Abstract
The goal of the present experiments was to transfer the chromosomes of Solanum sitiens (syn. Solanum rickii) into cultivated tomato (Lycopersicon esculentum). By crossing an allotetraploid L. esculentum × Solanum sitiens hybrid to sesquidiploid L. esculentum × S. lycopersicoides, a trigenomic hybrid (2n+14=38) was obtained. Analysis of the latter by GISH (genomic in situ hybridization) indicated it contained a full set of 12 S. sitiens chromosomes, plus two extras from S. lycopersicoides. This and other complex hybrids were pollinated with Lycopersicon pennellii-derived bridging lines to overcome unilateral incompatibility. A total of 40 progeny were recovered by embryo rescue, including diploids and aneuploids (up to 2n+8). In order to determine the origin of chromosomes and the location of introgressed segments, progeny were genotyped with RFLP markers. S. sitiens-specific markers on all chromosomes, except 6 and 11, were detected in the progeny. Several S. sitiens chromosomes were transmitted intact, either through chromosome addition (i.e., trisomics) or substitution (i.e., disomics). Recombination between S. sitiens and L. esculentum was detected on most chromosomes, in both diploid and aneuploid progeny. A monosomic alien addition line for S. sitiens chromosome 8 was identified, and the extra chromosome was stably transmitted to approximately 13% of the backcross progeny. This study demonstrates the feasibility of gene transfer from S. sitiens to L. esculentum through chromosome addition, substitution, and recombination in the progeny of complex aneuploid hybrids.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Wild relatives of many crop plants have been important resources for plant breeding (Harlan 1976; Tanksley and McCouch 1997; Jarvis and Hodgkin 1999). Cultivated tomato (Lycopersicon esculentum) is cross compatible to varying degrees with each of the nine wild Lycopersicon species, which have been valuable sources of economic traits such as disease and insect resistance, fruit quality and environmental-stress tolerance (Rick and Chetelat 1995). The much larger Solanum genus (approximately 1,250 spp, Nee 1999) includes four tomato-like nightshades, Solanum lycopersicoides, Solanum sitiens, Solanum juglandifolium and Solanum ochranthum (Rick 1988). Their morphology, ecology, distribution and crossing-relationships all suggest these species represent two pairs of sister taxa, of which the first (S. lycopersicoides and S. sitiens) are most closely related to Lycopersicon (Rick 1988; Child 1990). Crosses between S. lycopersicoides and S. sitiens result in fertile interspecific hybrids, while only S. lycopersicoides is cross compatible with L. esculentum (Rick 1951, 1979; Pertuzé et al. 2002). However, S. sitiens can be indirectly hybridized with cultivated tomato using L. esculentum × S. lycopersicoides derivatives as a bridge (see below).
First discovered by Johnston (1929), S. sitiens was described again by Correll (1961) as Solanum rickii, a synonymy pointed out by Marticorena and Quezada (1977). Collection-site information from genebank accessions and herbarium specimens indicate that S. sitiens is restricted to a small area of the Atacama Desert in northern Chile, a region of extreme aridity. Populations of S. sitiens are found only in a narrow altitudinal belt (about 2,500–3,000 meters above sea level) in a minor cordillera (Cord. de Domeyko) of the Andes. Precipitation in this region is strongly dependent on elevation, and is generally <5 cm/year below 3,000 m (Alpers and Brimhall 1988). Tolerance of extreme aridity, most-likely exceeding that found in any Lycopersicon, can therefore be inferred from conditions in the native habitat of S. sitiens (Rick 1988). Other useful traits, such as low temperature tolerance and/or disease resistances, are likely to be discovered when this species is more thoroughly evaluated.
The genome of S. sitiens is mostly colinear with that of Lycopersicon (Pertuzé et al. 2002). The only large-scale rearrangement is a paracentric inversion encompassing most of the long arm of chromosome 10. The S. sitiens configuration on 10L is shared with its sibling species S. lycopersicoides, as well as several other Solanaceous species, including potato, eggplant and pepper (Livingstone et al. 1999; Doganlar et al. 2002). On the basis of this inversion, as well as chromosome affinities during meiosis of interspecific and intergeneric hybrids, we recognize two genomes among cultivated tomato and its immediate wild relatives: the L genome, shared by all Lycopersicon spp., and the S genome of S. sitiens and S. lycopersicoides.
Direct hybridizations between S. sitiens (genome herein designated Ss) and L. esculentum (Le) were unsuccessful in either direction (Rick 1979, 1988). Although this sexual incompatibility can be avoided by somatic hybridization, the only fusion products reported to-date were apparently sterile (O'Connell and Hanson 1986). In the case of S. lycopersicoides (Sl), treatment of the intergeneric hybrid (LeSl) with colchicine resulted in allotetraploids (LeLeSlSl) with sufficient pollen fertility for further backcrosses to diploid tomato. This resulted in sesquidiploid hybrids, containing two genomes of L. esculentum and one of S. lycopersicoides (LeLeSl, Rick et al. 1986). During meiosis of LeLeSl, the S. lycopersicoides chromosomes are mostly unpaired and are eliminated. As a result, this genotype serves as a convenient donor of the L. esculentum genome, with greater compatibility to S. sitiens (DeVerna et al. 1990). From LeLeSl×SsSs crosses, the first diploid LeSs hybrids were obtained. Chromosome doubling produced allotetraploid hybrids (LeLeSsSs), which exhibited preferential pairing between homologous chromosomes and significant pollen fertility (DeVerna et al. 1990).
The objectives of the present experiments were to transfer the chromosomes of S. sitiens into cultivated tomato and to monitor recombination between their homeologous genomes. The LeLeSsSs allotetraploid hybrid provided a means to derive sesquidiploid, aneuploid and recombinant diploid derivatives, in theory similar to what was accomplished previously for S. lycopersicoides. Special attention was given to the development of monosomic alien addition lines and recombinant diploids, as these are particularly useful in breeding programs and genetic studies.
Materials and methods
Plant material and pollinations
The following stocks used in this study were obtained from the Tomato Genetics Resource Center (TGRC), Department of Vegetable Crops, University of California, Davis: L. esculentum cv 'Vendor Tm-2a'; L. pennellii accession LA0716, collected by Donovan Correll near Atico, Arequipa, Peru; S. lycopersicoides LA1964, collected by Charles Rick, 5 km above Palca, Tacna, Peru; and S. sitiens LA1974, collected by Carlos Ochoa at Chuquicamata, Region II (Antofagasta), Chile. Clones of intergeneric hybrids, including an allotetraploid (LeLeSsSs, plant GH2754-4x) and sesquidiploid (LeLeSl, plant GH266) were provided by Joe DeVerna, formerly at the Campbell Soup Company.
L. pennellii-derived bridging lines were used to overcome unilateral incompatibility of L. esculentum × S. sitiens hybrids. Effective bridging lines were selected in segregating populations using RFLP markers for loci on chromosomes 1, 6 and 10 determining compatibility of pollen with L. esculentum × S. lycopersicoides hybrids (Chetelat and DeVerna 1991). In addition, the compatibility reactions of bridging lines were evaluated phenotypically by observation of pollen tube growth in pistils of a putative sesquidiploid hybrid (plant 90L4190-1). At 48 h post-pollination, pistils were removed, fixed and stained with aniline blue to reveal pollen tubes by epifluorescence microscopy as described by Martin (1959). At least three pistils were observed per cross. A cross was judged compatible if pollen tubes reached the ovaries. Conversely, if pollen tube growth was arrested in the style (usually the upper half) the cross was considered incompatible.
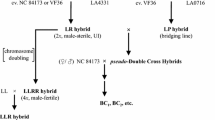
Selected bridging lines were used as male parents in crosses to 90L4190-1 and aneuploid derivatives (Fig. 1). Later progenies were backcrossed to L. esculentum cv Vendor-Tm-2a whenever possible, unless prevented by unilateral incompatibility, in which case the bridging lines were employed as pollen parents for an additional generation. All pollinations were performed in the greenhouse (approximately 25°C by day, 15°C by night) at UC-Davis.
Prior to cross pollination, each S. sitiens derivative was evaluated for pollen viability (PV). Pollen was collected from up to four flowers per plant, and each flower was squashed in a drop of acetocarmine (1% in 50% glacial acetic acid) and observed under the microscope. The percentage of stainable (i.e., viable) pollen grains was estimated from at least 100 grains per flower, and values averaged across the replicate flowers. Plants with high PV, inserted stigmas and/or evidence of fruit set from self-pollination were emasculated prior to controlled pollinations. However, on plants with low PV, shrunken anthers and exserted stigmas, emasculation were not performed. The number of controlled pollinations and resulting fruit set were recorded for each cross.
Embryo culture
Embryo culture was used to improve the survival rate of S. sitiens derivatives. At approximately 30 to 45 days after pollination, fruit were surface-sterilized with 70% alcohol (2 min) and 25% bleach (10 min), rinsed and opened under sterile conditions. Swollen ovules were dissected, and embryos plated either on the HLH medium (Neal and Topoleski 1983) if poorly developed (e.g., heart or torpedo stages), or on Gamborg's B-5 medium with minimal organics (Sacks et al. 1997) if they were more advanced stages (e.g., walking stick to mature). Embryos on HLH medium were transferred to B-5 1–2 weeks later. The number of embryos or seeds per fruit was recorded for each cross. Once large enough, plants were transplanted to soil, acclimatized, then transferred to the greenhouse.
Chromosome observations
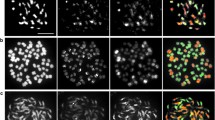
Chromosome counts of S. sitiens derivatives were performed by the acetocarmine squash method (Khush and Rick 1963). Immature flower buds were collected, dissected to remove calyx and corolla, and fixed in 3:1 95% EtOH : glacial acetic acid with FeCl3. Fixative was replaced after the first 1–2 h, buds were fixed overnight, then transferred to 70% EtOH, and stored at 4°C. Anthers were squashed in a drop of acetocarmine and chromosomes were observed under the microscope using phase-contrast optics. Chromosomes were counted and their pairing relationships were analyzed from meiocytes at diakinesis or metaphase-I. In order to help determine the number and origin of chromosomes in a putative sesquidiploid (90L4190-1), genomic in situ hybridization (GISH) was performed on mitotic and meiotic chromosomal preparations as described (Ji and Chetelat 2003).
Marker analysis
RFLP, isozyme, and morphological markers were used to genotype the S. sitiens derivatives. A total of 148 RFLP probes combined with 5–6 restriction enzymes (REs) were evaluated for polymorphism between the four species involved in this study (L. esculentum, L. pennellii, S. sitiens and S. lycopersicoides). From these, 142 loci were polymorphic between at least the Lycopersicon and the Solanum species, with one or more REs. RFLP probes were chosen based on their known map locations in tomato (Tanksley et al. 1992) and S. sitiens / S. lycopersicoides (Pertuzé et al. 2002), as well as the level of polymorphism that they provided. RFLP analysis, including DNA isolations, digestions, electrophoresis, blotting and hybridizations were performed as previously described (Pertuzé et al. 2002). DNA was digested with the following restriction enzymes: EcoRI, EcoRV, HindIII, XbaI, DraI and PstI. Each S. sitiens derivative was genotyped at a minimum of 48 RFLP loci distributed amongst all 12 chromosomes. Each informative marker was polymorphic between the Lycopersicon and the Solanum species, and in most cases also distinguished between S. sitiens and S. lycopersicoides (Fig. 2). In addition, a subset of plants were scored for up to 11 isozyme markers, according to previously described protocols (Chetelat et al. 1997).
Genetic map of markers used to genotype S. sitiens derivatives. The approximate positions of marker loci and centromeres are from the tomato RFLP map (Pillen et al. 1996), with additional loci (in parentheses) from Chetelat et al. (2000). The location of a paracentric inversion on chromosome 10 (solid segment, from Pertuzé et al. 2002) that distinguishes S. sitiens and S. lycopersicoides from the Lycopersicon spp., is also shown. Markers in bold were scored for most derivatives, others only in a subset (see Fig. 4)
Results
Synthesis of aneuploid and diploid derivatives
The allotetraploid L. esculentum × S. sitiens hybrid (LeLeSsSs) was crossed as pollen parent to a S. lycopersicoides sesquidiploid (LeLeSl) to obtain a putative S. sitiens sesquidiploid (LeLeSs, plant 90L4190-1) by embryo culture (Fig. 1). This plant was the source of all other S. sitiens derivatives. Initial chromosome counts by acetocarmine staining indicated plant 90L4190-1 was approximately triploid (2n, ≅36). Since three species were involved in its generation, genomic in situ hybridization (GISH) was used to distinguish chromosomes of L. esculentum from those of the two Solanum spp., and to determine the precise chromosome number (see Fig. 3). The results of GISH on meiocytes at diakinesis or metaphase indicated this plant had 24 Lycopersicon and 14 Solanum chromosomes (2n+14=38). From the pedigree of 90L4190-1, the origins of these nightshade chromosomes could be inferred: a full set of S. sitiens chromosomes transmitted from the LeLeSsSs allotetraploid, plus two S. lycopersicoides chromosomes inherited from the LeLeSl sesquidiploid. RFLP analysis confirmed this hypothesis and permitted identification of each extra S. lycopersicoides chromosome (see below).
Genomic in situ hybridization of a putative sesquidiploid plant (90L4190-1) showing its chromosome constitution. A Mitotic chromosome spread showing 14 Solanum (red) and 24 L. esculentum (blue) chromosomes, with the nucleolar organizers (NOR) marked (arrow for Solanum, arrowheads for L. esculentum). B Meiotic cell at diakinesis in which the 14 Solanum chromosomes (red) form two bivalents (arrows), one trivalent involving the L. esculentum homeologues (arrowhead), and nine univalents. C Meiotic cell at metaphase-I in which the 14 Solanum chromosomes are present as two bivalents (arrows), two trivalents with their respective L. esculentum homeologues (arrowheads), and eight univalents
This 2n+14 plant was used as the female parent to generate additional aneuploids with reduced numbers of chromosomes by pollination with L. pennellii-derived bridging lines. A total of more than 8,000 flowers were pollinated, yielding five plants by embryo culture (Table 1). Only one of these progeny (plant 93L9463-3, 2n+9) survived long enough to produce additional derivatives. The 2n+9 plant was more fecund, yielding a total of 34 plants from about 600 pollinations with the L. pennellii-derived bridging lines (Table 1), constituting most of the progeny analyzed in the present experiments. Among the progeny of 93L9463-3 was another higher-order aneuploid (2n+8, 98L8983-1), from which additional progeny were obtained.
The pollen viability of these three higher-order aneuploids was so low (2.5%, 0.0% and 12% PV for the 2n+14, 2n+9 and 2n+8 plants, respectively), that they could only be used as female parents. As a result, the L. pennellii-derived bridging lines were required to overcome unilateral incompatibility in all but one of them (98L8983-1). The greater fecundity of the 2n+9 than the 2n+14 aneuploid may have been due to greater female fertility or a weakened incompatibility response in the former. To distinguish these possibilities, the female fertility of 90L4190-1 was evaluated by pollination with pure L. pennellii (Table 1). This wild species was assumed to have the maximum number of compatibility factors (i.e., the greatest bridging ability) required to overcome the unilateral incompatibility of this 2n+14 hybrid. The yield of progeny per pollination was approximately 2% using pure L. pennellii (Table 1). In contrast, the L. pennellii-derived bridging lines yielded progeny at a rate of only 0.06%, which suggests that the bridging lines did not carry all the necessary pollen-compatibility loci.
The bridging lines had been selected for their ability to cross to F1 L. esculentum × S. lycopersicoides and derivatives in early backcrosses from L. pennellii to L. esculentum. Selection for compatibility with S. sitiens derivatives was only performed in the last backcross-generation, by which time one or more compatibility gene(s) may have been eliminated. In support of this hypothesis, earlier generation (BC3) bridging plants selected for compatibility with 90L4190-1 resulted in a higher-progeny yield (Table 1).
In addition to incomplete pollen compatibility, a low rate of embryo survival reduced the efficiency of these crosses. A total of nearly 350 embryos were rescued from these three aneuploids following pollination with bridging lines or L. esculentum. Of these, only 40 viable plants were recovered, a success rate of only about 12%. This low yield was apparently due to a strong tendency of cultured embryos to abort in vitro, rather than a lack of culturable embryos. A large number of cultured embryos failed to develop—many were abnormally small or deformed, or were surrounded by a degenerate endosperm. Most embryos that did produce viable plants were well-developed, later-stage embryos (i.e., walking stick or later) at the time of culture.
Ploidy and fertility of S. sitiens derivatives
Chromosomes counts were performed on most of the progeny of 90L4190-1 and 93L9463-3 (Table 2). For some plants only approximate chromosome numbers could be determined by cytology, either because they produced too few flowers, were sterile, or the right meiotic stages were not observed. For several plants, chromosome numbers were inferred from RFLP marker genotypes and/or morphology. Most of the derivatives originated from the 2n+9 aneuploid, and therefore had nine or fewer Solanum chromosomes. Diploids were the most common category (37% of all derivatives), followed by the trisomics (26%), with higher numbers of extra chromosomes becoming increasingly rare (Table 2).
Fecundity of the derivatives was inversely correlated with the chromosome number (Table 2). In general, a greater number of extra Solanum chromosomes was associated with lower pollen fertility and seed production. Diploids had the highest pollen fertility (average 57.6% PV), although some were quite sterile, followed by 2n+1 (34.5%) and 2n+2 (18%). As a result of their low pollen viability, the aneuploids generally failed to produce seeds from self pollinations. Fortunately, a majority of the aneuploids had sufficient female fertility to produce seed following pollination with L. esculentum or the bridging lines.
Marker analysis and chromosome transmission
More than 700 probe × RE combinations (148 probes and 5–6 REs) were analyzed to find polymorphisms among L. esculentum, L. pennellii, S. sitiens and S. lycopersicoides. Almost all probes differentiated the Lycopersicon from the Solanum spp. with at least one of the REs analyzed, but only a subset of these probes were polymorphic between the two Lycopersicon (77%) or the two Solanum (53%) congeners. Three-way polymorphisms, which distinguished the Lycopersicon spp. from S. sitiens and S. lycopersicoides were even less common (23% of probes). Based on this survey information, the S. sitiens derivatives were genotyped with RFLP markers, and a few isozymes and morphological loci, spanning most of the genome (Figs. 2 and 4). The marker genotypes indicated which Solanum chromosomes or segments thereof were transmitted to each progeny, and their species of origin.
Graphical genotypes of S. sitiens derivatives based on marker analysis. Plant numbers of parental hybrids are underlined and their progeny are indented to indicate their relationships. Estimates of ploidy are based on chromosome counts and/or marker data. Chromosomes are oriented with the short arm to the left. The order and approximate positions of marker loci used in this study are represented in the header row. The subset of markers scored in each plant are indicated by tick marks on the corresponding chromosomes. L. esculentum chromosomes, if present, are shown below any Solanum or recombinant chromosomes. In a given marker interval, the genotype of each plant is color-coded according to the schema presented in the legend
GISH analysis of the 2n+14 derivative (90L4190-1) showed consistent bivalent formation between two pairs of Solanum chromosomes, as well as ten Solanum univalents (Fig. 3). Therefore, this plant was expected to show marker genotypes indicating a total of 14 Solanum chromosomes in addition to a complement of 24 L. esculentum chromosomes. The results of the marker analysis (Fig. 4) were consistent with this prediction, and indicated a complete set of 12 S. sitiens chromosomes and two intact S. lycopersicoides chromosomes (numbers 4 and 10). Additionally two L. esculentum chromosomes had recombinant S. lycopersicoides segments.
Marker analysis and chromosome counts of 93L9463-3 suggested this plant was trisomic for chromosomes 1, 2, 3, 4, 5, 7, 8, 9 and 10 (Fig. 4). Intact S. sitiens chromosomes 1, 2, 3, 7 and 8 were indicated by the marker data. All the other extra chromosomes (4, 5, 9 and 10) were recombinant. In addition, chromosome 12, present in the disomic condition, was also recombinant for S. sitiens and L. esculentum segments.
Among all the descendents of the 2n+14 plant, S. sitiens chromosomes 1, 2, 3, 7, 8 and 12 were transferred intact to at least one derivative, in most cases along with segments from other chromosomes (Fig. 4). Chromosomes 4, 5, 9, 10 and 12 were represented only as recombinant segments of different sizes. Only S. sitiens chromosomes 6 and 11 were not transmitted in any form to the progeny. S. sitiens markers from chromosome 2 were transmitted at a much higher rate (69%) than those on all other chromosomes. Considering both the S. sitiens and the S. lycopersicoides alleles together, more than 80% of the progeny showed Solanum markers on chromosome 2. Also remarkable was the high level of recombination on this chromosome evident in the progeny genotypes: only one plant (00L3076-5) inherited an intact S. sitiens chromosome 2. Furthermore, the latter plant was diploid, indicating the S. sitiens chromosome had substituted for one of its L. esculentum homeologues, probably as a result of pairing and chiasma formation.
S. sitiens chromosomes 1 and 12 were also transmitted at relatively high frequencies: about 30% of the progeny carried some S. sitiens alleles on each of these chromosomes. Chromosome 1 was transmitted more often as recombinant segments than as the parental chromosome. In the case of chromosome 12 (and number 5), the 2n+9 plant carried a recombinant S. sitiens segment on one arm of the chromosome; this segment was transmitted intact, undergoing little further recombination in the progeny. S. sitiens chromosomes 3, 7 and 9 were transmitted to approximately 20% of the derivatives, both in the form of recombinant segments and intact chromosomes. Chromosomes 8 and 10 were transmitted at a low rate (13%), the former always intact, and the latter chromosome always recombinant for S. lycopersicoides and S. sitiens segments.
Trisomics
Among the S. sitiens derivatives, seven putative trisomic plants were identified by chromosome counts and morphology (Table 2). Only two plants (00L2568-2 and 95L2026-1) carried intact extra S. sitiens chromosomes (numbers 1 and 8, respectively) and hence constituted monosomic alien addition (MA) lines. MA-1 was highly sterile and incompatible with L. esculentum pollen, and could not be transmitted via seed. Fortunately, a recombinant diploid (00L3073-1) carried a nearly intact S. sitiens chromosome 1, and could be backcrossed to L. esculentum (however only as the male parent, see below). Many other trisomics carried recombinant Solanum chromosomes, as well as additional S. sitiens markers elsewhere in the genome, primarily on chromosome 2.
Diploids
Of the nine putative diploids analyzed with RFLP markers, all but one carried some S. sitiens markers as a result of recombination or chromosome substitution (Fig. 4). In different proportions, S. sitiens chromosomes 1, 2, 3, 8 and 12 were represented among the diploids. In two plants (00L3076-5 and 99L1138-1), intact S. sitiens chromosomes 2 or 8 were substituted for one of the L. esculentum homeologues. As with the aneuploid progeny, alleles from S. sitiens chromosome 2 were transmitted far more than any other, being present in five out of the eight diploid plants. The average pollen viability of the diploid plants was higher than for any of the aneuploid derivatives and all produced seed, either by backcrosses to L. esculentum or by selfing (Table 2).
Genetic basis of bridging ability
Almost all the S. sitiens derivatives carried L. pennellii alleles near previously mapped pollen-compatibility loci on chromosomes 1, 6 and 10 (Chetelat and DeVerna 1991). L. pennellii markers on chromosomes 1 and 6 were transmitted to all the progeny of the 2n+14 and 2n+9 hybrids, suggesting that loci on these chromosomes were required to overcome unilateral incompatibility of the S. sitiens hybrids. In addition, L. pennellii markers on chromosomes 7 and 10 were transmitted at a high rate, indicating that important compatibility factors are also located on these chromosomes. However, some L. pennellii alleles may have been transmitted through the female parent, independent of selection for bridging ability, because the 2n+9 hybrid was already heterozygous for L. pennellii alleles on several chromosomes. As a result, homozygosity was observed for L. pennellii markers among some progeny of this S. sitiens hybrid. Plants with fewer chromosomes from either of the Solanum species tended to have a weakened unilateral incompatibility, and most produced backcross seed following pollinations with pure L. esculentum (Table 2). The only plants that produced neither self nor backcross seeds were those that had three or more Solanum chromosomes (or segments thereof), or plants that carried S. sitiens markers near the S locus on chromosome 1 (Fig. 2).
A monosomic addition for chromosome 8
To study the transmission of the extra S. sitiens chromosome in more-advanced backcross progeny, a monosomic alien addition line for chromosome 8 (MA-8) was backcrossed as female parent to L. esculentum. Sixty backcross plants were genotyped with three RFLP markers (TG176, TG624 and TG294) covering the S. sitiens chromosome. The RFLP data revealed eight putative trisomic plants (13.3%) that were heterozygous for all three S. sitiens markers and showed morphological features of S. lycopersicoides MA-8 (Chetelat et al. 1998): white, dialytic anthers (genes Wa and Dl s), exserted stigmas, leaves curved downward at the margins and small, whitish-green fruits. The trisomic nature of these plants was confirmed by relative-band intensity on autoradiograms and chromosome counts on selected individuals (data not shown). All other plants were presumably diploid, and no plants with recombinant segments were found.
Chromosome pairing in diploids and aneuploids
The frequency of various chromosome pairing configurations was estimated for many of the derivatives (Table 2). Though based on a limited number of meiocytes in some cases, the results suggest a relatively normal meiosis in the diploids, whereas substantial pairing failure was observed in the aneuploids. In the former, chromosomes at diakinesis or metaphase-I of pollen mother cells tended to form 12 bivalents. This included diploids with relatively large chromosome segments from S. sitiens (e.g., 00L3076-5 and 99L1239-1). However, occasional univalents were detected in two diploid progeny, one of which contained a substituted S. sitiens chromosome 8 (99L1138-1), the other a large introgression on chromosome 12 (00L2568-1). Chromosomes of the trisomics formed primarily 12 bivalents and one univalent at meiosis. However, many also formed trivalents at a substantial rate, indicating recombination between homeologous chromosomes. Meiosis in the higher-order aneuploids (2n+2 or greater) was disrupted to a greater extent, with fewer bivalents, and more univalents and trivalents observed.
Discussion
The present study represents the first successful gene transfer from S. sitiens to cultivated tomato. This was achieved through use of a pseudo-sesquidiploid (2n+14) plant carrying a full set of S. sitiens chromosomes (plus 2 from S. lycopersicoides) in the background of L. esculentum. Analysis of meiosis in this intergeneric hybrid using GISH cytology showed that chromosomes of the two-nightshades pair readily with one another, and less frequently with their L. esculentum homeologues. The same tendency for preferential pairing among homologous chromosomes was observed in the allotetraploid hybrid (LeLeSsSs, DeVerna et al. 1990), as well as in true sesquidiploids representing S. lycopersicoides (LeLeSl, Rick et al. 1986). From the latter hybrids, we obtained monosomic additions (Chetelat et al. 1998) and recombinant diploids (Rick et al. 1986), suggesting that similar stocks might be synthesized for S. sitiens.
Unilateral incompatibility
Strong unilateral incompatibility (UI) observed in the 2n+14 hybrid towards the pollen of L. esculentum necessitated use of bridging lines derived from L. pennellii. UI is generally observed in crosses between self-incompatible species (i.e., S. sitiens or its hybrids in this case) used as female parents, and related self-compatible taxa (i.e., L. esculentum) used as pollen donors (see review by De Nettancourt 1977). Incompatibility can be avoided in such combinations by using the self-compatible species as female parent. However, in the present study, pollen viability of the 2n+14 hybrid and many of its aneuploid derivatives was so low that they were not functional pollen parents. Therefore, the only means of producing progeny was to develop suitable bridging lines.
However, the bridging lines used in this study were only partially compatible with the 2n+14 hybrid. Whereas the yield of progeny using the backcrossed bridging lines was quite low, pollinations with pure L. pennellii were much more successful. Therefore, female fertility of the 2n+14 hybrid did not appear to be the limiting factor in determining success of this cross. The use of earlier generation L. pennellii-derived bridging lines increased the progeny recovery rate, presumably because they contained a greater number of pollen-compatibility factors. Noteworthy in this regard was the high transmission rate observed for L. pennellii markers on chromosome 7. This may indicate a compatibility locus important in crosses to S. sitiens hybrids that was not detected in our earlier studies with S. lycopersicoides (Chetelat and DeVerna 1991). Therefore, selection of more-effective bridging lines in early backcross generations could substantially improve prospects for gene transfer from S. sitiens to tomato.
UI was weakened or eliminated in most of the derivatives with fewer extra chromosomes. This will simplify elimination of residual genetic material from L. pennellii, since backcrosses to L. esculentum are productive, even in derivatives with relatively low male fertility. However, UI persisted in 2n and 2n+1 plants (00L3073-1 and 00L2568-2, respectively) containing most or all of S. sitiens chromosome 1, which includes the S-locus controlling specificity of the self-incompatibility system (Tanksley and Loaiza-Figueroa 1985). Similarly, the S. lycopersicoides monosomic addition for chromosome 1 was also incompatible with L. esculentum pollen (Chetelat et al. 1998). In the case of the diploid derivative (00L3073-1), pollen fertility was sufficient to allow backcrossing as male parent to L. esculentum. However, this solution would not be effective for the trisomic individual (00L2568-2), since male transmission of extra chromosomes is relatively low in tomato due to strong gametophytic selection against n+1 pollen (Khush 1973).
Aneuploidy
Progeny from the 2n+14 hybrid included aneuploids with up to nine extra chromosomes. This far-exceeds the maximum aneuploidy (2n+4) observed previously in a much-larger progeny array (459 plants) obtained from the S. lycopersicoides sesquidiploids (Chetelat et al. 1998). This discrepancy might be related to the slightly higher ploidy of the intergeneric hybrid representing S. sitiens (2n+14) than S. lycopersicoides (2n+12). Regular association in the former hybrid between the two pairs of nightshade chromosomes would result in more-frequent transmission than expected from a true sesquidiploid. However, this provides only a partial explanation, since most of the S. sitiens derivatives were descendants of the 2n+9 plant.
The relatively low success rate of crosses involving these S. sitiens hybrids implies a strong selection at pre-fertilization and/or post-syngamic stages. In this context, our results are all the more surprising, since selection would normally favor gametes or zygotes with fewer extra chromosomes. In the progeny of tomato triploids, for example, a maximum number of three extra chromosomes was observed, and this category of aneuploids represented less than 1% of the total (Rick and Barton 1954). The fact that embryo rescue in the present experiments was performed relatively late in development may have allowed weak embryos (i.e., those with higher ploidy levels) to mature. This interpretation is consistent with our observations that most of the viable embryos were more advanced (walking sticks or later) at the time of culture.
Of the aneuploid progeny, the trisomics are potentially the most useful as they are more fertile and may include monosomic addition lines. The original goal was to produce a complete set of monosomic additions for each of the 12 S. sitiens chromosomes. However, the rate of plant recovery from the 2n+14 plant was extremely low. As a result, certain chromosomes were not transmitted. Although a greater number of progeny were obtained from the 2n+9 plant, it contained only a partial set of S. sitiens chromosomes. This further-limited opportunities for transmission of some chromosomes.
Intact S. sitiens chromosomes 1, 2, 3, 7, 8 and 12 were identified in the aneuploid progeny. Although all 2n+1 plants had S. sitiens markers on more than one chromosome, their relatively high fecundity should enable recovery of true monosomic additions in later generations. This has already been accomplished for MA-8, the first monosomic addition isolated in this study. In contrast, S. sitiens chromosome 1 could not be transmitted due to its pollen sterility and incompatibility. These results are identical to our experience with the S. lycopersicoides monosomic additions, wherein MA-8 was among the first to be recovered (DeVerna et al. 1987) and MA-1 was impossible to maintain for the same reasons (Chetelat et al. 1998).
In progeny of MA-8, the extra S. sitiens chromosome transmitted well but did not recombine. The frequency of trisomic progeny, though somewhat lower than observed for the corresponding S. lycopersicoides chromosome (13% vs 22%, Chetelat et al. 1998), was sufficient for maintenance purposes. However, no recombinant chromosomes were found, either among the diploid or the trisomic plants. Although the population size was not large (n=60), the results do suggest a much lower rate of recombination in the backcrossed monosomic addition than in the earlier generation aneuploids. A similar suppression of recombination following introgression into L. esculentum was observed for the S. lycopersicoides monosomic additions (Ji and Chetelat 2003). For this reason, selection for recombinant genotypes would be more efficient in the earliest descendants of the intergeneric hybrid.
Evidence of recombination
All but one of the diploids carried genetic material from S. sitiens, usually in the form of recombinant chromosome segments. Extensive recombination was also detected in the aneuploid progeny. The frequency of recombinant genotypes in these derivatives appears to be at least as high as that observed previously in similar stocks containing S. lycopersicoides chromosomes in tomato (Rick et al. 1988; Chetelat et al. 1998). Evidence for chromosome substitution was also detected, as one plant contained an intact chromosome 8. Heterozygous substitution lines of this type are also useful sources of recombinants. Since they lack homologous partners, homeologous chromosomes recombine at relatively high frequencies (Ji and Chetelat 2003).
Although recombination was detected on all chromosomes except 6 and 11, the frequency of recombinant genotypes was highest for chromosome 2. This could indicate a greater degree of homology between the genomes on this chromosome, or selection for recombinant gametes. However, a more-likely explanation is that the 2n+9 aneuploid, from which most of the derivatives were obtained, itself contained a recombinant chromosome 2 with a large S. lycopersicoides segment. As a result, this chromosome could pair and recombine in homologous regions on both the L. esculentum and the S. sitiens homeologues.
Future prospects
Finally, we conclude that prospects are good for transmission of a wide range of variation found in S. sitiens. Despite the relatively limited number of progeny obtained, our results demonstrate the feasibility of gene transfer via aneuploids and recombinant diploids. With additional crosses and marker-assisted selection, a more complete series of monosomic additions could be obtained. Such pre-bred stocks would provide a convenient means for preserving the genome of S. sitiens in a more accessible form, for chromosomal assignment of dominant genes identified in the nightshade, and eventual transfer of useful traits into horticultural tomatoes. From progeny of the recombinant diploids, homozygous introgression lines (ILs) could be selected. Libraries of ILs have already been synthesized for the L. pennellii and L. hirsutum genomes (Eshed and Zamir 1995; Monforte and Tanksley 2000), and have many advantages for breeding and genetic studies (Zamir 2001). Recent synthesis of an IL library for S. lycopersicoides (Chetelat and Meglic 2000; Canady 2002) suggests similar genotypes for S. sitiens would be viable. Thus, our results so far bode well for expanding the gene pool of cultivated tomato to include this interesting desert nightshade.
References
Alpers CN, Brimhall GH (1988) Middle Miocene climatic change in the Atacama Desert, northern Chile: evidence from supergene mineralization at La Escondida. Geol Soc Am Bull 100:1640–1656
Canady MA (2002) Development and genetic studies of a Solanum lycopersicoides introgression line library. PhD Thesis, University of California at Davis
Chetelat RT, DeVerna JW (1991) Expression of unilateral incompatibility in pollen of Lycopersicon pennellii is determined by major loci on chromosomes 1, 6 and 10. Theor Appl Genet 82:704–712
Chetelat RT, Meglic V (2000) Molecular mapping of chromosome segments introgressed from Solanum lycopersicoides into cultivated tomato (Lycopersicon esculentum). Theor Appl Genet 100:232–241
Chetelat RT, Rick CM, Cisneros P, Alpert KB, DeVerna JW (1998) Identification, transmission, and cytological behavior of Solanum lycopersicoides Dun. monosomic alien addition lines in tomato (Lycopersicon esculentum Mill.). Genome 41:40–50
Chetelat RT, Cisneros P, Stamova L, Rick CM (1997) A male-fertile Lycopersicon esculentum × Solanum lycopersicoides hybrid enables direct backcrossing to tomato at the diploid level. Euphytica 95:99–108
Child A (1990) A synopsis of Solanum subgenus Potatoe (G. Don) D'Arcy [Tuberarium (Dun.) Bitter (s.l.)] Fedes Rept 101:209–235
Correll DS (1961) Four new Solanums in section Tuberarium. Wrightia 2:133–141
De Nettancourt D (1977) Incompatibility in angiosperms. In: Frankel R, Gall GAE, Grossman M, Linskens HF, de Zeeuw D (eds) Monographs on theoretical and applied genetics. Springer-Verlag, Berlin Heidelberg New York
DeVerna JW, Chetelat RT, Rick CM (1987) Cytogenetic, electrophoretic, and morphological analysis of progeny of sesquidiploid Lycopersicon esculentum—Solanum lycopersicoides hybrids × L. pennellii. Biol Zent bl 106:417–428
DeVerna JW, Rick CM, Chetelat RT, Lanini BJ, Alpert KB (1990) Sexual hybridization of Lycopersicon esculentum and Solanum rickii by means of a sesquidiploid bridging hybrid. Proc Natl Acad Sci, USA 87:9496–9490
Doganlar S, Frary A, Daunay M-C, Lester RN, Tanksley SD (2002) A comparative genetic linkage map of eggplant (Solanum melongena) and its implications for genome evolution in the Solanaceae. Genetics 161:1697–1711
Eshed Y, Zamir D (1995) An introgression line population of Lycopersicon pennellii in the cultivated tomato enables the identification and fine mapping of yield-associated QTL. Genetics 141:1147–1162
Harlan JR (1976) Genetic resources in wild relatives of crops. Crop Sci 16:329–333
Jarvis DI, Hodgkin T (1999) Wild relatives and corp cultivars: Detecting natural introgression and farmer selection of new genetic combinations in agroecosystems. Mol Ecol 8:S159-S173
Ji Y, Chetelat RT (2003) Homeologous pairing and recombination in Solanum lycopersicoides monosomic addition and substitution lines of tomato. Theor Appl Genet 106:979–989
Johnston IM (1929) A new Chilean plant and some nomenclatural changes. Revista Chilena de Historia Natural 25–27
Khush GS (1973) Cytogenetics of aneuploids. Academic Press, New York, USA
Khush GS, Rick CM (1963) Meiosis in hybrids between Lycopersicon esculentum and Solanum pennellii. Genetica 33:167–183
Livingstone KD, Lackney VK, Blauth JR, Van Wijk RIK, Kyle Jahn M (1999) Genome mapping in Capsicum and the evolution of genome structure in the Solanaceae. Genetics 152:1183–1202
Marticorena C, Quezada M (1977) Notas sobre Solanum. Bol Soc Biol Concepcion 51:153–157
Martin FW (1959) Staining and observing pollen tubes in the style by means of fluorescence. Stain Technol 34:125–128
Monforte AJ, Tanksley SD (2000) Development of a set of near-isogenic and backcross recombinant inbred lines containing most of the Lycopersicon hirsutum genome in a L. esculentum genetic background: a tool for gene mapping and gene discovery. Genome 43:803–813
Neal CA, Topoleski LD (1983) Effects of the basal medium on growth of immature tomato embryos in vitro. J Am Soc Hort Sci 108:434–438
Nee M (1999) Synopsis of Solanum in the New World. In: 285–333
O'Connell MA, Hanson MR (1986) Regeneration of somatic hybrid plants formed between Lycopersicon esuclentum and Solanum rickii. Theor Appl Genet 72:59–65
Pertuzé RA, Ji Y, Chetelat RT (2002) Comparative linkage map of the Solanum lycopersicoides and S. sitiens genomes and their differentiation from tomato. Genome 45:1003–1012
Pillen K, Pineda O, Lewis CB, Tanksley SD (1996) Status of genome mapping tools in the taxon Solanaceae. In: 282–308
Rick CM (1951) Hybrids between Lycopersicon esculentum Mill. and Solanum lycopersicoides Dun. Proc Natl Acad Sci, USA 37:741–744
Rick CM (1979) Biosystematic studies in Lycopersicon and closely related species of Solanum. In: 667–678
Rick CM (1988) Tomato-like nightshades: affinities, autecology, and breeders opportunities. Econ Bot 42:145–154
Rick CM, Barton DW (1954) Cytological and genetical identification of the primary trisomics of the tomato. Genetics 39:640–666
Rick CM, Chetelat RT (1995) Utilization of related wild species for tomato improvement. Acta Hort 412:21–38
Rick CM, DeVerna JW, Chetelat RT, Stevens MA (1986) Meiosis in sesquidiploid hybrids of Lycopersicon esculentum and Solanum lycopersicoides. Proc Natl Acad Sci USA 83:3580–3583
Sacks EJ, Gerhardt LM, Graham EB, Jacobs J, Thorrup TA, St Clair DA (1997) Variation among 41 genotypes of tomato (Lycopersicon esculentum Mill.) for crossability to L. peruvianum (L.) Mill. Ann Bot 80:469–477
Tanksley SD, Loaiza-Figueroa F (1985) Gametophytic self-incompatibility is controlled by a single major locus on chromosome 1 in Lycopersicon peruvianum. Proc Natl Acad Sci USA 82:5093–5096
Tanksley SD, McCouch SR (1997) Seed banks and molecular maps: unlocking genetic potential from the wild. Science 277:1063–1066
Tanksley SD, Ganal MW, Prince JP, de Vicente MC, Bonierbale MW, Broun P, Fulton TM, Giovanonni JJ, Grandillo S, Martin GB, Messeguer R, Miller JC, Miller L, Paterson AH, Pineda O, Roder M, Wing RA, Wu W, Young ND (1992) High-density molecular linkage maps of the tomato and potato genomes. Genetics 132:1141–1160
Zamir D (2001) Improving plant breeding with exotic genetic libraries. Nature Rev 2:983–989
Acknowledgements
The authors thank Joe DeVerna and Brenda Lanini for providing intergeneric hybrids and bridging lines, and the TGRC staff for seed of the wild species and for maintenance of greenhouse stocks. Steve Tanksley provided RFLP probes and Carlos Quiros helpful comments on the manuscript. This research was supported in part by grants from the California Tomato Commission.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by J.S. Heslop-Harrison
Rights and permissions
About this article
Cite this article
Pertuzé, R.A., Ji, Y. & Chetelat, R.T. Transmission and recombination of homeologous Solanum sitiens chromosomes in tomato. Theor Appl Genet 107, 1391–1401 (2003). https://doi.org/10.1007/s00122-003-1384-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00122-003-1384-z