Abstract
High-molecular-weight glutenin subunits (HMW-GS) are important determinants of wheat dough quality as they confer visco-elastic properties to the dough required for mixing and baking performance. With this important role, the HMW-GS alleles are key markers in breeding programs. In this work, we present the use of a PCR marker initially designed to discriminate Glu1 Bx7 and Glu1 Bx17 HMW-GS. It was discovered that this marker also differentiated two alleles, originally both scored as Glu1 Bx7, present in the wheat lines CD87 and Katepwa respectively, by a size polymorphism of 18 bp. The marker was scored across a segregating doubled-haploid (DH) population (CD87 × Katepwa) containing 156 individual lines and grown at two sites. Within this population, the marker differentiated lines showing the over-expression of the Glu1 Bx7 subunit (indicated by the larger PCR fragment), derived from the CD87 parent, relative to lines showing the normal expression of the Glu1 Bx7 subunit, derived from the Katepwa parent. DNA sequence analysis showed that the observed size polymorphism was due to an 18 bp insertion/deletion event at the C-terminal end of the central repetitive domain of the Glu1 Bx 7 coding sequence, which resulted in an extra copy of the hexapeptide sequence QPGQGQ in the deduced amino-acid sequence of Bx7 from CD87. When the DH population was analysed using this novel Bx7 PCR marker, SDS PAGE and RP HPLC, there was perfect correlation between the Bx7 PCR marker results and the expression level of Bx7. This differentiation of the population was confirmed by both SDS-PAGE and RP-HPLC. The functional significance of this marker was assessed by measuring key dough properties of the 156 DH lines. A strong association was shown between lines with an over expression of Bx7 and high dough strength. Furthermore, the data demonstrated that there was an additional impact of Glu-D1 alleles on dough properties, with lines containing both over-expressed Bx7 and Glu-D1 5+10 having the highest levels of dough strength. However, there was no statistically significant epistatic interaction between Glu-B1 and Glu-D1 loci.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Polymeric glutenins and monomeric gliadins are the major storage proteins in the wheat endosperm and endow wheat flour with its unique visco-elastic properties essential in end-product quality and bread-making ability (Payne and Lawrence 1983). Glutenin polymers are comprised of both high- and low molecular-weight glutenin sub-units (HMW-GS and LMW-GS respectively). The HMW-GS are encoded by paired genes designated as "x" and "y" types at three loci, Glu-A1, Glu-B1 and Glu-D1, present on the long arms of chromosome 1A, 1B and 1D respectively (Payne et al. 1987). The LMW-GS are encoded by genes at the Glu-A3, Glu-B3 and Glu-D3 loci on the short arms of chromosome 1A, 1B and 1D, respectively (Gupta and Shepherd 1990). Although the HMW-GS are less abundant than the LMW-GS, they have a major impact on dough quality, are amenable to relatively straightforward analysis and therefore have been used extensively as markers in breeding programs (Cornish et al. 2001a).
SDS-PAGE (sodium dodecyl sulphate polyacrylamide gel electrophoresis) is an important and efficient method for profiling HMW-GS in different wheat lines. However, difficulties arise when different subunits of the same molecular weight and mobility cannot be differentiated, as indicated by D'Ovidio and Anderson (1994) and Lafiandra et al. (1994). Some of the ambiguity encountered with SDS-PAGE has been overcome using RP-HPLC analysis, where HMW-GS of near identical molecular-weight and electrophoretic mobility, such as Ax2 and Ax2*, could be readily resolved according to their elution times (Marchylo et al. 1992). However, in the case of alleles of the Glu-B1-encoded subunit Bx7, only a small difference in electrophoretic mobility exists between subunit Bx7 and Bx7* (Bx7 being slightly larger than Bx7*) and, additionally, these subunits cannot be differentiated on the basis of elution time using RP-HPLC (Marchylo et al. 1992). Quantitative analysis of these two subunits in Canadian cultivars using RP-HPLC, revealed that the proportion of subunit Bx7 relative to the total amount of HMW-GS was significantly higher than for subunit Bx7*. Subunit Bx7* was found in combination with subunit By9 (as for Katepwa, one of the parent lines used in this study), subunit By8, or a variant designated as subunit By8*, whereas subunit Bx7 was found in combination with By8 or By8* only (Marchylo et al. 1992).
The balance between the viscous and the elastic properties of dough determine its strength during mixing and the baking process. In many breeding programs, high dough strength is used as a predictor of good quality bread wheat. Dough strength has been attributed largely to the type of allele present at the Glu-D1 locus; the HMW-GS allelic pair Dx2+Dy12 (Glu-D1 a) has been found to give weaker, inferior doughs relative to wheat containing the allelic pair Dx5+Dy10 (Glu-D1 d) (Payne et al. 1987). The increased dough strength of the Dx5+Dy10 subunit pair is thought to be due to an additional cysteine residue present in Dx5 (Anderson and Green 1989) and was demonstrated in in vitro experiments incorporating bacterially expressed Dx5 and Dy10 polypeptides into the glutenin polymer (Békés et al. 1994), and most recently in vivo, by the functional analysis of independent doubled-haploid populations established both in Australia (Békés et al. 2001) and Canada (Radovanovic et al. 2002). It has also been observed that cultivars with an over-expression of Glu-B1 Bx7 tend to have improved dough strength (Lukow et al., 1992; Marchylo et al. 1992; D'Ovidio et al. 1997). This association has recently been shown for a range of Australian and North American cultivars and lines (Butow et al. 2002; Radovanovic et al. 2002), and in old Hungarian varieties and landraces (Júhasz et al. 2000) assessed for functional dough properties. Greater dough strength was indicated by increased mixing times (MT), maximum resistance to extension (Rmax) and decreased resistance breakdown (RBD, Békés et al. 2001). Dough extensibility was also increased in cultivars showing an increased expression of Glu1Bx7, although this appeared to be a more complex trait involving other parameters such as LMW-GS and gliadin composition (Gupta et al. 1994; Cornish et al. 2001b).
The aim of this current research was to define at the molecular level, the nature of allelic differences amongst previously undifferentiated Bx7 alleles. This information has allowed us to investigate the linkage between the Bx7 structural gene and expression level, and to characterize associations between distinct Bx7 alleles and wheat flour dough quality parameters. A combination of molecular and biochemical approaches have been used in order to achieve this objective.
Materials and methods
Materials
Doubled-haploid lines (n=156 progeny lines) were derived from a CD87 × Katepwa F1 cross and subsequent wheat × maize doubled-haploid system; plants grown at Wongan Hills and Horsham were produced by the National Wheat Molecular Marker Program in Australia (NWMMP) (Kammholz et al. 2001). The parents of this population were chosen for their contrasting physiological and functional features, specifically the high extensibility of CD87 as compared to that of Katepwa. The HMW-GS genotypes of the parent lines are: CD87—Glu-A1b (Ax2*), Glu-B1b (Bx7+By8*), Glu-D1a (Dx2+Dy12), and Katepwa—Glu-A1b (Ax2*), Glu-B1c (Bx7+By9), Glu-D1d (Dx5+Dy10).
Wheat DNA extraction
Genomic DNA was extracted from 3–6 day old hypocotyls (10 mg) of germinating seeds or from 50 mg of flour for the DH lines using a rapid isolation technique (Stewart and Via 1993).
PCR analysis of Glu-B1 sub-alleles
PCR was performed in a reaction volume of 20 μl using 20 ng of genomic DNA, 1 U of HotStar Taq DNA polymerase (Qiagen), in 1 × PCR buffer (Qiagen, containing 1.5 mM of MgCl2) 200 μM of dNTP mix and 10 ρmol each of the following Bx primers: forward 5′-CGCAACAGCCAGGACAATT-3′, and reverse 5′-AGAGTTCTATCACTGCCTGGT-3′ (Ma et al. 2003). Amplification conditions for the PCR reaction were an initial cycle at 95°C for 5 min, followed by 38 cycles of; 95°C for 30 s, 58°C for 30 s and 72°C for 1 min, followed by a final extension at 72°C for 5.25 min. PCR products were visualised as described by Rampling et al (2001). Genescan analysis of PCR products of the whole DH population, was carried out with the addition of the 0.04 μlL/PCR reaction dUTP-labelled with the fluorescent dye, R110 (Applied Biosystems). The PCR product was run on an ABI 377 sequencer and the data subsequently processed using Genescan 3.1 software (Rampling et al. 2001).
Sequencing of polymorphic PCR fragment
The 670-bp Bx7 PCR fragment obtained from PCR amplification of the CD87 parent line was purified from Agarose gel (QIAquick Gel Extraction Kit, Qiagen); 0.01 μl of the resulting purified gel-band extract was re-amplified using the original Bx PCR primers as described above, except that only 20 PCR cycles were used. For each sequencing reaction, 1 μl of the re-amplified PCR product was incubated with one unit of shrimp alkaline phosphatase and 2.5 U of exonuclease 1 (both from USB) in 1 × PCR buffer (6-μl final volume of Qiagen, containing 1.5 mM of MgCl2) at 37°C for 15 min. Enzymes were denatured at 80°C for 15 min and the reaction placed on ice prior to addition of DNA sequencing mix [6 μl of Big Dye terminator (Applied Biosystems) and 3.2 pmoles of sequencing primer] to give a final volume of 20 μl; 30 cycles of sequencing were performed with an annealing temperature of 58°C. Sequence was obtained for both strands using the Bx Forward and Bx Reverse primers as used for the original PCR amplification. These sequences were aligned using Pileup (GCG package, WebANGIS) and used to search the Genbank NR database (Blast-N).
SDS-PAGE analysis of HMW-GS composition
Crushed grain (25 mg) was initially extracted with 200 μl of 50% n-propanol by vortexing and centrifugation to remove the gliadin fraction. Glutenins were extracted from the resulting pellet with 100 μl of 80 mM of Tris-HCl buffer pH 8.0, containing 1% dithiothreitol (DTT). This extract was then treated with 100 μl of 1.4% 4-vinylpyridine to alkylate the subunits and prevent the re-formation of disulphide bonds. The extracts were subjected to one-dimensional electrophoresis and stained as described in Cornish et al. (2001a).
RP-HPLC analysis of HMW-GS
Following milling of the grain, the flour was extracted sequentially for gliadins and glutenins, and the HMW-GS composition was analysed by reversed phase HPLC (RP-HPLC) (Marchylo et al. 1989) as modified by Larroque et al. (2001).
The % mole of Bx protein present was calculated using the following Mr for each HMW-GS: Ax2*, 86 309; Bx7, 82 865; Dx2, 87 022; Dx5, 88 128; By9, 73 518; Dy10, 67 476; and Dy12, 68 650 Da respectively (Shewry et al. 1989). The deduced amino-acid sequence of the HMW-GS genes in cv Glenlea (Cloutier and Lukow 2001) were used for estimation of the molecular weights of Bx7* and By8 (83 017 and 75 017 Da respectively).
Dough-quality testing
A two-gram Mixograph (Rath et al. 1990) was used to evaluate functional dough properties of the DH lines. The water absorption of each was estimated by the Approved Method (AACC 1995) using flour protein and moisture content. Mixing tests were performed in duplicate and the following key parameters were measured: mean time to peak-dough development [mixing time, MT (s)]; height of mixograph trace at peak resistance {peak resistance, PR [arbitrary units (au)]}; percentage decrease in dough resistance 3 min after the peak [resistance breakdown, RBD (%)].
Extensigraph measurements were carried out (single repeats of duplicate samples) on 1.7 g dough pieces to determine the maximum resistance of the dough to extension [Rmax (N)] (D'Ovidio et al. 1997) and dough extension before severing [Ext. (m)], calculated with custom software (Rath et al. 1994).
Statistical analysis
A general linear model was applied to the data sets. A two-way analysis of variance (AVOVA, at two-level factors) and regression analysis were carried out using MSUSTAT v. 4.1 (Richard E. Lund, Montana State University, Bozeman, Mont.) in order to test the effects of HMW-GS encoded at Glu-B1 and Glu-D1 on dough quality parameters.
Results and discussion
Expression levels of the Bx7 sub-unit
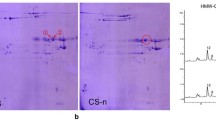
A preliminary investigation comparing expression levels of Glu1Bx7 by RP-HPLC and SDS PAGE in different cultivars showed that the cultivar CD87 gave a similar RP-HPLC profile to Glenlea, a Canadian cultivar with a high-expression Glu1Bx7 allele and possessing high dough strength (Marchylo et al. 1992). In CD87, although the expression level of Bx7 as a function of % HMW-GS was not as high as that of Glenlea, Bx7 was expressed to consistently higher levels than in the reference line Katepwa (Fig. 1, upper traces). RP-HPLC analysis of progeny from a DH population generated from an F1 cross between CD87 × Katepwa revealed a range of expression levels in the progeny, with some DH lines having three times the relative amount of Bx7 within the total HMW-GS fraction (Fig. 1, lower traces).
RP-HPLC chromatograms showing HMW-GS profiles of the cultivars Glenlea (a), CD87 (b) and Katepwa (c). Sample profiles from the CD87 × Katepwa double-haploid population indicate the range of Bx expression levels (as a function of %HMW-GS) in lines containing Bx7CD+By8* (d) and Bx7*+By9 (e) progeny lines
Discrimination of Bx7 alleles at the DNA sequence level
The Bx PCR marker developed by Ma et al. (2003) was found to be a useful tool that could be used to assist in the discrimination of Bx7 alleles that differ in expression level. The marker produced two bands for cultivars containing Glu1Bx7, with fragment sizes of approximately 650 and 750 bp, and a single band of approximately 675 bp for cultivars such as Sunstate used here, containing Glu1Bx17 (Fig. 2a). The two PCR products produced from the Glu1Bx7 gene are due to the presence of two priming sites for the forward Bx primer in the central repetitive domain of this gene (Ma et al. 2003). The cultivar Katepwa (Bx7*, normal Bx7 expression level) yielded two bands of approximately 650 bp and 750 bp, whereas the cultivar CD87 (over-expression of Bx7) gave two bands of approximately 670 and 770 bp. The Bx7-containing cultivars, Glenlea, Red River, Chara and Kukri, all shown by RP-HPLC to exhibit an over expression of the Glu1Bx7 subunit (Butow et al. 2002) were tested with the Bx7 marker and found to give identical-sized PCR products to CD87, while other Bx7 subunit-containing cultivars, Janz, Frame, Meering and Cheyenne, that did not show over-expression of Bx7, gave the same pattern as Katepwa (Fig. 2b). Although a specific PCR marker amplifying a 2,373 bp fragment of Glu1Bx7 has been reported (Ahmad 2000) it has not been shown to identify cultivars showing an over-expression of the Bx7 subunit.
PCR assay for the Bx HMW-GS gene used for Bx7-typing of lines from the CD87 × Katepwa DH population. Lanes labelled from left- right: (1) GeneRuler 50 bp ladder; (2–10) progeny lines for the DH population, those marked with an asterisk (*) indicate progeny lines producing larger PCR fragments (18 bp for both amplicons); (11) Sunstate (Bx17, fragment at ~675 bp); (12) CD87 parent (Bx7CD, fragments ~670 and 770 bp); (13) Katepwa parent (Bx7*, fragments ~650 bp and 750 bp; (14) nil DNA control; (15) GeneRuler 50-bp ladder. b PCR assay for the Bx HMW-GS gene used to differentiate cultivars showing normal and over-expression of Bx7. Cultivars with normal Bx7 expression: Janz, Meering, Frame, Krichauff and Cheyenne all have fragments of 650 bp and 750 bp. Cultivars with high Bx7expression levels: Chara, Glenlea, Red River and Kukri all have fragments of 670 bp and 770 bp. Lanes 1 and 12 show the 50-bp ladder and lane 11 is a "nil DNA" control
DNA sequence analysis of the major (670 bp) Bx7 PCR product from CD87 and comparison with the Bx7 sequence from the cultivar Cheyenne (Genbank X13927, Anderson and Green 1989) revealed the presence of an 18 bp duplication in the CD87 sequence after nucleotide 18,245 of the Cheyenne database sequence (Fig. 3). This results in a duplication of the hexapeptide sequence QPGQGQ towards the C-terminal end of the central repetitive domain of the Bx7 deduced amino-acid sequence. One additional single nucleotide polymorphism (SNP) was observed in the CD87 sequence relative to the Cheyenne sequence, a C to G change at position 18,242 of the sequence. Results of a BLAST search against the Genbank EST database using the sequence of the CD87 Bx7 PCR fragment, revealed that undefined EST sequences derived from developing seeds of Glenlea also carry the same 18 bp duplication and SNP (e.g. Genbank BQ252352, S. Cloutier 2002). This BLAST search also picked up the same duplication and SNP for undefined EST sequences in a wheat cDNA library of the cultivar Chinese Spring (e.g. Genbank BJ299809, T. Shin-i 2002). Furthermore, a similar 18 bp duplication and SNP is also present in the Glu-1Bx20 sequence from T. turgidum subsp. durum (Shewry et al. 2002, Genbank AJ437000), although in this case, the duplication is imperfect (Fig. 3). The sequence identity between the 670 bp CD87 Bx7 sequence and the durum Bx20 sequence was found to be 98%. The association being between the Bx7 over-expression phenotype and the DNA sequence polymorphism.
DNA sequence alignment illustrating the sequence duplication in the T. aestivum CD87 Bx7 and T. turgidum subsp. durum Bx20 sequences compared to Bx7 from T. aestivum Cheyenne. Gaps are indicated by dashes (-) and sequence polymorphisms by bold type. The 18-nucleotide sequence tandemly duplicated in the CD87 Bx7 sequence is underlined. Numbering refers to the sequence of the Cheyenne Bx7 gene (Genbank X13927; Anderson and Greene 1989)
The CD87 × Katepwa DH progeny were scored using the Bx7 PCR marker and analysed quantitatively for the Bx7 expression level using RP-HPLC. The molar % of Glu1 Bx7, present in the HMW-GS fraction, was calculated for all the lines. The mean % Bx7 for CD87 Bx7-type lines was 13.25±0.78 (n=84) whereas Katepwa Bx7-type lines showed a mean value of 8.55±0.69 (n=72). Thus 46.2% of the population was found to be Glu-B1 c (i.e. Bx7*), and 53.8% exhibited CD87-type expression of Glu-B1. This analysis also enabled a clear separation of the doubled-haploid population according to the Bx7 type. An identical differentiation of the population was found from scoring with the Bx7 DNA marker, and when the allelic compositions of sample lines were measured by SDS PAGE and compared to respective mol % Bx values. Having ascertained that there was a wide range in expression levels of the Bx7 sub-unit, the RP-HPLC data was also used to calculate the distribution frequency of % Bx7 expression in flour (Fig. 4). The bimodal distribution shows "long tails" implying transgressive segregation within the DH population. This observation is suggestive of multiple gene effects and a phenomenon in which there is a main single gene underlying the expression, which is clear in this population, i.e. the Bx7 gene, which is expressed in two different forms. The variation within each distribution may be due to transcriptional factors or the effects of non-structural genes. It is interesting to note that the % Bx7 expression in the CD87 parent-line flour was considerably lower than that of the extreme value for its progeny, this is currently being investigated further. A clear double-peaked distribution was also obtained when the same population was grown at Horsham (data not shown), although there was a shift in the frequency distribution towards higher flour % Bx values for both Katepwa- Bx7-type and CD87-Bx7-type lines. Further evidence that the CD87 Bx7+By8* allele was different to other reference Bx7+By8 lines (such as the cultivar Janz), was also provided by RP-HPLC, whereby the By8* peak in CD87 and CD87 Bx7-like lines was eluted before the Dx peak, as shown for Glenlea (Marchylo et al. 1992). SDS-PAGE analysis of the DH population readily enabled the differentiation of Katepwa-like (Bx7*+By9) and CD87-like (Bx7+By8*) progeny due to the difference in mobility the By subunit. However, this technique was not sufficiently quantitative to allow discrimination of progeny on the basis of expression level. A two-fold difference in staining intensity of subunit Bx7 has been observed in cultivars showing over-expression of Glu1Bx7, for example in Red River (D'Ovidio et al. 1997), but in contrast, CD87 was not strongly over-stained and therefore differences in Bx7 expression could not be resolved by SDS-PAGE. The CD87 and Katepwa parents and DH lines (grown at both Wongan Hills and Horsham) were scored with the Bx PCR marker using both initial Agarose-gel analysis of the PCR products and subsequent confirmation of the 18 bp size shift between alleles using Genescan analysis. Figure 2a shows a sub- sample of the DH population subjected to analysis with the Bx PCR marker. DH lines producing two fragments of 670 and 770 bp were designated CD87 Bx7-like progeny, whereas lines with fragments of 650 and 750 bp were Katepwa Bx7-like progeny. Thus clear lines of evidence demonstrated that CD87 and its progeny lines were found to exhibit over-expression of Bx7 as shown by RP-HPLC and, together with allele discrimination using the PCR marker, this enabled an identical classification of the DH population.
Frequency distribution of Bx subunits in flour (normalised for protein content) in 156 doubled-haploid lines grown at Wongan Hills. Values of %Bx in flour for the parent lines, CD87 and Katepwa are shown with thick vertical arrows, and the standard error for values is represented by the small horizontal arrows
At this point we feel it is salient to address the issue of nomenclature of the Bx7 alleles described in this DH population. We have shown that differences in the HMW-GS cannot be based upon electrophoretic mobility alone but are fundamentally related to different DNA sequences, and therefore we will refer to the over-expressing CD87-type of Bx7 allele as Bx7CD. Whether or not this allele is in fact Glu-B1al, as designated for Glenlea by Marchylo et al. (1992), can only be clarified by full comparative sequencing.
Functional analysis of the DH population reveals the association of the Bx marker expression level and increased dough strength
The significance of the differentiation of the two types of Bx7 expression level became evident when 156 lines of the CD87/Katepwa DH population were tested for functional dough properties. A significant increase in dough strength (P<0.001) was associated with all the DH lines exhibiting an over-expression of Glu1 Bx7. The association of over-expression of Bx7 in Glenlea, Red River, Chara and Kukri with increased dough strength has also been shown previously using small-scale Mixograph and Extensigraph methods (Butow et al. 2002).
As it is known that the Glu-D1 locus also plays an important role in dough quality, we investigated the associations of genes and functional characters in the population according to the Glu-D1-encoded alleles, Glu-1a (i.e. Dx2+Dy10) and Glu-D1d (i.e. Dx5+Dy10), readily identifiable by SDS PAGE (Shewry et al. 1989). The DH population showed functional differentiation, according to the combination of the Glu-B1- and Glu-D1-encoded alleles (Fig. 5a and c). It was clearly demonstrated that those lines with Bx7CD+By8*, in combination with Dx5+Dy10, had significantly higher dough strength (Table 1, P<0.001), and indicated higher MT (~385 s) and Rmax (0.53 N) values and lower %RBD (<10% RBD).
Functional dough analysis relating to dough strength and extensibility of 156 lines of the CD87 × Katepwa DH population. The population was differentiated into four groups of allele combinations for Rmax vs Ext (a) and MT vs RBD (c) where symbols represent the following Glu-B1/Glu-D1 combinations: (■) Bx7*+By9/Dx5+Dy10; (❑) Bx7CD+By8*/Dx5+Dy10; (▲) Bx7*+By9/Dx2+Dy12; (Δ) Bx7CD+By8*/Dx2+Dy12. The same population was also graphed differently for two groups of allele combinations at Glu-D1: Rmax vs Ext (b) and MT vs RBD (d), where symbols represent the following Glu-D1 combinations: (+) Dx5+Dy10; (O) Dx2+Dy12
The association of Bx7CD+By8* with Dx2+Dy12 did not give a clear differentiation, and the functional results were similar to those obtained with lines with a Bx7*+By9/Dx5+Dy10 combination (Fig. 5a and c). These combinations of Glu-B1 and Glu-D1 encoded-alleles, resulted in functional data whereby the dough formed was weaker, as shown by the increased % RBD with the decreased MT and Rmax. The weakest and least-stable dough was given by lines with the Bx7*+By9/Dx2+Dy12 combination (Table 1, Fig. 6a and c). Table 1 highlights the significant differences between lines showing variation in dough quality; those lines with intermediate dough quality (Bx7 CD+By8*/Dx2+Dy12 and Bx7*+By9/Dx5+Dy10) only differed in their dough stability (% RBD), with the "Bx7 CD+By8*/Dx2+Dy12" lines showing significantly reduced dough stability relative to the Bx7*+By9/Dx5+Dy10 lines. The spread of data points within each of the four genotypes may be explained by the interactions of other factors such as the qualitative and quantitative effects of LMW-GS and gliadins, which also play a significant role in dough quality (Gupta et al. 1994; Cornish et al. 2001b).
Functional dough analysis relating to dough strength and extensibility of 156 lines of the CD87 × Katepwa DH population: (a) MT vs mol% Bx7 (b) Rmax vs mol% Bx7. Symbols represent the following Glu-B1 /Glu-D1 combinations: (■) Bx7*+By9/Dx5+Dy10; (❑) Bx7CD+By8*/Dx5+Dy10; (▲) Bx7*+By9/Dx2+Dy12; (Δ) Bx7CD+By8*/Dx2+Dy12
A recent study (Radovanovic et al. 2002) of a DH population established between a Glenlea-derived line and Karma, confirmed the importance of both "H7+8" (i.e. over-expressing Bx7) and "5+10" allelic combinations for the determination of dough strength; low intergenomic interactions between Glu-B1 and Glu-D1 were shown for SDS-sedimentation volume, total energy and band-width energy only. When the statistical interaction of Glu-B1 and Glu-D1 was tested within the current DH population, there was no significant interaction at either of the field sites (Table 2). This result is in accord with a recent comprehensive analysis of data from other Australian breeding programs (Eagles et al. 2002). The impact for breeders of the Bx PCR marker in differentiating the expression level of Glu1Bx7 (i.e. Bx7CD or Bx7*) is demonstrated in Fig. 5b and d in which the DH progeny are only delineated by their Glu-D1 alleles. Whereas SDS PAGE was clearly unsuitable to differentiate expression levels of Bx7 in the CD87 × Katepwa DH population, RP-HPLC can be used, but is time-consuming, expensive and requires dedicated equipment; moreover, both methods require an endosperm protein sample. The use of the Bx PCR marker therefore has the advantage for breeders of being amenable to routine analysis using DNA from any tissue. Scoring of the CD87 × Katepwa DH population used in this study using the Bx marker permitted the re-definition of the two sub-populations shown in Fig. 5b and d to that of four functionally different sub-populations shown in Fig. 5a and c respectively.
It has been surmised (Radovanovic et al. 2002) that increased gluten strength associated with the over-expression of Bx7 in Glenlea-derived lines was due to the amount of the x-type subunit and this was confirmed in the CD87 × Katepwa DH population used in our study by the analysis of mol% Bx7. A clear delineation between three groups of allele combinations is shown in Fig. 6a and b, whereby the strongest dough characteristics could be attributed to mol% Bx values ranging from 35 to 50%. Such values were found in Bx7CD+By8*/Dx5+Dy10 lines and also in Bx7CD+By8*/Dx2+Dy12 lines, which gave lower MT and Rmax values, similar to those of Bx7*+By9/Dx5+Dy10 lines. The poorest strength was given by lines with a Bx7*+By9/Dx2+Dy12 combination where the mol% Bx was between 25 and 35%. As an extension of the statistical analysis enabling a grouping of dough quality attributes with allele type (Table 1, 2 and 3), we established a correlation matrix between dough quality parameters and the molar % Bx and By (calculated from RP-HPLC data) for each Glu-D1 combination respectively (Tables 4 and 5). Overall, there was a significant interaction between mol% By, mol% Bx and Bx/By ratio, and dough-strength parameters to a greater extent in Glu1 Dx5+Dy10 lines (Table 4), with a highly significant relationship between mol% Bx rather than mol% By or the Bx/By ratio. Similarly, the most-significant correlations in Glu1Dx2+Dy12 lines was found between mol% Bx and MT and Rmax (Table 5), although the correlations were lower than for Glu1 Dx5+Dy10 lines. Within both the Glu1 Dx5+Dy10 lines and Glu1Dx2+Dy12, there was a highly significant correlation between mol% Bx and MT (r=0.679*** and 0.7264*** respectively), RBD (r=−0.6624*** and −0.4912** respectively), and with Rmax (r=0.8065*** and 0.6347*** respectively).
Environmental effects
The effects of genotype on dough strength were generally similar for the two field sites at which the CD87 × Katepwa DH population was grown. However, the hotter, drier climate of Wongan Hills produced increased MT and Rmax values (377 s and 0.508 N respectively) for Bx7*+By9/Dx5+Dy10 lines as compared to those values recorded in the more temperate climate at Horsham (315 s and 0.447 N respectively) for the same lines. It is interesting to note that the average %Bx in the flour (normalized for total protein content) of Bx7*+By9/Dx5+Dy10 lines was higher in the Horsham samples (0.77%) compared to the Wongan Hills samples (0.74%). A different pattern was shown by the other three combinations of Glu-B1- and Glu-D1-encoded alleles, whereby dough strength was greater in all cases for lines grown at Horsham. The flour mol %Bx for these three genotypes was also greater in lines grown at Horsham than at Wongan Hills (Table 1).
Extensibility
The DH lines exhibiting over-expression of Glu1 Bx7 showed significantly lower extensibility (P<0.01) than lines with a normal expression level of Glu1 Bx7 (Fig. 5a, Table 3). Furthermore, extensigraph measurements showed a significant separation between Bx7CD+By8*/Dx5+Dy10 lines with high dough strength (Rmax>0.3 N) and the poorest extensibility (Ext<0.13 m), and the other allelic combinations (Table 1, Fig. 5a). There was a negative correlation between extensibility and mol% Bx, although this effect was less significant for the Glu1 Dx2+Dy12 lines. A significant correlation with peak resistance and mol% By, mol% Bx and Bx/By ratio was only shown with Glu1 Dx5+Dy10 lines (Table 4), indicating the importance of genetic background effects. Similarly, mol% By correlated significantly with extensibility in Glu1 Dx5+Dy10 lines only.
In summary, dough strength and stability were highest in the Bx7 CD+By8*/Dx5+Dy10 lines whereas extensibility was lowest. This statistical analysis conclusively supports the hypothesis of the linkage between over-expression of Glu1Bx7 and enhanced dough strength.
Conclusions
We have demonstrated that despite the ostensibly similar SDS-PAGE electrophoretic mobility of the Bx7 sub-unit of CD87 to that of Bx7 from other Bx7+By8-containing cultivars, a size polymorphism exists in its Bx PCR product as in the Bx7CD allele. The subsequent identification of the sequence difference in the Bx PCR fragment enabled the distinction of this cultivar as one exhibiting over-expression of the Bx7 allele.
The use of the PCR marker to differentiate two types of the Glu1 Bx7 allele that differed in expression levels enabled a more definitive grouping of the DH population segregating for Glu1 Bx7*+By9 and Glu1 Bx7CD+By8*. Subsequent to this finding, three independent measurements of Bx7 allelic composition (SDS-PAGE), Bx7 type (using the Bx PCR marker) and mol% Bx (RP-HPLC) showed a perfect correlation. The PCR marker discriminates by revealing an insertion/deletion of an 18 bp sequence. Database searches revealed that two non over-expressing cultivars, a hexaploid wheat (Chinese Spring) and a durum wheat, both contained the same insertion as identified in CD87. These observations lead us to conclude that the 18 bp duplication is not causal for over-expression or increased dough strength and that it is therefore reasonable to conclude that the over-expression of Bx7 is linked to dough strength. Whether the observed over-expression of Bx7 in CD87 is due to a duplication of the Bx7 gene, as suggested for Red River and TAA36 (Lukow et al 1992; D'Ovidio et al. 1997), an improved translational mechanism (Marchylo et al. 1992), or other mechanism, is currently being investigated. Furthermore, either over-expression of Glu1 Bx7 is the determinate of the altered functional properties, or the intrinsic differences between the CD87 and Katepwa Glu-B1 alleles produce functionality changes resulting in increased dough strength.
Using the Bx7 PCR marker, in combination with SDS PAGE and RP-HPLC results, an assessment was made of the functional dough properties of progeny lines at two independent sites grouped according to four allelic combinations at Glu-B1 and Glu-D1. Despite the evidence that the character of the Glu-B1 and Glu-D1-encoded alleles are both important determinants of dough strength, there was no statistically significant B × D inter-genomic or epistatic interaction for the parameters measured.
Thus we have discovered a valuable molecular tool for following the segregation of quality characteristics and a powerful method for controlling dough quality parameters in breeding programs. For example, this work pinpoints potential pre-breeding material with the desirable traits of high dough strength and moderate extensibility.
The results from this work bear importance, also regarding the genetic mapping of the CD87 × Katepwa DH population due to the re-designation of CD87 and resulting DH lines as over-expressing Bx7. In retrospect, this can be deemed as an initial inaccuracy in polymorphism scoring, and could well be one source of segregation distortion, as discussed by Kammholz et al. (2001). The development of this new marker now creates the opportunity for an improved method of polymorphism scoring in populations derived from CD87, and should be evaluated for use in other populations containing high-expressing Bx7 alleles, bearing in mind that the 18 bp duplication should not be assumed to be an unambiguous marker for the over-expression of the Bx7 allele.
References
Ahmad MA (2000) Molecular marker-assisted selection of HMW glutenin alleles related to wheat bread quality by PCR-generated DNA markers. Theor Appl Genet 101:892–896
American Association of Cereal Chemists (1995) Approved Methods of AACC, 9th edn. Methods 54–40A, approved November 1995. The Association: St. Paul, Minnesota
Anderson OD, Green FC (1989) The characterisation and comparative analysis of high-molecular-weight glutenin genes from genomes A and B of a hexaploid bread wheat. Theor Appl Genet 77:689–700
Békés F, Gras PW, Gupta RB (1994) Mixing properties as a measure of reversible reduction and oxidation of doughs. Cereal Chem 71:44–50
Békés F, Gras PW, Anderssen RS, Appels R (2001) Quality traits of wheat determined by small-scale dough testing methods. Aust J Agric Res 52:1325–1338
Butow BJ, Gras PW, Haraszi R, Békés F (2002) The effects of different salts on mixing and extension parameters on a diverse group of wheat cultivars using 2-g mixograph and extensigraph methods. Cereal Chem 79:823–826
Cloutier S, Lukow OM (2001) Cloning the genes responsible for dough strength of CWES variety Glenlea. http://res2.agr.ca/Winnipeg/biotech_sspg.htm
Cornish GB, Skylas DJ, Siriamornpun S, Békés F, Larroque OR, Wrigley CW, Wooton M (2001a) Grain proteins as markers of genetic traits in wheat. Aust J Agric Res 52:1161–1171
Cornish GB, Békés F, Allen HM, Martin DJ (2001b) Flour proteins linked to quality traits in an Australian doubled haploid wheat population. Aust J Agric Res 52:1339–1348
D'Ovidio R, Anderson OD (1994) PCR analysis to distinguish between alleles of a member of a multigene family correlated with wheat bread-making quality. Theor Appl Genet 88:759–763
D'Ovidio R, Masci S, Porceddu E, Kasarda D (1997) Duplication of the high-molecular-weight glutenin subunit gene in bread wheat (Triticum aestivum L.) cultivar Red River 68'. Plant Breeding 116:525–531
Eagles HA, Hollamby GJ, Gororo NN, Eastwood RF (2002) Estimation and utilisation of glutenin gene effects from the analysis of unbalanced data from wheat breeding programs. Aust J Agric Res 53:367–377
Gupta RB, Shepherd KW (1990) Two-step one-dimensional SDS-PAGE analysis of LMW subunits of glutenin. I. Variation and genetic control of the subunits in hexaploid wheats. Theor Appl Genet 80:65–74
Gupta RB, Singh NK, Cornish GB, Palmer GA, Békés F, Rathjen AJ (1994) Allelic variation at glutenin subunit and gliadin loci, Glu-1, Glu-3 and Gli-1, of common wheats. I. Its additive and interaction effects on dough properties. J Cereal Sci 19:9–17
Juhász A, Larroque OR, Tamás L, Vida Gy, Zeller FJ, Békés F, Bedo Z (2000) Biochemical and molecular genetic background of the traditional Bankút breadmaking quality. In: Wootton M, Batey IL, Wrigley CW (eds) Cereals 2000 Proc 50th RACI Cereal Chemistry Conference. RACI, Surfers Paradise, Australia, pp 699–702
Kammholz SJ, Campbell AW, Sutherland MW, Hollamby GJ, Martin PJ, Eastwood RF, Barclay I, Wilson RE, Brennan PS, Sheppard JA (2001) Establishment and characterisation of wheat genetic mapping populations. Aust J Agric Res 52:1079–1088
Lafiandra D, D'Ovidio R, Margiotta B (1994) Studies of high-molecular-weight glutenin subunits and their encoding genes. In: Henry RJ, Ronalds JA (eds) Improvement of cereal quality by genetic engineering. Plenum Press, New York, pp 105–111
Larroque OR, Békés F, Wrigley CW, Rathmell WG (2001) Analysis of gluten proteins in grain and flour blends by RP-HPLC. In: Shewry PR, Tatham AS (eds) "Wheat Gluten". Royal Society of Chemistry, Cambridge, UK, pp 136–139
Lukow OM, Forsyth SA, Payne PI (1992) Over-production of HMW glutenin subunits coded on chromosome 1B in common wheat, Triticum aestivum. J Genet Breed 46:187–192
Ma W, Zhang W, Gale KR (2003) Multiplex-PCR typing of high-molecular-weight glutenin subunit alleles in wheat. Euphytica (in press)
Marchylo BA, Kruger JE, Hatcher DW (1989) Quantitative reverse-phase high performance liquid chromatographic analysis of wheat storage proteins as a potential quality prediction tool. J Cereal Sci 9:113–130
Marchylo BA, Lukow OM, Kruger JE (1992) Quantitative variation in high-molecular-weight glutenin subunit 7 in some Canadian Wheats. J Cereal Sci 5:29–37
Payne PI, Lawrence GJ (1983) Catalogue of alleles for the complex gene loci Glu-A1, Glu-B1 and Glu-D1 which code for high-molecular weight subunits of glutenin in hexaploid wheat. Cereal Res Commun 11:29–35
Payne PI, Nightingale MA, Krattinger AF, Holt LM (1987) The relationship between HMW glutenin subunit composition and breadmaking quality of British grown wheat varieties. J Sci Food Agric 40:51–65
Radovanovic N, Cloutier S, Brown D, Humphreys DG, Lukow OM (2002) Genetic variance for gluten strength contributed by high-molecular-weight glutenin proteins. Cereal Chem 79:843–849
Rampling LR, Harker N, Shariflou MR, Morell MK (2001) Detection and analysis systems for microsatellite markers in wheat. Aust J Agric Res 52:1131–1141
Rath CR, Gras PW, Wrigley CW, Walker CE (1990) Evaluation of dough properties from two grams of flour using the Mixograph principle. Cereal Foods World 35:572–574
Rath CR, Gras PW, Zhonglin Z, Appels R, Békés F, Wrigley C (1994) A prototype extension tester for two-gram dough samples. In: Panozzo JF, Downie PG (eds) "Proc 44th RACI Cereal Chemistry Conference", RACI: North Melbourne, pp 122–126
Shewry PR, Halford NG, Tatham AS (1989) The high-molecular-weight subunits of wheat, barley and rye: genetics, molecular biology, chemistry and role in wheat gluten structure and functionality. In: Oxford Surveys of Plant Molecular and Cell Biology Vol. 6, pp 163–219
Stewart CN, Via LE (1993) A rapid CTAB DNA isolation technique useful for RAPD fingerprinting and other PCR applications. Bio Techniques 15:748–749
Acknowledgements
We thank the NWMMP for providing the doubled-haploid samples, and D. Murray and A. Nikolov for their technical assistance. This work was carried out as part of the Graingene Research program; support from AWB Ltd. and GRDC is acknowledged. Graingene is a research consortium between the Australian Wheat Board, the CSIRO and the Grains Research and Development Corporation.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by J.W. Snape
Rights and permissions
About this article
Cite this article
Butow, B.J., Ma, W., Gale, K.R. et al. Molecular discrimination of Bx7 alleles demonstrates that a highly expressed high-molecular-weight glutenin allele has a major impact on wheat flour dough strength. Theor Appl Genet 107, 1524–1532 (2003). https://doi.org/10.1007/s00122-003-1396-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00122-003-1396-8