program and Statgraphics/Plus. The normality of the residuals and the homogeneity of the variances were also checked using Bartlett’s test. Using the framework map orders, QTL were detected using the methods of simple interval mapping (SIM) of Mapmaker/QTL (Lincoln et al. 1992) and composite interval mapping (CIM) of QTL Cartographer 1.13 (Basten et al. 1999) with model 6 of Zmapqtl and a LOD score threshold of 3.0. The advantage of CIM over SIM is that it can allow more than one QTL to be detected on a given chromosome and increases the precision of QTL location and effect (r 2) (Basten et al. 1999).
Results
Disease observations and tests
The observations of natural S. sclerotiorum attack on terminal buds (Table 1) confirmed that PAZ2 was quite susceptible (30%), whereas FU showed very few symptoms (5%). Attack of the F3 families varied from 0 to 100%, with about 1/4 (39/150) showing more symptoms than PAZ2. Differences between families were highly significant whereas there was no replication effect. Weather conditions in 2000 were such that of the 7,500 plants observed, disease symptoms destroyed the capitula of only seven plants. For the others, the symptoms dried and did not affect the S. sclerotiorum ascospore infections made on the capitula at flowering.
Conditions for the ascospore test were extremely favourable in 2000, and almost all plants showed symptoms, with no difference between the two parents (FU, 98%; PAZ2, 96%). However, some transgressive F3 families showed only 60% attack. The high level of attack led to a precise measurement of the latency index since this was based on the results of nearly all the plants in each replication, and the F genotype was greater for the second characteristic than for the first (% attack, 4.54; latency index, 7.97). However, for both characteristics, broad sense heritabilities were quite high (% attack, 0.64; latency index, 0.78).
The P. macdonaldii infections were made under controlled conditions. They confirmed previous observations that PAZ2 is less susceptible (mean note, 3.9) than FU (mean note, 6.4). As for the S. sclerotiorum reactions, there were transgressive F3 families, in this case, in both directions, more susceptible and more resistant. Heritability was very high (0.94), probably due to the fact that there were no environmental effects and the observations were made only 7 days after infection.
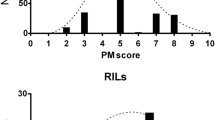
Frequency distributions of the phenotypes of the 150 F3 families for each resistance test are shown in Fig. 1. All traits showed continuous, unimodal and normal distributions (after transformation). The correlation coefficient calculated by phenotypic regression between the S. sclerotiorum latency index and the percentage attack was significant (r=−0.55). For families showing 100% attack, the latency indices varied from 0.73 to 1.47, whereas for the lower levels of attack (60–70%), the latency indices were at least 1.13. Percent attack was not correlated with reactions to terminal bud attack or P. macdonaldii. The latency index was weakly correlated with susceptibility to P. macdonaldii ( r=0.20) and the latter characteristic was also slightly negatively related to the terminal bud reaction (r=−0.20), so that there appears to be some opposition between resistances to S. sclerotiorum and P. macdonaldii.
Molecular analysis and map construction
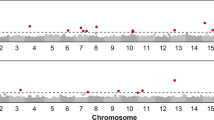
The complete map included one morphological (the apical branching gene b1), 90 RFLP and 125 AFLP markers. Segregation analysis was performed on the 150 F2 plants. Overall, a total of 216 markers were mapped onto 17 linkage groups, which were identified and noted using concordance of RFLP loci on the CARTISOL map (Gentzbittel et al. 1995, 1999) and one additional group noted A (Fig. 1). The total map length covered by all the 216 markers was 1,937.5 cM. The average density of markers was approximately one per 9.0 cM and the greatest length between two markers was 32 cM. Linkage group 1, with 13 markers, was the longest (186 cM) while linkage group 16 was the smallest with only 45.3 cM and eight markers.
Quantitative trait loci detection for resistance to S. sclerotiorum and P. macdonaldii
Quantitative trait loci that affected resistance of sunflower to both pathogens are presented in Table 2 and Fig. 2. QTL were designated as “att” (percentage of attack), “lat” (latency index), “bud” (terminal bud) and “bsr” (black stem resistance) followed by number of the linkage group carrying the QTL. SIM and CIM detected almost the same QTL and the LOD score values were similar. However, the percentages of variation explained r 2) were generally lower when calculated by CIM than by SIM (CIM mean effect of QTL, 9.22%; SIM mean effect, 14.22%), especially for S. sclerotiorum capitulum attack.
Genetic linkage map of the sunflower genome based on an intraspecific F2 population of a cross between inbred lines FU and PAZ2 at a LOD score of 4.0. Genetic distances are in Kosambi cM. On the left of each linkage group, the QTL detected are indicated in bold type. Linkage groups are those defined by Gentzbittel et al. (1995)
Seven QTL for capitulum resistance to S. sclerotiorum (three for percentage of attack and four for latency index) and seven others for resistance to S. sclerotiorum on terminal buds were identified. Analysis of resistance to P. macdonaldii revealed four QTL. Among the 18 QTL detected in this study, only eight were significant by CIM, and only two were significant by SIM.
For S. sclerotiorum attacks on terminal buds, the seven QTL were identified on linkage groups 1, 7–10, 14 and 15, each accounting for only 6–10% of the phenotypic variance. Except for the QTL on group 7, all alleles linked to resistance were from the parental line FU.
Three genetic regions were identified for resistance to ascospore penetration on capitula (percentage attack) on linkage groups 6, 7 and 9. These QTL explained from 9% to 20% of observed phenotypic variance and came from both FU and PAZ2. Four QTL were identified for disease development (latency index), located on linkage groups 6, 7, 9 and 17 with r 2 of 3–19%. Again both parents provided some favourable alleles. The two resistance characteristics showed close or identical chromosomal locations of the QTL on groups 6, 7 and 9 (Fig. 2), with similar relative magnitude effects.
Resistance to black stem involved four QTL on linkage groups 1, 3, 15 and 16, each explaining 5–16% of phenotypic variance. Two QTL came from each parent, with very similar levels of phenotype explanation, in contrast with the behaviour of the parental lines, since FU was more susceptible than PAZ2.
Discussion
The length of the map was in the range of those described recently by Flores Berrios et al. (2000a, b) using mainly AFLPs, and updated by Rachid Al-Chaarani et al. (2002), Bert et al. (2002), Mokrani et al. (2002) and Langar et al. (2003). It was also significantly greater than the RFLP maps described by Berry et al. (1995), Gentzbittel et al. (1995, 1999) and the integrated RFLP-AFLP map described by Gedil et al. (2001). When using dominant markers, mostly on F2 populations with the maximum likelihood method, estimations of the recombination rate between loci are not very precise. The standard deviations of the estimated distances may be as great as the distances and thus may affect local locus ordering and give some overestimation of distances between loci (Lorieux and Gonzales de Leon 1993).
The inheritance of resistance to S. sclerotiorum terminal bud attack was reported by Achbani et al. (1996) as quantitative, but under mostly additive control. Since cultivated sunflower genotypes show quite clear differences, from very susceptible to completely resistant, it has been thought that this characteristic may be less polygenic than resistance to capitulum attack. Thus, it was surprising to find many QTL, all with small effects, although of the six QTL with a LOD of more than 3.0 for CIM, only two had a LOD of more than 3.0 for SIM.
The favourable conditions for S. sclerotiorum capitulum attack in 2000, with the parental lines being attacked at a rate of more than 90% whereas in 1997 they had only shown 40% attack (Bert 2002), confirmed that resistance is not complete, with relatively resistant genotypes appearing to be susceptible when conditions favour the pathogen. The QTL for resistance to capitulum attack did not differ between CIM and SIM for the percentage attack rate, and with only two differences for latency index (group 6 CIM only, group 9 SIM only), they may therefore be considered as quite robust. Other than the linkage to branching for the branched parent PAZ2, the most important QTL is situated on group 6, also with PAZ2 as the favourable parent, FU contributing small effects on groups 9 and 17. Thus it is confirmed that the two lines provide some different alleles for resistance.
Table 3 summarises the QTL for S. sclerotiorum capitulum resistance found by our group in the six crosses between cultivated sunflower lines, all using the same linkage group nomenclature. At least 14 linkage groups have been shown to carry QTL, only groups 11 and 12 did not carry QTL; group 3 is not listed because significant QTL appeared in only one replication. These results confirm earlier field observations that resistance is polygenic and, not surprisingly, with a continuous range of variation in phenotypes (Vear and Tourvieille de Labrouhe 1988).
For each cross, the markers used depended on which were polymorphic between the parents, so that very often there were two or more common RFLP or gene markers which permitted identification and ordering of the linkage groups. Since, in addition, the confidence limits of the QTL were often quite wide, it was generally not possible to be sure whether the QTL from the different parental lines had the same position on a given linkage group. If it is assumed that there is only one QTL for resistance to S. sclerotiorum capitulum attack on each linkage group, the most frequent (for both percentage attack and latency index) is that on group 7, probably related to the branching phenotype. Other QTL which appeared quite frequently were situated on groups 9, 6 and 13. Group 17 carries QTL with favourable alleles from both SD and FU and it may be noted that these two sterility maintainer lines both provide favourable alleles on groups 9 and 17. QTL for percentage attack were found on group 8 in PSC8 and group 10 in PAC1 and PAC2. The large QTL on linkage group 1 appears to be specific to PAC1. The sunflower lines used in these studies were chosen because of their different origins but, although they are typical of modern cultivated sunflowers, they do not cover the whole range of genotypes used in breeding and it appears quite probable that other QTL exist in other sunflower lines.
It would be useful to know which of these QTL are the most important in providing a resistant phenotype. Since the earlier work only reported data generated by SIM, comparisons of the relative effects of the QTL can only use this calculation. The mean level of effects of the QTL identified in crosses studied over 1 year and in one location only was generally higher (FU × PAZ2, 23% (this paper); GH × PAC2, 44% (Mestries 1996) than that in the other crosses, studied over 2 years and/or in different locations (SD × PAC1, 14% (Mestries 1996; Gentzbittel et al. 1998 and unpublished); XRQ × PSC8, 13% (Bert et al. 2002); CP73 × SD, 22% (Gentzbittel et al., unpublished); CP73 × PAC1, 27 or 16% if the large QTL on group 1 from PAC1 [Gentzbittelet al. (1998) is excluded]. Thus, with the possible exception of this last QTL, it is not yet really possible to conclude which QTL have the largest effects and therefore would be the most useful in breeding. It will be necessary to observe plants with different combinations of favourable alleles from the different origins under the same experimental conditions to determine the QTL which explain the largest proportion of phenotypic variation and the best level of resistance which could be obtained by pyramiding them.
The QTL for capitulum resistance on linkage group 1 linked to a serine/threonine protein kinase gene (Gentzbittel et al. 1998) appears to be specific to PAC1, with a large effect in a cross with a very susceptible line, CP73 (52–60%) and a smaller effect in a cross with another resistant line, SD (15–16%). In the present cross, FU × PAZ2, there are small QTL for resistance to terminal bud attack (allele from FU) and to P. macdonaldii (allele from PAZ2) linked with this kinase. This kind of co-localisation could be due to physiological relationships between mechanisms of resistance to different pathogens. According to the sunflower genotype tested, the receptor of the kinase-like gene may confer resistance to different plant parts and different pathogens like S. sclerotiorum and P. macdonaldii. This gene may thus be important in general defence mechanisms but does not provide complete resistance. For S. sclerotiorum capitulum attack for example, the favourable allele in PAC1 needs to be combined with the main favourable alleles on other linkage groups.
Analyses of QTL associated with resistance to extension of S. sclerotiorum mycelium on sunflower capitula and on leaves were made by Mestries et al. (1998) from a cross between inbred lines GH and PAC2. Individual QTL explained between 9 and 48% of the phenotypic variability. One of these QTL appeared to be involved in resistance in both plant parts to S. sclerotiorum attack while the others appeared to be specific for resistance in one part of the plant. The same approach was undertaken in a comparative genetic analysis for resistance to S. sclerotiorum with another pathogen: D. helianthi from a cross between two other inbred lines XRQ and PSC8 (Bert et al. 2002) which revealed the existence of a multi-disease resistance QTL (MDR-QTL) common to resistance on leaves to both fungi.
For resistance to P. macdonaldii, a comparison can be made with resistance to S. sclerotiorum terminal bud attack, since again four small QTL were detected by CIM, two by SIM, and in spite of a broad sense heritability of 0.94, the percentage contribution to variability explained is small (only about 20%). This could be because the map is not saturated, however, this is in contrast to the study of Rachid Al-Chaarani et al. (2002) who also studied resistance to P. macdonaldii and showed seven QTL for resistance, which explained 92% of the phenotypic variation of the trait. However, both studies revealed QTL with moderate individual effects, comprising between 5 and 20%.
In conclusion, the main aim of this QTL determination was to permit comparison of QTL for S. sclerotiorum and P. macdonaldii resistances in different populations of cultivated sunflower to determine the QTL that marker assisted selection programmes could usefully combine. Breeding sunflowers for resistance to S. sclerotiorum and P. macdonaldii is complicated by the fact the two pathogens can attack several parts of the plant and the level of resistance may be different for each plant part (Castano et al. 1993). The QTL identified in this study could be usefully combined with some of S. sclerotiorum resistance QTL detected by Mestries et al. (1998) and Bert et al. (2002) and some of the resistance QTL detected by Rachid Al-Chaarani et al. (2002) in order to transfer favourable alleles into elite sunflower lines giving high yielding varieties.
References
Achbani EH, Vear F, Tourvieille De Labrouhe D (1996) Resistance of sunflower (Helianthus annuus L.) to terminal bud attack by Sclerotinia sclerotiorum (Lib.) de Bary. Eur J Plant Pathol 102:421–429
Basten CJ, Weir BS, Zeng ZB (1999) QTL Cartographer version 1.13: program in statistical genetics. Department of Statistics, North Carolina State University
Berry ST, Leon AJ, Hanfrey CC, Challis P, Bukholz A, Barnes SR, Rufeber GK, Lee M, Caligari PDS (1995) Molecular marker analysis of Helianthus annuus L. 2. Construction of an RFLP linkage map for cultivated sunflower. Theor Appl Genet 91:195–199
Bert PF (2002) Etude génétique et marquage moléculaire des facteurs de résistance aux principaux champignons pathogènes du tournesol (Helianthus annuus L.). PhD thesis, Université Clermont Ferrand II
Bert PF, Charmet G, Sourdille P, Hayward MD, Balfourier F (1999) A high-density molecular map for ryegrass (Lolium perenne) using AFLP markers. Theor Appl Genet 99:445–452
Bert PF, Jouan I, Tourvieille de Labrouhe D, Serre F, Nicolas P, Vear F (2002) Comparative genetic analysis of quantitative traits in sunflower ( Helianthus annuus L.) 1. Characterisation of QTL involved in resistance to Sclerotinia sclerotiorum and Diaporthe helianthi. Theor Appl Genet 105:985–993
Bert PF, Jouan I, Tourvieille de Labrouhe D, Serre F, Nicolas P, Vear F (2003) Comparative genetic analysis of quantitative traits in sunflower (Helianthus annuus L.) 2. Characterisation of QTL involved in developmental and agronomic traits. Theor Appl Genet 107:181–189
Castaño F, Vear F, Touvielle de Labrouhe D (1993) Resistance of sunflower inbred lines to various forms of attack by Sclerotinia sclerotiorum and relations with some morphological characters. Euphytica 68:85–98
Flores Berrios E, Gentzbittel L, Kayyal H, Alibert G, Sarrafi A (2000a) AFLP mapping of QTLs for in vitro organogenesis traits using recombinant inbred lines in sunflower (Helianthus annuus L.). Theor Appl Genet 101:1299–1306
Flores Berrios E, Gentzbittel L, Alibert G, Sarrafi A (2000b) Genotypic variation and chromosomal location of QTLs for somatic embryogenesis revealed by epidermic layers culture of recombinant inbred lines in the sunflower (Helianthus annuus L.). Theor Appl Genet 101:1307–1312
Gedil MA, Wye C, Berry S, Segers B, Peleman J, Jones R, Leon A, Slabaugh MB, Knapp SJ (2001) An integrated restriction fragment length polymorphism-amplified fragment length polymorphism linkage map for cultivated sunflower. Genome 44:213–221
Gentzbittel L, Vear F, Zhang Y-X, Bervillé A, Nicolas P (1995) A composite map of expressed sequences and phenotypic traits of the sunflower (Helianthus annuus L.) genome. Theor Appl Genet 90:1079–1086
Gentzbittel L, Mouzeyar S, Badaoui S, Mestries E, Vear F, Tourvieille D, Nicolas P (1998) Cloning of molecular markers for disease resistance in sunflower, Helianthus annuus L. Theor Appl Genet 96:519–525
Gentzbittel L, Mestries E, Mouzeyar S, Mazeyrat F, Badaoui S, Vear F, Tourvieille D, Nicolas P (1999) A composite map of expressed sequences and phenotypic traits of the sunflower (Helianthus annuus L.) genome. Theor Appl Genet 99:218–234
Hervé D, Fabre F, Flores Berrios E, Leroux N, Al Chaarani G, Planchon C, Sarrafi A, Gentzbittel L (2001) QTL analysis of photosynthesis and water status traits in sunflower (Helianthus annuus L.) under greenhouse conditions. J Exp Bot 52:1857–1864
Lander ES, Green P, Abrahamson J, Barlow A, Daly MJ, Lincoln SE, Newburg L (1987) Mapmaker: an interactive computer package for constructing primary genetic linkage maps of experimental and natural populations. Genomics 1:174–181
Langar K, Lorieux M, Desmarais E, Griveau Y, Gentzbittel I, Bervillé A (2003) Combined mapping of DALP and AFLP markers in cultivated sunflower using F9 recombinant inbred lines. Theor Appl Genet 106:1068–1074
Leon AJ, Lee M, Andrade FH (2001) Quantitative trait loci for growing degree days to flowering and photoperiod response in sunflower (Helianthus annuus L.). Theor Appl Genet 102:497–503
Lincoln E, Daly MJ, Lander ES (1992) Mapping genes controlling quantitative trait loci (QTL) using Mapmaker/QTL version1.1: a tutorial and reference manual, 2nd edn. Whitehead Institute Technical Report, Cambridge
Lorieux M, Gonzales de Leon D (1993) Mapping populations: a few guidelines for prospective developers. In: Proceedings of INIBAP workshop on biotechnology for banana and plantain, San Jose, Costa Rica, pp 46–51
Lu YH, Gagne G, Grezes-Besset B, Planchard P (1999) Integration of a molecular linkage group containing the broomrape resistance gene Or5 into an RFLP map in sunflower. Genome 42:453–456
Mestries E (1996) Recherche de QTL (Quantitative Trait Loci) impliqués dans la résistance du tournesol (Helanthus annuus L.) à la pourriture blanche, causée par Sclerotinia sclerotiorum (Lib.) de Bary. PhD thesis, Université Clermont Ferrand II
Mestries E, Gentzbittel L, Tourvielle de Labrouhe D, Nicolas P, Vear F (1998) Analyses of quantitative trait loci associated with resistance to Sclerotinia sclerotiorum in sunflowers (Helianthus annuus L.) using molecular markers. Mol Breed 4:215–226
Mokrani L, Gentzbittel L, Azanza F, Al-Chaarani G, Sarrafi A (2002) Mapping and analysis of quantitative trait loci for grain oil content and agronomic traits using AFLP and SSR in sunflower (Helianthus annuus L.). Theor Appl Genet 106:149–156
Penaud A, Peres A (1994) Phoma du tournesol. Oléoscope 15:37
Peres A, Regnault Y, Allard LM (1989) Sclerotinia sclerotiorum du bourgeon terminal du tournesol: Etude de la protection fongicide. In: A.N.P.P. 3rd international conference on plant diseases, Bordeaux, 3–5 December 1991, pp 401–405
Rachid Al-Chaarani G, Roustaee A, Gentzbittel L, Mokrani L, Barrault G, Dechamp-Guillaume G, Sarrafi A (2002) A QTL analysis of sunflower partial resistance to downy mildew (Plasmopara halstedii) and black stem (Phoma macdonaldii) by the use of recombinant inbred lines (RILs). Theor Appl Genet 104:490–496
Radwan O, Bouzidi MF, Vear F, Phillipon J, Tourvieille De Labrouhe D, Nicolas P, Mouzeyar S (2002) Identification of non-TIR-NBS-LRR markers linked to the Pl5/Pl8 locus for resistance to downy mildew in sunflower. Theor Appl Genet 106:1438–1446
Roustaee A, Barrault G, Dechamp-Guillaume G, Lesigne P, Sarrafi A (2000) Inheritance of partial resistance to black stem (Phoma macdonaldii Boerema L.) in sunflower. Plant Pathol 49:396–401
Stam P (1993) Construction of integrated genetic linkage maps by means of a new computer package: JoinMap. Plant J 3:739–744
Tang S, Yu JK, Slabaugh MB, Shintani DK, Knapp SJ (2002) Simple sequence repeat map of the sunflower genome. Theor Appl Genet 105:1124–1136
Tourvieille D, Vear F (1984) Comparaison de méthodes d’estimation de la résistance du tournesol à Sclerotinia sclerotiorum (Lib.) De Bary. Agronomie 4:517–525
Tourvieille D, Mestries E, Vear F (1996) Multilocal Sclerotinia sclerotiorum resistance tests. In: ISA symposium “Disease tolerance in sunflower”, Beijing, 13 June 1996, pp 71–79
Vear F, Tourvieille De Labrouhe D (1988) Heredity of resistance to Sclerotinia sclerotiorum in sunflower. II. Study of capitulum resistance to natural and artificial ascospore infections. Agronomie 8:503–508
Vear F, Tourvieille D, Castaño F (1992) Recurrent selection for sunflower capitulum resistance to attack by Sclerotinia sclerotiorum. In: Proceedings of 13th international sunflower conference, Pisa, pp 1275–1280
Vos P, Hogers R, Bleeker M, Reijans M, van de Lee T, Hornes M, Frijters A, Pot J, Peleman J, Kuiper M, Zabeau M (1995) AFLP: a new technique for DNA fingerprinting. Nucleic Acids Res 23:4407–4414
Acknowledgements
We would like to thank H. Bony, G. Joubert, J. Philippon, S. Roche, P. Walser, A. Jalte, H. Abd El-Wahab and the personnel of the INRA farms for their help with the disease resistance tests, F. Cambon for laboratory work, and M. Martignac for computer programmes. P.-F.B. received a INRA – CETIOM doctoral scholarship and the programme was supported by PROMOSOL and CETIOM. A grant for some equipment was provided by a State-Region contract “Qualité des Aliments”.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by C. Möllers
Rights and permissions
About this article
Cite this article
Bert, P.F., Dechamp-Guillaume, G., Serre, F. et al. Comparative genetic analysis of quantitative traits in sunflower (Helianthus annuus L.). Theor Appl Genet 109, 865–874 (2004). https://doi.org/10.1007/s00122-004-1701-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00122-004-1701-1