Abstract
Gametophytic self-incompatibility (GSI) in sweet cherry is determined by a locus S with multiple alleles. In the style, the S-locus codifies for an allele-specific ribonuclease (S-RNase) that is involved in the rejection of pollen that carries the same S allele. In this work we report the cloning and genomic DNA sequence analysis including the 5′ flanking regions of four S-RNases of sweet cherry (Prunus avium L., Rosaceae). DNA from the cultivars Ferrovia, Pico Colorado, Taleguera Brillante and Vittoria was amplified through PCR using primers designed in the conserved sequences of sweet cherry S-RNases. Two alleles were amplified for each cultivar and three of them correspond to three new S-alleles named S 23 , S 24 and S 25 present in 'Pico Colorado', 'Vittoria' and 'Taleguera Brillante' respectively. To confirm the identity of the amplified fragments, the genomic DNA of these three putative S-RNases and the allele S 12 amplified in the cultivar Ferrovia were cloned and sequenced. The nucleotide and deduced amino-acid sequences obtained contained the structural features of rosaceous S-RNases. The isolation of the 5′-flanking sequences of these four S-RNases revealed a conserved putative TATA box and high similarity among them downstream from that sequence. However, similarity was low compared with the 5′-flanking regions of S-RNases from the Maloideae. S 6 - and S 24 -RNase sequences are highly similar, and most amino-acid substitutions among these two RNases occur outside the rosaceous hypervariable region (RHV), but within another highly variable region. The confirmation of the different specificity of these two S-RNases would help elucidate which regions of the S-RNase sequences play a role in S-pollen specific recognition.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Sweet cherry (Prunus avium L), like other species of the Rosaceae, Solanaceae and Scrophulariaceae, exhibits stylar monofactorial gametophytic self-incompatiblity (GSI) (de Nettancourt 2001). The self-incompatibility reaction in this system is determined by a single locus (S) with multiple alleles, and fertilisation only takes place when the S allele in the haploid genome of the pollen grain is different from the two S alleles in the diploid tissue of the style. In the 1980s, it was observed that the S locus in the style of the Solanaceae codifies an S-specific ribonuclease (S-RNase) (McClure et al. 1989) and that this protein is responsible for the rejection in the style of pollen that carries the same S-allele (Huang et al. 1994; Lee et al. 1994; Murfett et al. 1994). Several studies carried out later in Rosaceae species, such as almond (Prunus dulcis) (Tao et al. 1997; Ushijima et al. 1998a), apple (Malus x domestica) (Sassa et al. 1994; Broothaerts et al. 1995), European pear (Pyrus communis) (Zuccherelli et al. 2002), Japanese apricot (Prunus mume) (Yaegaki et al. 2001), Japanese pear (Pyrus serotina) (Sassa et al. 1992, 1993; Ishimizu et al. 1998), sour cherry (Prunus cerasus) (Yamane et al. 2001) and sweet cherry (Tao et al. 1999a) have shown that stylar RNases similar to solanaceous S-RNases are involved in the incompatibility response and, thus, that a similar mechanism to that described in the Solanaceae operates in the Rosaceae to control GSI in the style.
The isolation and comparison of the cDNA of several S-RNases of almond, apple, Japanese pear and sweet cherry, has revealed a series of common structural features of rosaceous S-RNases. As in solanaceous S-RNases (Iorger et al. 1991) five conserved regions (C1, C2, C3, RC4 and C5) have been identified, although one of them (RC4) is specific to the Rosaceae and shows no homology to the corresponding region in the Solanaceae (Sassa et al. 1996, Ushijama et al. 1998a). The two hypervariable regions found in the Solanaceae (HVa and HVb), which are solid candidates for S-haplotype specificity (Matton et al. 1997), are represented in the Rosaceae by a unique hypervariable region (RHV) located in the same region as HVa (Ushijama et al. 1998a). Phylogenetic analysis of these S-RNases has also shown that the rosaceous S-RNases fall in two distinct groups that correlate with their subfamily classification, Maloideae and Prunoideae (Ushijama et al. 1998a; Ma and Oliveira 2002). Genomic DNA isolation of rosaceous S-RNases showed that in Japanese pear (Ushijama et al. 1998a,b), European pear (Zuccherellili et al. 2002), apple (Janssens et al. 1995) and almond (Tamura et al. 2000), there is a single intron inserted within the hypervariable region, while in sweet cherry (Tao et al. 1999c; Yamane et al. 2000) and other almond S-RNases (Ma and Oliveira 2001) an additional intron is found within the junction between the signal peptide and the mature protein.
The recent isolation of the S-RNase cDNAs of several S-alleles of sweet cherry (Tao et al. 1999a; Sonneveld et al. 2001; Wiersma et al. 2001) has allowed to characterise the S-allele constitution of sweet cherry cultivars by PCR analysis. Using the PCR primers derived from the conserved sequences of sweet cherry S-RNases (Tao et al. 1999a; Wiersma et al. 2001) we have screened two germplasm collections of sweet cherry cultivars to identify their S-allele genotypes (Wünsch and Hormaza, in press). Local Italian and Spanish sweet cherry cultivars have revealed the presence of at least three new sweet cherry S-alleles. In this work we report the genomic DNA sequence including the 5′ flanking regions of those three new S-RNases, as well as the sequence of the previously described allele S 12 . Since no 5′ flanking regions have been described in the Rosaceae, we also compare the 5′ flanking sequences of sweet cherry S-RNases with those available from the Maloideae, and discuss the results in terms of their implications for S-RNase evolution and pollen-pistil specific recognition.
Materials and methods
Plant material and isolation of genomic DNA
Several sweet cherry cultivars of known S-allele genotype, including 'Summit' (S 1 S 2 ), 'Bing' (S 3 S 4), 'Hedelfingen' (S 3 S 5), 'Hartland' (S 3 S 6) and 'Burlat' (S 3 S 9 ), were chosen to establish the fragment sizes of their corresponding S-RNases after amplification by PCR. Each one of these cultivars belongs to a different self-incompatibility group and their S-allele constitution has been confirmed in various works (Bošković and Tobutt 2001; Sonneveld et al. 2001; Wiersma et al. 2001). Four cultivars, Ferrovia, Pico Colorado, Taleguera Brillante and Vittoria, were chosen to sequence four new putative S-RNases, as some of the fragments amplified by PCR in these cultivars were different from those described previously (Tao et al. 1999a; Wiersma et al. 2001). All the plant material was collected from the SIA-DGA experimental orchards located at the Campus de Aula Dei in Zaragoza, Spain. The S-allele nomenclature used throughout this work follows that proposed by Hauck et al. (2001) and Tobutt at al. (2001).
Total genomic DNA for PCR analysis was isolated according to Hormaza (2002). The protocol was modified for the isolation of genomic DNA for 'genome walking' where 5 g of young leaves were ground in 40 ml of extraction buffer (100 mM of Tris-HCl, 1.4 M of NaCl, 20 mM of EDTA, 2% CTAB, 1% PVP and 0.1% NaHSO3) and 20 μl of 2-mercaptoethanol. The samples were then incubated at 65°C for 30 min, mixed with 20 ml of chloroform-isoamyl alcohol (24:1) and centrifuged at 6,000 g for 10 min. The upper phase was recovered and the nucleic acid was precipitated by adding 12 ml of cold isopropanol. The precipitate was recovered by centrifugation at 6,000 g for 5 min, washed in 10 ml of 10 mM ammonium acetate in 76% ethanol, dried overnight and resuspended in 1 ml of modified TE buffer (10 mM of Tris-HCl, 0.1 mM of EDTA, pH 8.0). Five-hundred microliters of the extracted genomic DNA were then further purified by extracting with an equal volume of phenol-chloroform-isoamyl alcohol (24:24:1), followed by another extraction with chloroform-isoamyl alcohol (24:1), and incubated with RNase (10 μl/ml) during 30 min at 37°C. The DNA was then ethanol-precipitated in 1/10 vol. of 3 M sodium acetate and 2.5 vol. of cold ethanol, washed with 70% ethanol, dried and resuspended in modified TE. The extracted DNA was quantified and diluted to 100 ng/μl with modified TE.
PCR amplification of S-alleles
The PCR to identify the S-alleles of the different cultivars analysed was carried out using the primer pairs PruT2-PruC4R and PruC2-PruC4R (Tao et al. 1999a), and SI19-SI20, SI31-SI32 (Wiersma et al. 2001). PCR reactions were performed in 20-μl volumes containing 20 mM of Tris-HCl, pH 8.4, 50 mM of KCl, 2 mM of MgCl2, 0.1 mM of each dNTP, 0.2 μM of each primer, 40 ng of genomic DNA, 0.45 units of Taq polymerase (Invitrogen, Carlsbad, Calif., USA) and a drop of mineral oil. Reactions were carried out on a PTC-100 (MJ Research, Watertown, Mass., USA) thermocycler using the following temperature profile: an initial step of 3 min at 94°C, 35 cycles of 1 min at 94°C, 1 min at 56°C and 2 min at 72°C, and a final step of 7 min at 72°C. PCR products were separated by electrophoresis in 2% agarose gels in 1×TBE buffer, stained with ethidium bromide and visualised under UV light. Band scoring was carried out using a standard 1-kb DNA ladder (Invitrogen). PCR for band excision was carried out using the same protocol described above, but in a 50-μl PCR reaction volume, and electrophoresis performed in 1×TAE buffer.
Cloning and sequencing of genomic DNA
The DNA fragments of 2,065, 2,315 and 1,003 bp obtained from 'Ferrovia', 'Taleguera Brillante' and 'Vittoria' after amplification with the primer combination PruT2-PruC4R, and the DNA fragment of 448 bp obtained from 'Pico Colorado' after amplification with the primer combination PruT2-SI32, were excised from the gel and the DNA extracted using Ultrafree-DA columns (Millipore, Bedford, Mass., USA), ethanol-precipitated, washed, dried and resuspended in sterile water. The purified fragments of 'Ferrovia', 'Taleguera Brillante' and 'Vittoria' were cloned into the vector 'pGEM-T Easy' (Promega, Madison, Wis., USA) following the manufacturer's instructions. Transformed colonies were selected and plasmid purification was carried out using the Plasmid Mini Kit (Qiagen, Valencia, Calif., USA). Plasmids with inserts of the expected size were selected by restriction digestion and PCR amplification using the universal M13 primers. Positive clones were sequenced on an 'ABI Prism 3700 DNA Analyzer' (Applied Biosystems, Foster City, Calif., USA). The purified fragment of 'Pico Colorado' was sequenced directly from the PCR product. To obtain the full sequence of the putative S-RNase genes, the 5′- and 3′-adjacent DNA ends of the four PCR fragments sequenced were isolated using the 'Universal Genome Walker Kit' (Clontech, Palo Alto, Calif., USA) and the 'Advantage Genomic Polymerase Mix' (Clontech) following the manufacturer's instructions. The new PCR fragments that included the complete S-RNase genes and the 5′ flanking regions were purified using Ultrafree-DA columns (Millipore), precipitated and sequenced as described above. Sequences were aligned using Clustal X (Thompson et al. 1997).
Results and Discussion
S-allele identification by PCR analysis
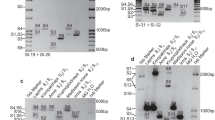
PCR amplification of the S-alleles S 1, S 2 , S 3 , S 4 , S 5 , S 6 and S 9 in the cultivars Summit, Bing, Hedelfingen, Hartland and Burlat, using the primer pairs PruT2-PruC4R, PruC2-PruC4R, SI19-SI20, SI31-SI32 and SI31-SI20, produced amplification fragments whose sizes were in agreement with those described by Tao et al. (1999a) and Wiersma et al. (2001). PCR amplification to identify the S-alleles of the cultivars Pico Colorado, Taleguera Brillante and Vittoria revealed two fragments for each cultivar: in 'Taleguera Brillante' the two fragments are of different size to the previously reported alleles; In 'Pico Colorado' and 'Vittoria' one corresponds to the known S-alleles S 6 and S 3 , respectively, and the other two show a different size from those described previously. Thus, one of the 'Taleguera Brillante' fragments and the fragment amplified in 'Vittoria' of different size from the known S-alleles were, respectively, 2,315- and 1,003-bp with primers PruT2-PruC4R, 1,834- and 599-bp with primers PruC2-PruC4R, 453- and 409-bp with primers SI31-SI32 and 1,906- and 671-bp with primers SI19-SI20. In 'Pico Colorado' only one fragment, corresponding to the allele S 6, could be distinguished using primer pairs PruT2-PruC4R, PruC2-PruC4R and SI19-SI20. However another fragment of 448 bp could be observed when using the primer pair PruT2-SI32. The fragments of sizes different from those described previously, amplified in the cultivars Vittoria, Pico Colorado and Taleguera Brillante, would represent three new S-alleles that were named S 23 , S 24 , S 25 respectively, following in consecutive order the nomenclature given to other sweet cherry S-alleles published and other sweet cherry alleles recently identified (K.Tobutt, personal communication). The alleles S 23 and S 24 correspond to the alleles temporarily named S R and S J found in the sweet cherry local Spanish cultivars Ramon Rachilla, Garrafal, Del Cardito, Pico Colorado Cirino and Virgo Juliana (Wünsch and Hormaza, in press).
PCR amplification of DNA from the cultivar Ferrovia, revealed a fragment that corresponds to the allele S 3 and another fragment that did not correspond to any of the fragments in our reference cultivars. However, Wiersma et al. (2001), using the primer pairs SI31-SI32 and SI19-SI20 in the cultivar Schneiders, described amplification fragments of 423- and 1,711-bp for the allele S 13 (now re-named S 12 , Tobutt et al. 2001), which agree with the sizes amplified with the same primer pairs in 'Ferrovia' and, consequently, the other fragment in 'Ferrovia' most likely corresponds to S 12 . This hypothesis was verified by amplification of the DNA of 'Schneiders' (kindly provided by K. Tobutt, East Malling, UK) with primer pairs PruT2-PruC4R and PruC2-PruC4R; the resulting fragments both in 'Ferrovia' and 'Schneiders' were 2,065 and 1,647-bp respectively.
Cloning and sequencing of genomic DNA of S 12 , S 23 , S 24 and S 25 RNases
In order to obtain the full sequences of the S 12 , S 23 , S 24 , and S 25 putative S-RNases, the partial genomic sequences obtained after PCR amplification with PruT2-PruC4R and PruT2-SI32 were used to isolate the 5′- and 3′-adjacent ends of the amplified fragments by genome walking. The complete genomic sequences obtained correspond to four S-RNases (DDBJ/EMBL/GenBank AY259112, AY259113, AY259114 and AY259915 respectively). A further confirmation that the S-RNase DNA sequence obtained in Ferrovia corresponds to the allele S 12 , is the presence of a XhoI restriction-site identified that produces two restriction fragments (625- and 1,086-bp) after amplification with SI19-SI20 and digestion as reported by Wiersma et al. (2001).
Similarly to other sweet cherry S-RNases (Tao et al. 1999c; Yamane et al. 2000), for all four S-alleles sequenced, S 12 , S 23 , S 24 and S 25 , two introns were found (Fig. 1). The first intron was inserted within the junction between the signal peptide and the mature protein while the second intron was found within the hypervariable region. The length of the first intron in the four S-alleles reported in this work, S 12 , S 23 , S 24 and S 25 , was 256-, 248-, 339- and 292-bp, respectively, while the length of the second was more variable, being 1,362-, 314-, 175- and 1,549-bp respectively. All the intron/exon splice junctions conserve the GT/AG consensus sequence found in the majority of plant introns and have a high AT content (66–73%) (Simpson and Filipowitcz 1996). The flanking exon sequences were highly conserved as well as the exon lengths; thus the length of the first exon was either 76 bp (for S 23 , S 24 and S 25 ) or 82 bp (for S 12 ), the length of the second exon was either 182 bp (for S 24 ) or 188 bp (for S 12 , S 23 and S 25 ) and the length of the third exon was 411 bp (for S 23 and S 24 ), 417 bp (for S 12 ) or 447 bp (for S 25 ). The deduced amino-acid sequences of the four S-alleles (Fig. 2) present a Ribonuclease T2 family conserved-domain (Sassa et al. 1996) and a putative signal peptide comprising approximately the 25 N-terminal amino-acid residues. The conserved regions (C1, C2, C3, RC4 and C5) defined by Ushijama et al. (1998a) in the S-RNases of other Rosaceae species were also conserved in these four S-RNases, and the hypervariable region (RHV) was also identified.
Alignment of the deduced amino-acid sequences of sweet cherry and other Prunus S-RNases. Amino-acid positions are numbered. Conserved residues are indicated by asterisks under the sequences. Gaps are marked by dashes. The five conserved regions and the variable region of the Rosaceae (Ushijama et al. 1998a) are underlined. Plant species from which each sequence was derived are represented by their initials, i.e. PA: Prunus avium, PC: Prunus cerasus, PD: Prunus dulcis, PM: Prunus mume
We isolated and sequenced DNA fragments of 721-, 739-, 281- and 307-bp upstream of the putative initiation codon for the S-RNases S 12 , S 23 , S 24 and S 25 respectively. These 5′ flanking sequences present a conserved putative TATA box (TATATAA) at −86, −84, −86 and −92-bp respectively from the 'A' of the putative ATG initiation codon (Fig. 3). In Japanese pear the putative TATA box was found 124 bp upstream from the putative initiation codon while the transcription initiation site was at −91 and/or −92 (Ushijama et al. 1998b). Thus, our results indicate that the 5′ untranslated region in sweet cherry (Prunoideae) is shorter than in the Maloideae, although the analysis of that region in other species of the Prunoideae will be needed to confirm if this situation is conserved in the subfamily. The characterisation of the flanking regions of three Japanese pear S-RNases and an apple S-RNase gene, evidenced that the 5′ flanking-sequences analysed did not contain any conserved sequence motifs other than the putative TATA box, indicating that the regulation of gene expression could be controlled either by an intragenic region or, alternatively, by a region located further upstream of the promoter region analysed (Ushijama et al. 1998b). The analysis of the 5′ flanking regions of the other three Japanese pear S-RNases by Norioka et al. (2001) revealed a highly similar 200-bp region upstream from the putative TATA box, that could be a candidate to contain a cis-element controlling the expression of Japanese pear S-RNases. In the S-RNases analysed in our work, sequence similarity was high in the 5′- flanking region downstream from the TATA box as 64 nucleotides were conserved in the four S-RNases, although similarity was lower upstream of the putative TATA box. The IA-like motif (Ficker et al. 1998) conserved in Japanese pear (Norioka et al. 2001) was not conserved in the sweet cherry sequences analysed, although a similar motif could be identified in the S 23 sequence at −254 bp. Other motifs identified in the Solanaceae that seem to be involved in the regulation of the S-RNase gene, like motif I and III (Kaufmann et al. 1991; Ficker et al. 1998), were not found in the sequences analysed. Alignment of the four sweet cherry and four Japanese pear 5′ flanking sequences (Fig. 3) showed overall little sequence identity in that region between sweet cherry (Prunoideae) and Japanese pear (Maloideae), confirming the phylogenetic differentiation in the S-RNases between those two subfamilies (Ma and Oliveira 2002).
Sequence alignment of 5′-flanking regions of sweet cherry S 12 -, S 23 -, S 24 - and S 25 -RNase genes and with the Japanese pear S 2-, S 3-, S 4- and S 5-RNase 5′-flanking sequences (Norioka et al. 2001). Conserved residues among sweet cherry, and among sweet cherry and Japanese pear sequences, are indicated by asterisks under the sequences. Gaps are marked by dashes. The nucleotides are numbered in relation to the 'A' of the putative ATG initiation codon numbered +1. The putative TATA box of sweet cherry and Japanese pear sequences (Norioka et al. 2001) are underlined. The IA-like (Ficker et al. 1998) sequence in Japanese pear (Norioka et al. 2001) and in sweet cherry S 23 - allele is double underlined
Comparison of S 12 , S 23 , S 24 and S 25 RNase sequences with sequences of other Prunus S-alleles
Sequence alignment of the deduced DNA coding region of S 12 , S 23 , S 24 and S 25 S-alleles with the cDNA sequences of the other six previously reported S-RNases from sweet cherry, S 1 , S 2 , S 3 , S 4 , S 5 (now re-named S 9 , Tobutt et al. 2001), and S 6 (DDBJ/EMBL/GenBank AB028153, AJ298311, AB010306, AB028154, AJ298314, AB010305) (Tao et al. 1999a,b; Sonneveld et al. 2001), and with the S-RNases of other Prunus species for which the full open reading-frame sequence is available, that is, almond alleles S a , S b , S c (DDBJ/EMBL/GenBank AB026836, AB011469, AB011470) (Ushijama et al. 1998a; Tamura et al. 2000); Japanese apricot alleles S 1 , S 7 , S f (DDBJ/EMBL/GenBank AB101438, AB101439, AB101437) (Tao et al. 2002) and sour cherry allele S a (DDBJ/EMBL/GenBank AB050393)(Yamane et al. 2001), revealed pairwise similarity scores ranging from 96% (sweet cherry S 6 –S 24 ) to 66% (sweet cherry S 9 , S 12 -almond S a ). Most sequence pairwise comparisons yielded maximum scores of 88%, S 6 and S 24 being exceptionally similar.
To evaluate the differences at the protein level (Fig. 2), the deduced amino-acid sequences of the alleles S 12 , S 23 , S 24 and S 25 were compared with the deduced amino-acid sequences of sweet cherry and the other Prunus species considered above. Alignment of the ten sweet cherry S-RNases revealed 112 conserved residues while comparison of the 17 Prunus S-RNases revealed only 82 conserved residues. The similarity at the amino-acid level amongst the 17 Prunus S-RNases ranged from 54% (sweet cherry S 23 - almond S a ) to 93% (sweet cherry S 6 –S 24 ). As noted by Tamura et al. (2000) and Ma and Oliveira (2002), the nucleotide sequence as well as the amino-acid sequence of S a of almond, consistently showed the lowest similarity scores when compared with other Prunus S-RNases. In general, higher intraspecific than interspecific pairwise amino-acid similarities were observed in this work, since the alleles S 12 , S 23 , S 24 , and S 25 are more similar to the sweet cherry S-RNases (71–93%) than to the other Prunus S-RNases (54–79%). However, it is interesting to note that both sweet cherry alleles S 2 and S 6 have lower similarity scores with sweet cherry allele S 3 (73% and 75%) than with the almond allele S b (84 and 82%). This result is expected under the hypothesis that S-RNase polymorphism in the Rosaceae took place before speciation (Ishimizu et al. 1998; Ushijama et al. 1998a).
S 6 and S 24 sequences have similar intron lengths and were exceptionally similar both at the nucleotide and amino-acid level indicating that those two alleles could be derived from a common ancestral allele or that one could have evolved from the other by several mutations. The fact that the S 6 allele is present in several sweet cherry genotypes (Bošković and Tobutt 2001; Sonneveld et al. 2001; Wiersma et al. 2001; Wünsch and Hormaza, in preparation) and that the S 24 allele has so far only been found in four cultivars of a particular area in Spain (Wünsch and Hormaza, in press) would sustain the later hypothesis. Those two alleles strikingly present the same amino-acid sequence in the RHV region while the greatest number of amino-acid substitutions accumulate within the 14 amino acids found right after the C1 region defined by Ushijama et al. (1998a) (Fig. 4). This region is also highly variable among all the sweet cherry and Prunus S-RNases analysed (Fig. 2), and in the solanaceous S-RNases this region contains two of the most-highly variable residues found outside the hypervariable regions HVa and HVb (Tsai et al. 1992). This variable region found between the C1 and C2 regions was defined as HV1 by Kheyr-Pour et al. (1990), upon comparison of the S-RNase sequences of Nicotiana alata. It is expected that the highest variable residues must be involved in allelic specificity (Tsai et al. 1992), and it has been shown that the hypervariable regions HVa and HVb of the solanaceous S-RNases play a role in the recognition of self-pollen (Matton et al. 1997), although other studies suggest that S pollen recognition may not be restricted to the HV regions (Verica et al. 1998). Highly similar pairs of S-RNases have been described in other species of the Rosaceae. Thus, Japanese pear S-RNases S 3 and S 5 show eight amino-acid substitutions in a stretch of 70 amino acids that include the HV region, which are enough to discriminate between S 3 and S 5 pollen (Ishimizu et al. 1998). However another pair of different S-RNases from Japanese pear of high similarity (S 1 –S 4 ) have the amino acid substitutions scattered throughout the sequence (Ishimizu et al. 1998). The results obtained in this work, together with the recent advances towards the isolation of the pollen S-gene (Ushijima et al. 2003) and combined with crosses made in the field, will help to confirm the regions in the S-RNase sequences of Rosaceae involved in allele-specific recognition between pollen and pistil.
Alignment of the deduced amino-acid sequences of S 6- and S 24-RNases from sweet cherry. Amino-acid positions are numbered. Conserved residues are indicated by asterisks under the sequences. Gaps are marked by dashes. The five conserved regions and the variable region of the Rosaceae are underlined (Ushijama et al. 1998a). The solanaceous variable region HV1 (Kheyr-Pour et al. 1990) is double underlined
References
Bošković R, Tobutt KR (2001) Genotyping cherry cultivars assigned to incompatibility groups, by analysing stylar ribonucleases. Theor Appl Genet 103:475–485
Broothaerts W, Janssens GA, Proost P, Broekaert F (1995) cDNA cloning and molecular analysis of two self-incompatibility alleles from apple. Plant Mol Biol 27:499–511
De Nettancourt D (2001) Incompatibility and incongruity in wild and cultivated plants, 2nd edn. Springer, Berlin Heidelberg New York
Ficker M, Kirch HH, Eijlander R, Jacobsen E, Thompson RD (1998) Multiple elements of the S 2 -RNase promoter from potato (Solanum tuberosum L.) are required for cell type-specific expression in transgenic potato and tobacco. Mol Gen Genet 257:132–142
Hauck N, Iezzoni AF, Yamane H, Tao R (2001) Revising the S-allele nomenclature in sweet cherry (Prunus avium) using RFLP probes. J Am Soc Hort Sci 126:654–660
Hormaza JI (2002) Molecular characterization and similarity relationships among apricot (Prunus armeniaca L.) genotypes using simple sequence repeats. Theor Appl Genet 104:321–328
Huang S, Lee HS, Karunanandaa B, Kao TH (1994) Ribonuclease activity of Petunia inflata S-proteins is essential for rejection of self-pollen. Plant Cell 6:1021–1028
Iorger TR, Gohlke JR, Xu B, Kao TH (1991) Primary structural features of the self-incompatibility proteins in the Solanaceae. Sex Plant Reprod 4:81–87
Ishimizu T, Shinkawa T, Sakiyama F, Norioka S (1998) Primary structural features of rosaceous S-RNases with gametophytic self-incompatibility. Plant Mol Biol 37:931–941
Janssens GA, Goderis IJ, Broekaert F, Broothaerts W (1995) A molecular method for S-allele identification in apple based on allele-specific PCR. Theor Appl Genet 91:691–698
Kaufmann H, Salamini F, Thompson RD (1991) Sequence variability and gene structure at the self-incompatibility locus of Solanum tuberosum. Mol Gen Genet 226:457–466
Kheyr-Pour A, Bintrim SB, Ioerger TR, Remy R, Hammond SA, Kao TH (1990) Sequence diversity of pistil S-proteins associated with gametophytic self-incompatibility in Nicotiana alata. Sex Plant Reprod 3:88–97
Lee HS, Huang S, Kao TH (1994) S-proteins control rejection of incompatible pollen in Petunia inflata. Nature 367:560–563
Ma RC, Oliveira MM (2001) Molecular cloning of the self-incompatibility genes S1 and S3 from almond (Prunus dulcis cv Ferragnès). Sex Plant Reprod 14:163–167
Ma RC, Oliveira MM (2002) Evolutionary analysis of S-RNase genes from Rosaceae species. Mol Gen Genomics 267:71–78
Matton DP, Maes O, Laublin G, Xike Q, Bertrand C, Morse D, Capadoccia M (1997) Hypervariable domains of self-incompatibility RNases mediate allele-specific pollen recognition. Plant Cell 9:1757–1766
McClure BA, Haring V, Ebert PR, Anderson MA, Simpson RJ, Sakiyama F, Clarke AE (1989) Style self-incompatibility gene products of Nicotiana alata are ribonucleases. Nature 342:955–957
Murfett J, Atherton T, Mou B, Gasser C, McClure BA (1994) S-RNase expressed in transgenic Nicotiana causes S-allele-specific pollen rejection. Nature 367:563–566
Norioka N, Katayama H, Matsuki T, Ishimizu T, Takasaki T, Nakanishi T, Norioka S (2001) Sequence comparison of the 5′ flanking regions of Japanese pear (Pyrus pyrifolia) S-RNases associated with gametophytic self-incompatibility. Sex Plant Reprod 13:289–291
Sassa H, Hirano H, Ikeshashi H (1992) Self-incompatibility related RNases in styles of Japanese pear (Pyrus serotina Rehd.). Plant Cell Physiol 33:811–814
Sassa H, Hirano H, Ikeshashi H (1993) Identification and characterization of stylar glycoproteins associated with self-incompatibility of Japanese pear, Pyrus serotina Rehd. Mol Gen Genet 241:17–25
Sassa H, Mase N, Hirano H, Ikeshashi H (1994) Identification of self-incompatibility related glycoproteins in styles of apple (Malus x domestica). Theor Appl Genet 89:201–205
Sassa H, Nishio T, Kowyama Y, Hirano H, Koba T, Ikeshashi H (1996) Self-incompatibility (S) alleles of Rosaceae encode members of a distinct class of the T2/S ribonuclease superfamily. Mol Gen Genet 250:547–557
Simpson GG, Filipowitcz W (1996) Splicing precursors to mRNA in higher plants: mechanisms, regulation and sub-nuclear organisation of the spliceosomal machinery. Plant Mol Biol 32:1–41
Sonneveld T, Robbins TP, Bošković R, Tobutt KR (2001) Cloning six cherry self-incompatibility alleles and development of allele-specific PCR detection. Theor Appl Genet 102:1046–1055
Tamura M, Ushijama K, Sassa H, Hirano H, Tao R, Gradziel TM, Dandekar AM (2000) Identification of self-incompatibility genotypes of almond by allele-specific PCR analysis. Theor Appl Genet 101:344–349
Tao R, Yamane H, Sassa H, Mori H, Gradziel TM, Dandekar AM, Sugiura A (1997) Identification of stylar RNases associated with gametophytic self-incompatibility in almond (Prunus dulcis). Plant Cell Physiol 38:304–311
Tao R, Yamane H, Sugiura A, Murayama H, Sassa H, Mori H (1999a) Molecular typing of S-alleles through identification, characterization and cDNA cloning for S-RNases in sweet cherry. J Am Soc Hort Sci 124:224–233
Tao R, Yamane H, Akira H (1999b) Cloning and nucleotide sequences of cDNA encoding S1- and S4-RNases (Accessions Nos. AB028153 and AB028154) from sweet cherry (Prunus avium L.). Plant Physiol 120:1207
Tao R, Yamane H, Akira H (1999c) Cloning of genomic DNA sequences encoding S1-, S3-, S4- and S6-RNases (Accessions Nos. AB031815, AB031816, AB031817, AB031818) from sweet cherry (Prunus avium L.). Plant Physiol 121:1057
Tao R, Habu T, Yamane H, Sugiura A (2002) Characterization and cDNA cloning for Sf-RNase, a molecular marker for self-compatibility, in Japanese apricot (Prunus mume). J Jpn Soc Hort Sci 71:595–600
Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The clustal X windows interface: flexible strategies for sequence multiple alignment aided with quality analysis tools. Nucleic Acids Res 25:4876–4882
Tobutt KR, Sonneveld T, Bošković R (2001) Cherry (in)compatibility genotypes-harmonization of recent results from UK, Canada, Japan and USA. Eucarpia Fruit Breeding Section Newslett 5:41–46
Tsai DS, Lee HS, Post LC, Kreiling KM, Kao TH (1992) Sequence of an S-protein of Lycopersicon peruvianum and comparison with other solanaceous S-proteins. Sex Plant Reprod 5:256–263
Ushijima K, Sassa H, Tao R, Yamane H, Dandekar AM, Gradziel TM, Hirano H (1998a) Cloning and characterization of cDNAs encoding S-RNases from almond (Prunus dulcis): primary structural features and sequence diversity of the S-RNases in Rosaceae. Mol Gen Genet 260:261–268
Ushijima K, Sassa H, Hirano H (1998b) Characterization of the flanking regions of the S-RNase genes of Japanese pear (Pyrus serotina) and apple (Malus x domestica). Gene 211:159–167
Ushijima K, Sassa H, Dandekar AM, Gradziel TM, Tao R, Hirano H (2003) Structural and transcriptional analysis of the self-incompatibility locus of almond: identification of a pollen-expressed F-box gene with haplotype-specific polymorphism. Plant Cell 15:771–781
Verica JA, McCubbin AG, Kao TH (1998) Are the hypervariable regions of S-RNases sufficient for allele-specific recognition pollen? Plant Cell 10:314–316
Wiersma PA, Wu Z, Zhou L, Hampson C, Kappel F (2001) Identification of self-incompatibility alleles in sweet cherry (Prunus avium L.) and clarification of incompatibility groups by PCR and sequencing analysis. Theor Appl Genet 102:700–708
Wünsch A, Hormaza JI (in press) Molecular evaluation of genetic diversity and S-allele composition of Spanish local sweet cherry (Prunus avium L.) cultivars. Genet Resour Crop Evol
Yaegaki H, Shimada T, Moriguchi T, Hayama H, Haji T, Yamaguchi M (2001) Molecular characterization of S-RNase genes and S-genotypes in the Japanese apricot (Prunus mume Sieb. et Zucc.). Sex Plant Reprod 13:251–257
Yamane H, Tao R, Murayama H (2000) Determining the S-genotypes of several sweet cherry cultivars based on PCR-RFLP analysis. J Hortic Sci Biotech 75:562–567
Yamane H, Tao R, Sugiura A, Hauck NR, Iezzoni AF (2001) Identification and characterization of S-RNases in tetraploid sour cherry (Prunus cerasus) J Am Soc Hort Sci 126:661–667
Zuccherelli S, Tassinari P, Broothaerts W, Tartarini S, Dondini L, Sansavini S (2002) S-Allele characterization in self-incompatible pear (Pyrus communis L.) Sex Plant Reprod 15:153–158
Acknowledgements
We gratefully thank M.A. Aranda and C. Fernández-Marco for help during the cloning experiments and M. Herrero for her helpful comments on this manuscript. A.W. was supported by a SIA-DGA fellowship and financial support for this work was provided by MCYT (Project Grant AGL2001-2414).
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by H.F. Linskens
Rights and permissions
About this article
Cite this article
Wünsch, A., Hormaza, J.I. Cloning and characterization of genomic DNA sequences of four self-incompatibility alleles in sweet cherry (Prunus avium L.). Theor Appl Genet 108, 299–305 (2004). https://doi.org/10.1007/s00122-003-1418-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00122-003-1418-6