Abstract
A genetic linkage map has been constructed for meadow fescue (Festuca pratensis Huds.) (2n=2x=14) using a full-sib family of a cross between a genotype from a Norwegian population (HF2) and a genotype from a Yugoslavian cultivar (B14). The two-way pseudo-testcross procedure has been used to develop separate maps for each parent, as well as a combined map. A total number of 550 loci have been mapped using homologous and heterologous RFLPs, AFLPs, isozymes and SSRs. The combined map consists of 466 markers, has a total length of 658.8 cM with an average marker density of 1.4 cM/marker. A high degree of orthology and colinearity was observed between meadow fescue and the Triticeae genome(s) for all linkage groups, and the individual linkage groups were designated 1F–7F in accordance with the orthologous Triticeae chromosomes. As expected, the meadow fescue linkage groups were highly orthologous and co-linear with Lolium, and with oat, maize and sorghum, generally in the same manner as the Triticeae chromosomes. It was shown that the evolutionary 4AL/5AL translocation, which characterises some of the Triticeae species, is not present in the meadow fescue genome. A putative insertion of a segment orthologous to Triticeae 2 at the top of 6F, similar to the rearrangement found in the wheat B and the rye R genome, was also observed. In addition, chromosome 4F is completely orthologous to rice chromosome 3 in contrast to the Triticeae where this rice chromosome is distributed over homoeologous group 4 and 5 chromosomes. The meadow fescue genome thus has a more ancestral configuration than any of the Triticeae genomes. The extended meadow fescue map reported here provides the opportunity for beneficial cross-species transfer of genetic knowledge, particularly from the complete genome sequence of rice.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Detailed genetic maps based on molecular markers have been developed in a number of crop plants, and are being used extensively in studies of genome organisation, for the dissection of complex traits, and in practical plant breeding. Ultimately, they make map-based cloning and marker-assisted selection possible. The development of tools and resources to conduct genomic research in forage grasses has so far lagged behind major crop species such as the cereals. Despite the fact that fodder crops like grasses and legumes provide the basis for economically important agricultural industries, to-date relatively few species have been studied in terms of genome analysis by genetic linkage mapping. So far marker-based genetic maps have been constructed only in alfalfa (Medicago sativa L.) (Brummer et al. 1993; Kiss et al. 1993; Echt et al. 1994; Kaló et al. 2000), tall fescue (Festuca arundinaceae Schreb.) (Xu et al. 1995), ryegrass (Lolium perenne L.) (Hayward et al. 1998; Bert et al. 1999; Jones et al. 2002a, 2002b), and meadow fescue (Festuca pratensis Huds.) (Chen et al. 1998). The meadow fescue map was a comparative RFLP map between meadow fescue and tall fescue, constructed by using 33 heterologous RFLP probes from tall fescue (Xu et al. 1991), and the total map length was 280.1 cM.
Meadow fescue (F. pratensis Huds.) is a diploid (2n=2x=14) obligate outbreeding forage grass species, with a gametophytic self-incompatibility system controlled by two loci designated S and Z (Lundqvist 1962). The DNA content is about 1.9 pg per haploid genome (Seal 1983), corresponding to a genome size of approximately 1.83×108 bp. As in other forage grasses the breeding of meadow fescue is hampered by a strong self-incompatibility system and severe inbreeding depression, that make the production of inbred lines and F1-hybrid varieties very difficult. The cultivars currently on the market are therefore synthetic populations based on a restricted number of clones, and this approach only utilises a small part of the potential heterosis. The strong self-incompatibility system also makes linkage mapping more complicated compared to plant species that can readily be inbred. One way of performing linkage mapping in such species is to utilise the two-way pseudo-testcross procedure (Ritter et al. 1990; Grattapaglia and Sederoff 1994; Hemmat et al. 1994). In this procedure the mapping family is based on the F1-progeny of a pair-cross between unrelated highly heterozygous individuals. If homozygous or near-homozygous testers are available, a one-way pseudo-test cross can be established by crossing heterozygous individuals and such testers (Ritter et al. 1990; DeSimone et al. 1997). In the present study we have used the two-way pseudo-testcross procedure to construct a genetic linkage map of meadow fescue. Construction of genetic linkage maps by this procedure is challenging since markers may vary in the number of segregating alleles (up to four combinations of alleles at a single locus, and with 16 combinations jointly at two loci). One or both parents may be heterozygous at a given locus, markers may be dominant or co-dominant, and usually the linkage phases of marker pairs are unknown (Maliepaard et al. 1997). This can lead to inaccuracy in the estimation of recombination frequencies. However, one advantage is that the recombination frequencies can be directly estimated for each parent separately, and this is not possible in classical F2-mapping populations. It is also expected that lower levels of segregation distortion will be observed in pseudo-testcross family structures, and consequently less complications in map construction, compared to progeny sets (such as F2 and BC1 structures) produced by enforced inbreeding (Tavoletti et al. 1996). The improvements in recent years of both theory and methods for constructing linkage maps in outbreeding species have made it possible to develop genetic linkage maps in outbreeders more efficiently than before (Ritter et al. 1990; Grattapaglia and Sederoff 1994; Maliepaard et al. 1997).
Meadow fescue belongs to the tribe Poeae, and is taxonomically closely related to Lolium. Oat belongs to the tribe Aveneae, which is in the same supertribe, Poodae, as Poeae. The third tribe in the subfamily Pooideae contains the Triticeae species wheat (Triticum), barley (Hordeum) and rye (Secale), and belongs to the supertribe Triticoideae. In the grass family there are two other subfamilies containing important crop species. The first is Ehrhartoideae to which rice belongs, and the second is Panicoideae, which comprises maize, Sorghum and pearl millet (Soreng and Davis 1998).
Comparative genetic maps show that chromosomal segment structure (orthology or conserved synteny) and marker order (colinearity) are conserved across species over substantial evolutionary distances (Gebhardt et al. 1991; Ahn et al. 1993; Bennetzen and Freeling 1993; Moore et al. 1995; Devos and Gale 1997), with the number of chromosomal and genetic differences between species generally increasing with evolutionary distance as demonstrated in the study of micro- colinearity in the Adh-1 region between maize, sorghum, rice and Arabidopsis (Tikhonov et al. 1999).
In the grass family genome relationships have been established between rice, wild rice (Zizania palustris) (Kennard et al. 1999), foxtail millet, sugar cane, sorghum, pearl millet, maize, the Triticeae cereals wheat, barley, and rye, and oat (for review see Gale and Devos 1998). Recently, Jones et al. (2002a) performed the first comparative mapping study between forage grasses and cereals by comparing Lolium with Triticeae, oat and rice. They found that the genetic maps of perennial ryegrass and the Triticeae cereals are highly conserved in terms of orthology and colinearity. Comparative genetics is especially important for 'minor crops' where little genomic information is available. Alignment of segments of chromosomes of such crops with chromosomal regions of species where detailed genetic information has been accumulated, e.g. forage grasses with rice, can be used to speed up the development of genetic linkage maps, and to rapidly identify chromosomal regions containing QTLs (quantitative trait loci) and genes of interest.
The aims of the present study were to develop an AFLP- and homologous RFLP-based core map of meadow fescue with the addition of heterologous RFLP anchor probes from cereals, and to study the syntenic relationships with other grass genomes. This paper presents the first detailed comparative genome study between meadow fescue, the Triticeae, rice, oat, Lolium, maize and sorghum.
Materials and methods
Construction of the mapping population
The mapping population was developed both for the construction of a linkage map and for the mapping of agronomically important traits such as frost-tolerance. Several pair-crosses were made between clones from the Norwegian population HF2 and clones from a Yugoslavian population B14, accession no. 1,700, by open-pollination in pollen-proof crossing chambers in 1995. Seeds were harvested on each parent plant separately, so for each cross there were two reciprocal full-sib families. Population HF2 originated from two generations of phenotypic selection for higher frost-tolerance, using artificial freezing tests, within the Norwegian cultivar 'Løken'. Strong selection response was observed even after one generation of selection (Larsen 1979). One of these pair-crosses, B14/16 (female)×HF2/7, was chosen as a mapping population, and 138 progeny were established after selfed progeny (3.5%) had been discarded based on the segregation of the isozyme locus Gpi-2.
RFLP markers
Homologous Festuca gDNA-probes
A protocol of Sharp et al. (1988) was used for the extraction of meadow fescue genomic DNA. Genomic DNA isolated from clone no. 3 of the Norwegian synthetic cultivar 'Salten' was used to establish a partial Pst1 genomic library. Fragments in the range 500–3,000 bp were cloned into the plasmid vector pUC18. A total of 217 fescue gDNA-probes (prefix IBF) with an average length of 0.8 kb were screened for polymorphism.
Heterologous cereal anchor probes
As part of the EU-project EGRAM (European Gramineae Mapping Project), two sets of anchor probes (198 probes) from wheat (PSR), barley (BCD), oat (CDO), rice (RZ, RGC, RGG, RGR), maize (CSU), sorghum (SbRPG), pearl millet (PSM) and meadow fescue (IBF) were assembled based on good cross-hybridisation ability in a range of grass species (http://jiio5.jic.bbsrc.ac.uk/egram/main.html). Another 48 anchor probes from wheat (PSR, WG), barley (BCD) and oat (CDO) were selected because they had been mapped in the ILGI (International Lolium Genome Initiative) perennial ryegrass reference mapping population (John Forster, Plant Biotechnology Centre, La Trobe University, Australia, personal communication 1998). In addition 21 RFLP probes from barley (ABG, BCD,), oat (CDO) and wheat (PSR, WG), that were located in chromosomal regions affecting frost tolerance, and ten dehydrin cDNA-clones (Dhn1–11) from barley (Close et al. 1993), were tested. A last group of 33 probes, also from wheat, barley and oat, were selected to complement uncovered regions.
RFLP screening and mapping
All RFLP procedures such as restriction enzyme digestion, gel electrophoresis, Southern transfer, probe labelling and filter hybridisation were as described by Devos et al. (1992). Since the mapping population was an F1 from a cross between heterozygous individuals, RFLP probes were screened for polymorphism using blots with DNA of five randomly selected progeny in addition to the parents, digested with 3–5 restriction enzymes (EcoRI, HindIII, DraI, EcoRV and BamHI). The filters used for screening the fescue probes also contained genomic DNA of the multiply heterozygous parent of the p150/112 perennial ryegrass reference population adopted by the ILGI collaborators (Jones et al. 2002a), and one Festulolium genotype.
AFLP markers and procedure
The AFLP procedure of Zabeau and Vos (1993) and Vos et al. (1995) was followed. The restriction enzymes PstI and MseI were used, with the following adapters and primers:
MseI adapters: 5′-GACGATGAGTCCTGAG/TACTCAGGACTCAT; MseI universal primer: M00=GATGAGTCCTGAGTAA; MseI+2 primer: M17=M00+CG; MseI+3 primers: M43=M00+ATA; M44=M00+ATC; M65=M00+GAG; M66=M00+GAT; M68=M00+GCC; M70=M00+GCT; M72=M00+GGC; M73=M00+GGG; M75=M00+GTA; M76=M00+GTC; M77=M00+GTG; M78=M00+GTT.
PstI adapters: 5′-CTCGTAGACTGCGTACATGCA/TGTACGCAGTCTAC; PstI universal primer: P00=GACTGCGTACATGCAG; PstI+3 primers: P63=P00+GAA; P64=P00+GAC; P65=P00+GAG; P66=P00+GAT; P76=P00+GTC, and P77=P00+GTG.
PCR reactions were carried out in a Peltier Thermal Cycler PTC-100 (MJ Research, Inc.), or a Perkin Elmer 9600. Radioactively labelled (33P) samples were separated by gel-electrophoresis on denaturing polyacrylamide gels (6% w/v). The fragment sizes of AFLP markers were determined by running a 30–330-bp ladder (Gibco-BRL) alongside the amplification products of the parents for all primer combinations. AFLP markers have been designated with the fragment size (bp) as a suffix.
Isozyme loci and procedure
Isozyme analysis using a starch-gel system was performed according to the method described by Rognli et al. (2000). Acp-2 (E.C.3.1.3.2), Est-4 (E.C. 3.3.1.4), Got-3 (E.C.2.6.1.1), Gpi-2 (E.C. 5.3.1.9) and Pgm-2 (E.C. 5.4.2.2) were stained as described by Wendel and Weeden (1989), and Dia-1 and Dia-2 (E.C.1.6.99) as described by Allendorf et al. (1977).
SSR loci and procedure
Eighteen Lolium SSRs (designated DJF) were kindly provided by Dr. Chris Taylor, The Danish Institute of Agricultural Sciences, Research Centre Flakkebjerg, Denmark. PCR reactions were carried out in a total volume of 20 μl containing approximately 60 ng of genomic DNA, 10 mM of TRIS pH 8.3, 1.5 mM of MgCl2, 50 mM of KCl, 0.2 μM (4 pmol) of each primer, 130 μM of each dNTP (Amersham Pharmacia Biotech), and 1.0 unit of Taq DNA polymerase (Roche Diagnostics GmBH). For all amplifications the following program was used: initial denaturation at 94°C for 2 min., followed by 30 cycles of 94°C for 1 min, 0.5°C/sec. down to annealing temperature, annealing temperature for 1 min, 0.5°C/sec. to 72°C, 72°C for 1 min, a final extension at 72°C for 4 min and ending with incubation at 10°C. Cycling was done in a DNA Engine Tetrad (MJ Research Inc.) or a Perkin Elmer 9600. Polymorphisms were resolved by analysing samples on 5% polyacrylamide sequencing gels (19:1 acrylamide:bis) containing 8 M urea and run under standard conditions. The amplification products were visualized by silver-staining, following the method of Bassam et al. (1991).
Scoring and mapping procedures
RFLP, AFLP and SSR markers were scored visually, and scoring was repeated at least twice to ensure accuracy. Markers were separated into those segregating in the paternal parent, the maternal parent, and those segregating in both parents. In a full-sib family there are seven possible segregation types, and the different segregation types of markers were identified and scored according to the notation given by Maliepaard et al. (1997).
Construction of the maps was accomplished by treating the segregation data as double-haploids (DH) for the parental maps, and as a cross pollinator (CP) for the combined map. Segregation analyses, grouping and linkage analyses were performed using JoinMap version 2.0 (Stam and van Ooijen 1995). In order to check the quality of the segregation data, the order of the markers in each linkage group was also verified using MAPMAKER 2.0 (Lander et al. 1987). All double-recombination events were re-checked on the original autoradiograms. Markers of segregation type ab×ab with an apparent 3:1 segregation ratio were first omitted from the mapping data. These markers segregate in both parents, but dominance leads to a 3:1 segregation, which limits their information content, and leads to inaccuracy in determining map positions. Grouping of markers into linkage groups was done using a LOD score threshold of 4.0 for all three maps. The grouping was tested and found to be consistent whether based on the combined data (CP) with only the fully informative markers (1:1:1:1), the combined data with all markers, or the two separate parental data sets (DH) with all markers. After having established stable linkage groups, the ab×ab 3:1 markers were added to the datasets, and the individual markers grouped to the same linkage groups both in the parental maps (lod 4.0) and in the combined map (lod 5.0). The ab×ab 3:1 markers and the ab×ab 1:2:1 markers were mapped only in the combined map, as there were large jumps in chi-square means when mapping of these markers was attempted in the parental maps. The mapping was performed using the Kosambi mapping function (Kosambi 1944) for calculation of map distances. The maps were drawn using MapChart 2.0 (Voorrips 2001).
Databases and publications consulted for comparative analyses
For comparative mapping, information was collected from GrainGenes (http://wheat.pw.usda.gov/index.shtml) and Gramene (http://www.gramene.org/), the integrated grass-genome map circle published by Devos and Gale (1997), and the comparative maps by Van Deynze et al. (1995a, b) and Sarma et al. (2000). In addition, information from the following publications were used: Triticeae and oat—ITMI proceedings from 1996 (McGuire and Qualset 1997); Lolium—Jones et al. (2002a) and Hayward et al. (1998); rice—Causse et al. (1994), Kurata et al. (1994), Harushima et al. (1998) and Wilson et al. (1999); maize—Davis et al. (1999) and S.A. Quarrie (unpublished data); and sorghum—Glaszmann et al. (1997), Boivin et al. (1999) and Ventelon et al. (2001).
We use the term 'orthology' to describe conserved segment structure. 'Orthology' is defined as the conservation of loci of common evolutionary ancestry between different species ('orthologous loci') (Keller and Feuillet 2000). We define a region to be orthologous if a segment containing orthologous loci is not interrupted by more than one non-orthologous locus. Genome coverage and orthology was calculated as in Van Deynze et al. (1995, b) as follows: genome coverage (%) = a/b×100, and genome orthology (%) = c/a×100 where a = the total map length (cM) represented in meadow fescue with probes mapped in the compared species, b = the total map length (cM) of meadow fescue, and c = the sum of the lengths (cM) of segments in meadow fescue conserved between meadow fescue and the compared species.
Results
Marker screening and mapping
A total number of 242 heterologous probes were screened, of which 53% were polymorphic, 38% were monomorphic, and 22% did not hybridise, or gave a very faint hybridisation signal (Table 1). About 60% of the fescue probes (IBF) were polymorphic in the mapping population for at least one of the five restriction enzymes used (data not shown). Hybridisation of IBF-probes to the Lolium P150/112 hybrid parent and the Festulolium showed that 88 and 94% of the probes, respectively, either had good or fair hybridisation signals. Seven fescue probes (IBF6, IBF33, IBF43, IBF95, IBF507, IBF520 and IBF540) did not hybridise to Lolium, but had good hybridisation signals on Festulolium, indicating that these probes are fescue-specific. Seventy five of the polymorphic fescue (IBF) probes were mapped. They detected 145 loci, of which 30 derived from the (15) fully informative ab×cd segregation class, were mapped also as single loci (ab×aa and aa×ab segregation classes) in the two parental maps (Table 2). The total number of IBF marker loci on the combined map is therefore 115. A total of 116 heterologous probes were used for mapping, and 184 loci, of which 50 were single loci from the ab×cd segregation class, were mapped. Eleven barley Dhn clones (Dhn1–11) were screened for polymorphism, and six of them (Dhn1, Dhn2, Dhn4, Dhn5, Dhn6 and Dhn10) were polymorphic. Dhn11 did not hybridise. All polymorphic dehydrin probes, except Dhn10, were mapped successfully (Table 2). The Dhn-probes all detected multiple copies. Dhn4 and Dhn5, and Dhn1 and Dhn2, showed the same banding patterns, and the corresponding loci were therefore designated Dhn4/5 and Dhn1/2, respectively. Twelve (67%) of the 18 Lolium SSRs could be amplified successfully but only two of them (DJF16 and DJF20) were polymorphic. The two SSR primer pairs identified five loci, of which two were single loci in the parental maps (Table 2). Details about the SSR primers can be found in Jensen et al., in preparation (2003).
A total number of 194 AFLP-loci were mapped, of which 46% are derived from segregation in the maternal parent (ab×aa), 42% from segregation in the paternal parent (aa×ab), and 12% from segregation in both parents (ab×ab) (Table 2).
The total number of marker loci on the maps is 550, of which 466 loci are on the combined maps since 84 loci are single loci from the ab×cd segregation class mapped in the parents only (Table 2). The 466 loci are distributed as follows regarding segregation types: 39% ab×aa, 42% aa×ab, 1% ab×ab (1:2:1), 9.0% ab×ab (3:1) and 9.0% ab×cd.
Map construction
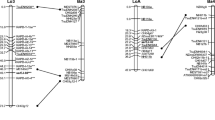
A maternal map (602.3 cM, 223 loci), a paternal map (581.5 cM, 237 markers), and a combined map (658.8 cM, 466 markers) were developed using the two-way pseudo-testcross procedure (Fig. 1). The parental maps were linked together using the 42 fully informative loci (ab×cd), and the parental and calculated combined maps are presented with the individual map lengths (cM) in Fig. 1. The average length of the linkage groups are 86.0, 83.0 and 94.1 cM for the maternal, paternal and combined maps, respectively, with average marker density of 2.7, 2.5 and 1.4 cM/marker, respectively.
Maternal (A), paternal (B), and combined (A×B) linkage maps of the "B14/16 × HF2/7" population of meadow fescue (F. pratensis Huds.) containing 332 homologous and heterologous RFLP loci, 194 AFLP loci, 14 Dhn loci, five isozyme loci, and five SSR loci. Map lengths (cM) are printed below each linkage group, and the linkage groups are displayed in the direction short arm→long arm from top to bottom following the Triticeae relationships. Loci marked with "***" have significantly distorted segregation ratios. All comparative markers and other markers segregating ab×cd or ab×ab (1:2:1) are printed in bold. In cases where putative functions of comparative RFLP probes are known, these have been indicated in parenthesis behind the probe name on the maps
Based on a comparison of the map locations of the heterologous cereal anchor probes in Festuca with the respective map locations on the consensus maps of Triticeae, the individual linkage groups of meadow fescue were designated 1F–7F in accordance with the orthologous Triticeae chromosomes. The combined maps of 1F, 5F and 7F are intermediate in length compared with the length of the respective parental maps, the combined map of 4F was shorter than both parental maps, while the combined maps of 2F, 3F and 6F are slightly longer than either of the respective parental maps (Fig. 1). This illustrates the difficulties in establishing perfect combined maps in a full-sib family of heterozygous parents. Construction of a combined map of 4F and to some extent also 7F, were difficult, whereas the other linkage groups could be established fairly easily. In addition, only one small linkage group could be established for the maternal 3F, consisting of two loci, Xibf568, which is fully informative and Xbcd1823, which is located on Triticeae group-3 chromosomes (Nelson et al. 1995b). For 2F no linkage group could be established for the maternal parent. However, Xcdo456, which is located at the end of Triticeae 2L (Nelson et al. 1995a) and is fully informative, maps to its orthologous position in meadow fescue. Xcdo36 has a map location on Triticeae group-2 chromosomes, but did not group to any of the linkage groups in Fescue. However, when this marker was added manually to the 2F maternal and combined maps, it did map to its orthologous position relative to Triticeae (linkage between these two markers is indicated as a dotted line on the maternal 2F map in Fig. 1).
Comparative mapping
There are two choices when it comes to comparing our meadow fescue map with maps of other grasses. Either to make a consensus map of the parental maps, or to use the combined map. Even though there were some problems of integrating the parental maps of 4F, in particular, the combined maps were chosen since the locations and order of markers on the different maps were very similar (Fig. 1). Conserved syntenic relationships between the meadow fescue linkage groups and the maps of Lolium, Triticeae, oat, rice, maize and sorghum are shown in Fig. 2.
Festuca orthology with Lolium (L), Triticeae (T), oat (O), rice (R), maize (M) and sorghum (S). Festuca linkage groups with the order of comparative marker loci are shown at the left-hand side of boxes representing the orthologous chromosomes of the other species in terms of chromosomal regions orthologous with the respective Festuca linkage group. The chromosomes have different colours for each orthologous chromosome and are numbered 1–12 for Lolium, Triticeae, rice and maize, and with letters A–J for oat and sorghum. Hatched boxes with two colours in maize indicate orthology with two homologous maize chromosomes. Chromosome arms are printed in the segments orthologous to Triticeae chromosomes
Lolium
The number of loci on the meadow fescue map with equivalent map locations in perennial ryegrass was 46, representing a genome coverage of 62% (Fig. 2). Two markers, BCD1823 on 3F and CDO365 on 5F, had non-orthologous positions in Fescue relative to Lolium following the definition of orthology used here. These markers map on LG2 in Lolium (Jones et al. 2002a). In addition, markers RGC390 on 7F and ABLPG94 on 5F did not have corresponding positions. In Lolium RGC390 is located on LG3 and ABLPG94 has loci on LG1, 4 and 7 (Hayward et al. 1998; Jones et al. 2002a). Only the corresponding position of ABLPG94 on LG7 is indicated in Fig. 2. The positions of the BCD1823 and RGC390 in Fescue are consistent with the expected conserved genome relationships with Triticeae and rice, while the position of CDO365 in Fescue is as expected relative to rice but not relative to Triticeae (Fig. 2).
Triticeae
The combined Fescue maps contain 117 loci with known map locations in the Triticeae. Of the 72% of the Fescue genome that was covered by Triticeae markers, 94% was orthologous and a high degree of orthology was observed for linkage groups 1F, 2F, 3F, 5F and 7F (Fig. 2). Within these linkage groups only seven loci, i.e. Xpsr153 (1F), Xcdo365 and Xpsr78b (5F), and Xabg387, Xpsr312, Xbcd131 and Xbcd1779a (7F), were in non-orthologous positions (Fig. 2). In the case of PSR78 and BCD1779, they each detected two loci in Fescue, and the other loci mapped in orthologous positions on 1F and 2F, respectively. Probe ABG387 has map locations on several Triticeae chromosomes (1, 3, 4, 5 and 6), while BCD131 has only been mapped in the wheat B-genome (3B). Chromosome 4F carries a segment orthologous to Triticeae group-5L chromosomes adjacent to the region that is orthologous to Triticeae 4S. The loci Dhn4 and Dhn5 are also located at the end of 4F instead of on 6F as expected (Choi et al. 2000). In addition to these rearrangements, two probes (CDO678 and ABG453) have loci on 4F that are non-orthologous relative to Triticeae. In 6F, a segment with possible orthology to the Triticeae group-2 chromosomes is located distal to the segment orthologous to the short arm of the Triticeae group-6 chromosomes. One of the probes in this segment is CDO678, which detects two loci in Fescue (Xcdo678a on 4F, Xcdo678b on 6F). The fragment mapped on 6F is probably the same as the one mapped on Triticeae group-2 chromosomes. The segments of 6F is also interrupted by two loci, Xpsr601, with expected location on 1F and Xcdo476, which is on the expected linkage group, but in a non-colinear position.
The other cereal species
The relationship between the genomes of the Triticeae and oat, rice, maize and sorghum are well documented (see Devos and Gale 1997). Since we have established the orthologous relationship between the Fescue genome and the Triticeae genomes, it follows that the same relationship that exists between these cereal genomes and the Triticeae genomes, also should exist between the Fescue genome and the other cereal genomes.
Based on the known relationships between the Triticeae and oat (Van Deynze et al. 1995b) the following orthologous relationships would be expected between Fescue and oat: 1F-A, 2F-B and C, 3F-C and G, 4F and 5F-E and F, 6F-A, B and G and 7F-D. The 48 loci with map locations in oat represented a genome coverage of only 48%, of which 90% was orthologous between oat and Fescue. The results show that all linkage groups, except 2F and 5F, were orthologous. In 2F two probes, i.e. CDO57 (oat E) and BCD1779 (oat D), have non-orthologous positions. However, two loci were mapped for each of these probes in Fescue, and the 2F loci may not correspond to the previously mapped loci in oat. There is some evidence for this for BCD1779, where the second locus, Xbcd1779b, maps in an orthologous position on 7F and oat D. In 5F, markers CDO365 (oat B) and CDO590 (oat A) are in non-orthologous positions, and the same is the case for BCD1823 and CDO57 on 3F and 7F, respectively.
The 78 loci with map locations in rice represented a genome coverage of 50%, of which 74% were orthologous. The expected relationships between Fescue and rice following those established between Triticeae and rice and Triticeae and Fescue would be: 1F-R5 and 10, 2F-R4 and 7, 3F-R1, 4F-R3, 2 and 11, 5F-R9, 11 and 12, 6F-R2 and 4 and 7F-R6 and 8. In general these relationships are confirmed in the present Fescue map (Fig. 2). However, a number of markers have non-orthologous map positions relative to rice, i.e. Gpi-2 (1F, R6), Xpsr115 (4F, R1), Xcdo105a (3F, 5R), Xcdo400 (5F, R2) and Xpsr601, Xcsu148, Xcdo497 and Xsbrpg360 on 6F that have rice locations on R9, R4, R7 and R9, respectively. A segment of 7F contains five markers that have previously been mapped on different rice chromosomes, Xpsr129 (R4), Xbcd131 (R7), Xbcd1779b (R9), Xrgc390 (R8) and Acp-2 (R12). The rearrangements in Festuca relative to wheat, i.e. segments of wheat 5L and 6L in 4F and wheat 2L in 6F, which lead to the expected presence of R2 on 4F, R4 on 6F and the lack of R3 on 5F, were verified in the case of 5F, which contains no markers with R3 location. The other rearrangements could not be verified due to the lack of common markers in these rice and Fescue segments.
The number of loci with map locations in maize was 89, and the genome coverage and total orthology were 62 and 89%, respectively. The expected orthologous relationships between Fescue and maize should be: 1F,-M1/5 and M6/8, 2F-M2/10 and M2/7, 3F-M3/8, 4F-M1/5 and M1/9, 5F-M2/7, M1/5 and M3/10, 6F-M4/5 and 7F-M6/9 and M1/5. These relationships were confirmed to a large extent (Fig. 2). Only 12 probes had non-orthologous positions based on the expected relationship with maize, i.e. SbRPG851, SbRPG854 and SbRPG360 on 1F, 2F and 6F, respectively, CDO504, CDO520 and RZ141 on 4F, CDO365, CDO590 on 5F, DHN1/2 on 5F and 6F and CDO678, CSU148 and CSU184 on 6F. Both DHN1/2 and RZ141 detect multiple loci in Fescue and RZ141 detects at least one position on 4F (Xrz141d) that is orthologous between Fescue and maize. For DHN1/2, it is difficult to establish orthology since these two dehydrins co-segregate completely in Fescue, and only DHN1 has been mapped in maize.
Only 33 loci were common between Fescue and Sorghum. This gave a genome coverage of only 23%, which makes the comparison between these genomes very incomplete. Based on the known relationships between the genome of Sorghum and the other cereals, the following relationships could be expected between Fescue and Sorghum: 1F-C and E, 2F-B and F, 3F-G, 4F-C, 5F-C, F, I and J, 6F-D and 7F-A and H. However, only 19 of the 33 common loci were found in orthologous positions. The expected orthology was confirmed for chromosomes 5F, 6F and 7F, while only 4 of the ten loci on 4F mapped to orthologous positions in Fescue and sorghum.
Colinearity across species
Complete or nearly complete colinearity of loci within regions conserved relative to Lolium, Triticeae, oat and rice was observed for all Fescue chromosomes except 4F. Some of the observed lack of colinearity is probably due to inaccurate mapping. In a full-sib family one or both parents may be heterozygous, markers may be dominant or codominant and the linkage phases of marker pairs are unknown (Maliepaard et al. 1997). Marker pairs will therefore provide different amounts of information for the estimation of recombination frequencies and linkage phases, and mapping in full-sib families is therefore less accurate than in populations such as F2, RILsor BCs. In addition, low recombination frequencies and differences in recombination between the two parents, sometimes lead to ambiguous map orders in the combined map. As the combined maps were used to estimate the extent of colinearity across species, it is expected that at least in some cases disruption of colinearity is due to ambiguous marker orders. For example, the non-colinearity observed for some of the markers on the combined 4F map could be the result of an imprecise placement of the large number of tightly linked markers on this linkage group (0.7 cM/marker on the combined map) and may not reflect a real disruption of colinearity. However, it is clear that chromosome 4F carries a segment with homology to wheat chromosome 5DL and this corresponds to the entire rice chromosome 3. Markers Xcdo365 and Xrz141a (map locations on rice 11) are located on top of Xcdo344 and Xcdo459 (map locations on rice 12) on 5F. Physical mapping has shown that on Triticeae 5S, the segment orthologous to rice 12 is located distally, and the segment with orthology to rice 11 more proximal to the centromere (Sarma et al. 2000). This indicates a putative inversion in Festuca, which unfortunately cannot be verified in Lolium since only one locus, Xcdo349, with map location on rice 12, and none from rice 11, was mapped on Lolium chromosome 5.
Duplicated loci
Duplicated loci were observed for seven probes that mapped to different linkage groups. These were CDO57, BCD1779 and PSR150, which all map to both 2F and 7F, CDO105 on 1F and 3F, PSR78 on 1F and 5F, PSR2021 on 5F and 6F, and CDO678 on 4F and 6F. In addition, more than one locus detected by the same probe were observed within a single linkage group for Dhn4 and Dhn5 and CDO1395 in 4F, CDO395 in 6F, and CDO347 and CDO780 in 7F.
Renaming of the Lolium linkage groups based on comparison between the Lolium and Fescue maps
Both cytogenetic studies, which demonstrate extensive recombination between homoeologous chromosomes in interspecific hybrids between Festuca and Lolium, and the present comparison of the linkage maps demonstrate the close evolutionary relationship between these species. The old nomenclature used to identify chromosomes and linkage groups in Lolium needs revision following the establishment of dense comparative linkage maps. In the recently published Lolium map the linkage groups were named according to the Triticeae nomenclature (Jones et al. 2002a), and compared with the first Lolium map published by Hayward et al. (1998). They suggested that Hayward LG2 corresponds to their LG3, and Hayward LG5 to LG4. Our Festuca map supports that Hayward LG5 is present to LG4 based on the location of Pgm-2, and further suggests that Hayward LG6 is present to LG1 (locations of Gpi-2, Xablmc106 and Xablmc107), Hayward LG3 is present to LG2 (location of Xablmc80), Hayward LG4 is present to LG5 (location of Xablmc127), and Hayward LG1 is present to LG7 (locations of Xablgp94 and Xcdo105).
Discussion
The degree of polymorphism detected by heterologous probes was approximately equivalent to the results reported for Lolium (48%) (Jones et al. 2002a). Because there are both cDNA- and gDNA-probes among the heterologous probes, and most of these were pre-screened for hybridisation ability, it is difficult to interpret the degree of hybridisation and polymorphism in Festuca in terms of evolutionary relationship between species. However, the results indicate that the degree of hybridisation and polymorphism were highest with probes originating from the most closely related species in taxonomic terms, perennial ryegrass and oat.
The expected length of linkage maps is approximately 100 cM per haploid chromosome, which corresponds to approximately two chiasmata per chromosome. The Lolium map of Jones et al. (2002a) has an average length per linkage group of 116 cM. Our combined Festuca maps have an average length per chromosome of 94.1 cM, and the map lengths of the linkage groups are fairly similar in both parental maps, except for the incomplete maternal 2F and 3F maps. An average length of 94.1 cM corresponds to approximately 1.9 chiasmata per chromosome. Simonsen (1975) studied the chiasma formation at metaphase-I in 50 genotypes of the Norwegian cultivar 'Løken', and observed a mean number of chiasmata per chromosome ranging from 1.5 to 1.96. The map constructed using the present mapping family indicates that both parents have normal levels of recombination. The marker density of the map is high, on average 1.4 markers per cM in the combined map, but there are also some gaps. More markers need to be added in order to establish whether the present map gives a complete coverage of the meadow fescue genome.
The problem of establishing maps of 2F and 3F for the maternal parent B14/16 is difficult to explain. Two possible explanations for this phenomenon could be that the parent B14/16 is heterozygous for inversions in the chromosomes corresponding to 2F and 3F, or that these chromosomes are homozygous at nearly all the RFLP-loci that should map there. Recombination is drastically reduced in chromosomes with inversions, and a selective recovery of non-crossover chromosomes in viable progeny will be the result both for para- and peri-centric inversions. Investigations of chromosome configurations during meiosis in the cultivar 'Løken', the origin of the HF2/7 parent, have shown an unusually high frequency of individuals heterozygous for chromosomal aberrations, i.e. translocations (22%) and paracentric inversions (42%) (Simonsen 1975). It is therefore possible that high frequencies of inversions are common in F. pratensis, and that inversions may also be present in the Yugoslavian population B14. In order to confirm this hypothesis, cytological investigations of the early stages of meiosis of B14/16 have to be undertaken. On the other hand, a number of the RFLP-probes with map locations on Triticeae group 2 and 3 chromosomes displayed a similar segregation pattern in the meadow fescue mapping family, i.e. one monomorphic fragment present in the female parent B14/16 and two fragments in the male parent HF2/7 which segregated in the progeny. This would make it impossible to estimate recombination among these markers in the female parent.
Meadow fescue chromosome 4F has a more-ancestral configuration compared to wheat chromosome 4D
Several probes located on Triticeae group-5 chromosomes map to meadow fescue 4F. These are Xbcd450, Xbcd1103, Xcdo504, Xcdo520, Xrgr3226, Xpsr115 and Xwg644 (Liu et al. 1992; Xie et al. 1993; Galiba et al. 1995; Nelson et al. 1995c; Sarma et al. 1998). Triticeae group-5 is known to be orthologous to segments of rice 12, 11, 9 and 3 (Moore 1995; Van Deynze et al. 1995a), and these probes belong to the part of Triticeae group-5 that is orthologous to rice chromosome-3 (Sarma et al. 2000). This suggests that the entire rice-3 part of Triticeae 5L is located on meadow fescue 4F, and that 4F is thus completely orthologous to rice chromosome-3. The present Lolium map carries only one locus from that region of rice-3, but its location distally on Lolium chromosome 4 indicates that the group-4 chromosomes in Lolium have the same configuration (Jones et al. 2002a). The configuration of this part of the genome of both Fescue and Lolium is thus more closely related to rice than to the Triticeae, and that suggests that this particular rice, Lolium and Fescue chromosomal-arrangement is more ancestral than that of the Triticeae.
The 4AL/5AL translocation is not present in meadow fescue
The location of Xwg114 on 4F is interesting. Xwg114 is located on 5AL, 4BL and 4DL in wheat, and is reciprocally diagnostic for the 4AL/5AL translocation event in the A-genome of wheat (Devos et al. 1995). The location of Xwg114 on 4F indicates that the 4AL/5AL translocation is not present in Fescue, and thus that the configuration of this part of the Fescue genome is similar to the Triticeae B, D and H genomes. The absence of the 4AL/5AL translocation also holds for Lolium, as Xcdo20 on LG4 of Jones et al. (2002a), with locations on wheat 5A and 4B (Nelson et al. 1995c), which indicates absence of the translocation.
Orthologous Triticeae 2 on top of 6F
Three probes (CDO395, BCD1095 and CDO678), which detect four loci on 6F, indicate that a segment orthologous to the Triticeae group-2 chromosomes is located on the top of 6F. Although, initially, there were problems splitting LG6 and LG2 in the female parent B14/16 mainly because of the problems encountered in establishing the maternal 2F map, we are confident that our results are correct. The three heterologous probes in question are all fully informative, and Xcdo395 is also located on the top of LG6 in Lolium (Jones et al. 2002a). Similar reciprocal translocations involving chromosome arms 2S and S have also been detected in the wheat B and the rye R genomes (Devos et al. 1993). However the number of common markers in this region are too few to postulate that reciprocal translocation is identical among the three genomes. The more precise mapping of the 2/6 breakpoints in the rye, fescue and B genomes is required to resolve this.
Mapping of Dehydrin genes
Dhn4/5a mapped to 6F, and Dhn1/2d, e and f mapped to 5F. These results coincide with previously reported positions for Dhn4, Dhn5 and Dhn1, which map to barley chromosomes 6H and 5H, respectively (Pan et al. 1994; Choi et al. 2000). However, Dhn1 also detected loci on 6F, whereas only Dhn3, Dhn4, Dhn5 and Dhn7 map to 6H in barley (Choi et al. 2000). Dhn5c, f, g and h mapped to 4F, while in barley only Dhn6 maps to 4H (Choi et al. 2000). However, the positions of Dhn4 and Dhn5 on the distal end of maize 1/5, the orthologous chromosomes of Triticeae 4, could support the location of these dehydrins on 4F. Dhn6 mapped to fescue LG6. All the dehydrins tested had multiple copies in meadow fescue due to the cross-hybridisation between different dhn genes, which have large conserved regions. The use of gene-specific PCR-primers, as developed and tested in barley by Choi et al. (2000), might overcome this problem.
Conclusions
The extended meadow fescue map reported here demonstrates high orthology and colinearity with the Triticeae genomes and Lolium, and orthology with oat, rice, maize and sorghum in more or less the same manner as these crops are orthologous with Triticeae. Festuca exhibits the rearrangements which are common to all Pooideae so far analysed, i.e. linkage blocks with orthology to R8 is inserted into R6 to give 7F, and R10 into R5 to give 1F. These evolutionary translocations appear to define this group of grasses (Gale et al. 2001). However, it is clear that the fescue genome differs from the various Triticeae genomes in a number of ways. Colinearity of the rice chromosome 3 as a single linkage block in 4F, rather than being distributed over homoeologous group 4 and 5 chromosomes as in all Triticeae genomes so far analysed, is one such difference. Lolium and oat show the same arrangement, which is clearly the more-primitive chromosomal configuration within the Poaceae. Thus the divergence of the Avena/Lolium/Festuca lineage from the common ancestor with Triticeae clearly pre-dates divergence within the Triticeae. Our results support the current estimates of the time of divergence of the Triticeae from oat, being 25 my (Gaut 2002), and the divergence of wheat and barley at 10–13 my ago (Wolfe et al. 1989; Gaut 2002). These estimates, which are based on the evolution of chloroplast genes, are also supported by estimates based on nuclear genes with divergence of Lolium (Lolium rigidum) at 35 my, and barley at 11 my (Huang et al. 2002). However, whether the rearrangement within the segment with orthology with R3 is the defining marker for the Triticeae group must await further study. The absence of the 4L/5L translocation, which is displayed by A, R and U genomes of wheat, Secale cereale and Aegilops umbellulata, respectively, is also significant. There has been some discussion as to whether the intact homoeologous group-4 chromosome or the translocated chromosome represents the ancestral configuration (Devos et al. 1995). The absence of the translocation in the fescue genome strongly suggests that the intact chromosome is the more primitive.
References
Ahn S, Anderson JA, Sorrells ME, Tanksley SD (1993) Homologous relationships of rice, wheat and maize chromosomes. Mol Gen Genet 241:483–490
Allendorf FW, Mitchell N, Ryman N, Stahl G (1977) Isozyme loci in brown trout (Salmo trutta L.). Detection and interpretation from population data. Hereditas 86:179–190
Bassam BJ, Caetano-Anolles G, Gresshoff PM (1991) Fast and sensitive silver staining of DNA in polyacrylamide gels. Anal Biochem 196:80–83
Bennetzen JL, Freeling M (1993) Grasses as a single genetic system—genome composition, colinearity and compatibility. Trends Genet 9:259–261
Bert PF, Charmet G, Sourdille P, Hayward MD, Balfourier F (1999) A high-density molecular map for ryegrass (Lolium perenne) using AFLP markers. Theor Appl Genet 99:445–452
Boivin K, Deu M, Rami J-F, Trouche G, Hamon P (1999) Towards a saturated sorghum map using RFLP and AFLP markers. Theor Appl Genet 98:320–328
Brummer EC, Bouton JH, Kochert G (1993) Development of an RFLP map in diploid alfalfa. Theor Appl Genet 86:329–332
Causse M, Fulton TM, Cho YG, Ahn SN, Chunwongse J, Wu K, Xiao J, Yu Z, Ronald PC, Harrington SB, Second GA, McCouch SR, Tanksley SD (1994) Saturated molecular map of rice genome based on inter-specific backcross population. Genetics 138:1251–1274
Chen C, Sleper DA, Johal GS (1998) Comparative RFLP mapping of meadow and tall fescue. Theor Appl Genet 97:255–260
Choi D-W, Koag MC, Close TJ (2000) Map locations of barley Dhn genes determined by gene-specific PCR. Theor Appl Genet 101:350–354
Close TJ, Fenton RD, Moonan F (1993) A view of plant dehydrins using antibodies specific to the caboxyl-terminal peptide. Plant Mol Biol 3:279–286
Davis GJ, McMullen MD, Baysdorfer C, Musket T, Grant D, Staebell M, Xu G, Polacco M, Koster L, Melia-Hancock S, Houchins K, Chao S, Coe Jr EH (1999) A maize map standard with sequenced core markers, grass genome reference points and 932 expressed sequenced tags (ESTs) in a 1,736-locus map. Genetics 152:1137–1172
DeSimone M, Morgante M, Lucchin M, Parrini P, Marocco A (1997) A first linkage map of Cichorium intybus L. using a one-way pseudo-testcross and PCR-derived markers. Mol Breed 3:415–425
Devos KM, Gale MD (1997) Comparative genetics in the grasses. Plant Mol Biol 35:3-15
Devos KM, Atkinson MD, Chinoy CN, Liu C, Gale MD (1992) RFLP based genetic maps of the homoeologous group-3 chromosomes of wheat and rye. Theor Appl Genet 83:931–939
Devos KM, Millian T, Gale MD (1993) Comparative RFLP maps of the homoeologous group-2 chromosomes of wheat, rye and barley. Theor Appl Genet 85:784–792
Devos KM, Dubcovsky J, Dvorák J, Chinoy CN, Gale MD (1995) Structural evolution of wheat chromosomes 4A, 5A and 7B, and its impact on recombination. Theor Appl Genet 91:282–288
Echt CS, Kidwell KK, Knapp SJ, Osborn TC, McCoy TJ (1994) Linkage mapping in diploid alfalfa (Medicago sativa). Genome 37:61–71
Gale MD, Devos KM (1998) Comparative genetics in the grasses. Proc Natl Acad Sci USA 95:1971–1974
Gale MD, Moore G, Devos KM (2001) Rice—the pivotal genome in cereal comparative genetics. In: Goode JA, Chadwick D (eds) Rice biotechnology: improving yield, stress tolerance and grain quality. John Wiley and Sons Ltd, Chichester, pp 46–58
Galiba G, Quarrie SA, Sutka J, Morgounov A, Snape JW (1995) RFLP mapping of vernalization (Vrn1) and frost resistance (Fr1) on chromosome 5A of wheat. Theor Appl Genet 90:1174–1179
Gaut BS (2002) Evolutionary dynamics of grass genomes. Tansley review no. 132. New Phytol 154:15–28
Gebhardt C, Ritter E, Barone A, Debener T, Walkemeier B, Shachtschabel U, Kaufmann H, Thompson RD, Bonierbale MW, Gana MW, Tanksley SD, Salamini F (1991) RFLP maps of potato and their alignment with the homologous tomato genome. Theor Appl Genet 83:49–57
Glaszmann JC, Dufour P, Grivet L, D'Hont A, Deu M, Paulet F, Hamon P (1997) Comparative genome analysis between several tropical grasses. Euphytica 96:13–21
Grattapaglia D, Sederoff R (1994) Genetic linkage maps of Eucalyptus urophylla using a pseudo-testcross mapping strategy and RAPD markers. Genetics 137:1121–1137
Harushima Y, Yano M, Shomura A, Sato M, Shimano T, Kuboki Y, Yamamoto Y, Nagamura Y, Kurata N, Khush G, Sasaki T (1998) A high-density rice genetic linkage map with 2,275 markers using a single F2 population. Genetics 91:618–494
Hayward MD, Forster JW, Jones JG, Dolstra O, Evans C, McAdam NJ, Hossain KG, Stammers M, Will J, Humphreys MO, Evans GM (1998) Genetic analysis of Lolium. I. Identification of linkage groups and the establishment of a genetic map. Plant Breed 117:451–455
Hemmat M, Weeden NF, Manganaris AG, Lawson DM (1994) A molecular marker linkage map for apple. J Hered 85:4-11
Huang S, Sirikhachornkit A, Faris JD, Su X, Gill BS, Haselkorn R, Gornicki P (2002) Phylogenetic analysis of the acetyl-CoA carboxylase and 3-phosphoglycerate kinase loci in wheat and other grasses. Plant Mol Biol 48:805–820
Jensen LB, Muylle H, Roldán-Ruiz I, De Riek J, Vosman B, Arens P, Taylor C, Lübberstedt T, Barre P (2003) A mapped SSR reference set in Lolium perenne (in preparation)
Jones ES, Mahoney NL, Hayward MD, Armstead IP, Jones JG, Humphreys MO, King IP, Kishida T, Yamada T, Balfourier F, Charmet G, Forster JW (2002a) An enhanced molecular marker-based genetic map of perennial ryegrass (Lolium perenne L.) reveals comparative relationships with other Poaceae genomes. Genome 45:282–295
Jones ES, Dupal MP, Dumsday JL, Hughes LJ, Forster JW (2002b) An SSR-based genetic linkage map for perennial ryegrass (Lolium perenne L.). Theor Appl Genet 105:577–584
Kaló P, Endre G, Zimáneyi L, Csanádi G, Kiss GBC (2000) Construction of an improved linkage map of diploid alfalfa (Medicago sativa). Theor Appl Genet 100:641–657
Keller B, Feuillet C (2000) Colinearity and gene density in grass genomes. Trends Plant Sci 5:246–251
Kennard W, Phillips R, Porter R, Grombacher A (1999) A comparative map of wild rice (Zizania palustris L. 2n=2x=30). Theor Appl Genet 99:793–799
Kiss GB, Csanadi G, Kalman K, Kaló P, Okresz L (1993) Construction of a basic genetic map for alfalfa using RFLP, RAPD, isozyme and morphological markers. Mol Gen Genet 238:120–137
Kosambi DD (1944) The estimation of map distances from recombination values. Ann Eugen 12:172–175
Kurata N, Nagambura Y, Yamamoto Y, Harushima Y, Sue N, Wu J, Antonio BA, Shomura A, Shimizu T, Lin S-Y, Inoue T, Fukuda A, Shimano T, Kuboki Y, Toyama T, Miyamoto Y, Kirihara T, Hayasaka K, Miyayo K, Monna L, Zhong HS, Tamura Y, Wang Z-X, Momma T, Umehara Y, Yano M, Sasaki T, Minobe Y (1994) A 300-kilobase interval genetic map of rice including 883 expressed sequences. Nature Genet 8:365–372
Lander ES, Green P, Abrahamson J, Barlow A, Daly MJ, Lincoln SE, Newburg L (1987) MAPMAKER: an interactive computer package for constructing primary genetic linkage maps of experimental and natural populations. Genomics 1:174–181
Larsen A (1979) Freezing tolerance in grasses: variation within population and response to selection. Meld Norg Landbruk Høgsk 58:1–28
Liu CJ, Atkinson MD, Chinoy CN, Devos KM, Gale MD (1992) Non-homoeologous translocations between group 4, 5 and 7 chromosomes within wheat and rye. Theor Appl Genet 83:305–312
Lundqvist A (1962) The nature of the two-loci incompatibility system in grasses. II. Number of alleles at the incompatibility loci in Festuca pratensis Huds. Hereditas 48:169–181
Maliepaard C, Jansen J, van Ooijen JW (1997) Linkage analysis in a full-sib family of an outbreeding plant species: overview and consequences for application. Genet Res 70:237–250
McGuire PE, Qualset CO (eds) (1997) Progress in genome mapping of wheat and related species: joint proceedings of the 5th and 6thpublic workshops of the International Triticeae Mapping Initiative, 1–3 September 1995, Norwich UK, and 30–31 August 1996, Sidney, Australia. Report No.18. University of California Genetic Resources Conservation Program, Davies California, USA
Moore G (1995) Cereal genome evolution: pastoral pursuits with 'lego' genomes. Curr Opin Genet Dev 5:717–724
Moore G, Devos KM, Wang Z, Gale MD (1995) Cereal genome evolution—grasses, line up and form a circle. Curr Biol 5:737–739
Nelson JC, Van Deynze AE, Autrique E, Sorrells ME, Lu YH, Merlino M, Atkinson M, Leroy P (1995a) Molecular mapping of wheat. Homoeologous group 2. Genome 38:516–524
Nelson JC, Van Deynze AE, Autrique E, Sorrells ME, Lu YH, Negre S, Bernhard M, Leroy P (1995b) Molecular mapping of wheat. Homoeologous group 3. Genome 38:525–533
Nelson JC, Sorrells ME, Van Deynze AE, Lu YH, Atkinson MD, Bernhard M, Leroy P, Faris JD, Anderson JA (1995c) Molecular mapping of wheat: major genes and rearrangements in homoeologous groups 4, 5 and 7. Genetics 141:721–731
Pan A, Hayes PM, Chen F, Chen THH, Blake T, Wright S, Karsai I, Bedö Z (1994) Genetic analysis of the components of winter hardiness in barley (Hordeum vulgare L.). Theor Appl Genet 89:900–910
Ritter E, Gebhardt C, Salamini F (1990) Estimation of recombination frequencies and construction of RFLP linkage maps in plants from crosses between heterozygous parents. Genetics 125:645–654
Rognli OA, Nilsson N-O, Nurminiemi M (2000) Effects of distance and pollen competition on gene flow in the wind-pollinated grass Festuca pratensis Huds. Heredity 85:550–560
Sarma RN, Gill BS, Sasaki T, Galiba G, Sutka J, Laurie DA, Snape JW (1998) Comparative mapping of the wheat chromosome 5A Vrn-A1 region with rice and its relationship to QTL for flowering time. Theor Appl Genet 97:103–109
Sarma RN, Fish L, Gill BS, Snape JW (2000) Physical characterization of the homoeologous group-5 chromosomes of wheat in terms of linkage blocks, and physical mapping of some important genes. Genome 43:191–198
Seal AG (1983) DNA variation in Festuca. Heredity 50:225–236
Sharp PJ, Kresi M, Shewry PR, MD Gale (1988) Location of β-amylase sequence in wheat and its relatives. Theor Appl Genet 75:286–290
Simonsen Ø (1975) Cytogenetic investigations in diploid and autotetraploid populations of Festuca pratensis Huds. Hereditas 79:73–108
Soreng RJ, Davis JI (1998) Phylogenetics and character evolution in the grass family (Poaceae): simultaneous analysis of morphological and chloroplast DNA restriction-site character sets. Bot Rev 64:1-85
Stam P, van Ooijen JW (1995) Joinmap version 2.0: software for the calculation of genetic linkage maps. CPRO-DLO, Wageningen, The Netherlands
Tavoletti S, Veronesi F, Osborn TC (1996) RFLP linkage map of an alfalfa meiotic mutant based on an F1 population. J Hered 87:167–170
Tikhonov AP, SanMiguel PJ, Nakajima Y, Gorenstein NM, Bennetzen J, Avramova Z (1999) Colinearity and its exceptions in orthologous adh regions of maize and sorghum. Proc Natl Acad Sci USA 96:7409–7414
Van Deynze AE, Nelson JC, O'Donoughue LS, Ahn SN, Siripoonwiwat W, Harrington ESY, Braga DP, McCouch SR, Sorrells ME (1995a) Comparative mapping in grasses. Oat relationships. Mol Gen Genet 249:349–356
Van Deynze AE, Nelson JC, Harrington SN, Yglesias ES, Braga D, McCouch SR, Sorrells ME (1995b) Comparative mapping in grasses. Wheat relationships. Mol Gen Genet 248:744–754
Ventelon M, Deu M, Garsmeur O, Doligez A, Ghesquiére A, Lorieux M, Rami JF, Glaszmann JC, Grivet L (2001) A direct comparison between the genetic maps of sorghum and rice. Theor Appl Genet 102:379–386
Voorrips RE (2001) MapChart version 2.0: windows software for the graphical presentation of linkage maps and QTLs. Plant Research International, Wageningen, The Netherlands
Vos P, Hogers R, Blecker M, Rijans M, van der Lee T, Hornes M, Frijters A, Pot J, Poleman J, Kuiper M, Zabeau M (1995) AFLP: a new technique for DNA fingerprinting. Nucleic Acids Res 23:4407–4414
Wendel JF, Weeden FF (1989) Visualisation and interpretation of plant isozymes. In: Solitis DE, Soltis PS (eds) Isozymes in plant biology. Chapman and Hall, London, UK, pp 1–45
Wilson WA, Harrington SE, Woodman WL, Lee M, Sorrells ME, McCouch SR (1999) Inferences on the genome structure of progenitor maize through comparative analysis of rice, maize and the domesticated Panicoids. Genetics 153:453–473
Wolfe KH, Gouy M, Yang Y-W, Sharp PM, Li W-H (1989) Date of the monocot-dicot divergence estimated from chloroplast DNA sequence data. Proc Natl Acad Sci USA 86:6201–6205
Xie DX, Devos KM, Moore G, Gale MD (1993) RFLP-based genetic maps of the homoeologous group-5 chromosomes of bread wheat (Triticum aestivum L.). Theor Appl Genet 87:70–74
Xu WW, Sleper DA, Hoisington DA (1991) A survey of restriction fragment length polymorphisms in tall fescue and its relatives. Genome 34:686–692
Xu WW, Sleper DA, Chao S (1995) Genome mapping of polyploid tall fescue (Festuca arundinaceae Schreb.) with RFLP markers. Theor Appl Genet 91:947–955
Zabeau M, Vos P (1993) Selective restriction fragment amplification: a general method for DNA fingerprinting. European Patent Application number: 92402629.7, Publication number 0 534 858 A1
Acknowledgments
The seed of B14, accession no. 1700, was kindly provided by Dr. Valeria Negri, University of Perugia, Italy. Dr. P. Stephenson, Comparative Genetics Unit, John Innes Centre, UK, and Dr. J.C. Glaszmann, CIRAD, Montpellier, France, developed the EGRAM anchor probe sets, and provided the following probes: PSR (wheat cDNA PSR50-200, wheat gDNA PSR300-1300), RZ (rice cDNA), RGC (rice cDNA), RGG (rice gDNA), CSU (maize cDNA), SbRPG (sorghum cDNA) and PSM (pearl millet gDNA). The BCD (barley cDNA), CDO (oat cDNA), WG (wheat gDNA) and ABG (wheat gDNA) clones were provided from the Grain Genes probe repository (http://wheat.pw.usda.gov/ggpages/probes/index.html), Dr. Olin Anderson, USDA-ARS-WRRC, US. The ABLMC (Lolium multiflorum coleoptile cDNA) and the ABLGP (L. perenne gDNA) clones were provided by Dr. John Forster, La Trobe University, Australia, and Dr. Timothy Close, UC Riverside, USA, kindly provided the dehydrin clones. Dr. Fred Eickmeyer, SZ Steinach, Germany, provided the mapping data for the isozyme locus Acp-2. We gratefully acknowledge all probe contributors, and Øyvind Jørgensen, Kristin Bye Olsen and Torleiv Veum for excellent technical assistance. We thank Prof. Mike Gale for support and many useful discussions and suggestions in preparing this manuscript. This study was supported by the EU-project EGRAM (European Gramineae Mapping Project (contract No. BI04-CT97-2220), and grant no. 110733/112 from the Research Council of Norway.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by P. Langridge
Rights and permissions
About this article
Cite this article
Alm, V., Fang, C., Busso, C.S. et al. A linkage map of meadow fescue (Festuca pratensis Huds.) and comparative mapping with other Poaceae species. Theor Appl Genet 108, 25–40 (2003). https://doi.org/10.1007/s00122-003-1399-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00122-003-1399-5