Abstract.
Phytate (myo-inositol-1,2,3,4,5,6-hexakisphosphate, InsP6) is the most abundant P-containing compound in plants, and an important anti-nutritional factor, due to its ability to complex essential micro-nutrients, e.g. iron and zinc. Analysis of natural variation for InsP6 and Pi accumulation in seeds and leaves for a large number of accessions of Arabidopsis thaliana, using a novel method for InsP6 detection, revealed a wide range of variation in InsP6 and Pi levels, varying from 7.0 mg to 23.1 mg of InsP6 per gram of seed. Quantitative trait locus (QTL) analysis of InsP6 and Pi levels in seeds and leaves, using an existing recombinant inbred line population, was performed in order to identify a gene(s) that is (are) involved in the regulation of InsP6 accumulation. Five genomic regions affecting the quantity of the InsP6 and Pi in seeds and leaves were identified. One of them, located on top of chromosome 3, affects all four traits. This QTL appears as the major locus responsible for the observed variation in InsP6 and Pi contents in the Ler/Cvi RIL population; the Ler allele decreases the content of both InsP6 and Pi in seeds and in leaves. The InsP6/Pi locus was further fine-mapped to a 99-kb region, containing 13 open reading frames. The maternal inheritance of the QTL and the positive correlation between InsP6 and total Pi levels both in seeds and in leaves indicate that the difference in InsP6 level between Ler and Cvi is likely to be caused by a difference in transport rather than by an alteration in the biosynthesis. Therefore, we consider the vacuolar membrane ATPase subunit G, located in the region of interest, as the most likely candidate gene for InsP6/Pi.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The major form in which phosphorus occurs in plants is myo-inositol-1,2,3,4,5,6-hexakisphosphate, commonly referred to as phytic acid or InsP6 (Lott et al. 2000). This highly negatively charged compound forms a mixed salt with various mineral cations, e.g. potassium, magnesium, iron and zinc. It is generally assumed that the major role of InsP6 in plants is to act as a storage form for Pi and probably also for cations, and is most abundant in seeds and fruits. The availability of Pi in soil is positively correlated with the level of InsP6 accumulated in seeds of soybean (Raboy and Dickinson 1987) and Brassica (Lickfett et al. 1999). Since InsP6 is the major form of phosphorus in seeds, total phosphorus and InsP6 levels in seeds are usually positively correlated (Raboy 2001).
Due to its mineral-binding capacity, InsP6 affects the nutritional quality of food; minerals, when bound to InsP6, are not or are hardly absorbed in the intestine and are largely excreted, resulting in iron and zinc deficiencies, especially in developing countries, when food is mainly seed-based. Monogastric animals only very inefficiently take up phosphorus from InsP6, and hence additional Pi is added to their feed, resulting in P-pollution.
Decreasing the level of InsP6 in seeds is one of the recent strategies to improve iron and zinc availability in food (Lucca et al. 2001). In maize (Zea mays), barley (Hordeum vulgare), rice (Oryza sativa) and soybean (Glycine max), mutants with lowered InsP6 levels (low phytic acid; lpa) have been isolated (Raboy and Gerbasi 1996; Larson et al. 1998, 2000; Raboy et al. 2000; Wilcox et al. 2000). In these mutant lines, a negative correlation was observed between InsP6 and free Pi levels. The explanation is that a smaller portion of total seed phosphorus is converted into InsP6, the total amount of phosphorus in the seeds remaining unaltered. In some mutants also lower inositol-phosphates (mainly InsP3 and InsP5) were found. Mutant lines with a block in InsP6 metabolism in the developing seed appear to have a reduced seed weight, which is leading to reduced yields as well. This reduction in seed dry weight might be because of reduction in starch accumulation that results from the disturbance of phosphorus or because of disruption in inositol or inositol-phosphate metabolism found in lpa seeds (reviewed by Raboy 2001).
Except its effect on seed weight it now appears that InsP6 probably serves several cellular functions as well: as a major pool in inositol phosphate pathways (Safrany et al. 1999) and as a second messenger ligand (Sasakawa et al. 1995). Furthermore, it may play a role in DNA double-strand break repair (Hanakahi et al. 2000), RNA export from the nucleus (York et al. 1999), ATP metabolism (Safrany et al. 1999) and in phosphorus and mineral storage (Raboy 1997). A recent study indicates the role of InsP6 in the physiological response of guard cells to ABA as well (Lemtiri-Chlieh et al. 2000).
In light of these numerous functions for InsP6 in cells generally, and in seeds specifically, it is somewhat surprising that the lpa mutants perform rather well under field conditions. Clearly the levels of InsP6 typical for normal seeds are not essential for seed functioning.
In eukaryotic cells, InsP6 is synthesised by sequential phosphorylation of myo-inositol and/or partly via phosphatidylinositol phosphates (Loewus and Murthy 2000; Raboy 2001). However, relatively little is known about plant genomic sequences encoding functions in the late part of the pathway, playing a direct role in seed InsP6 accumulation and mobilisation (Raboy 2001). Only recently, Hitz et al. (2002) reported that a defective myo-inositol-1-phosphate synthase decreased the amount of InsP6 in soybean seeds. Furthermore, it is known that during germination InsP6 is broken down by phytase enzymes, releasing myo-inositol and mineral contents. There probably are multiple phytases in seed and pollen, with different patterns of expression and regulation, the molecular biology and genetics of which is relatively undeveloped (Loewus and Murthy 2000). There is significant progress in the study of microbial phytase genes (Maugenest et al. 1997).
In the present study we describe a genetic approach to unravel InsP6 and free phosphorus accumulation in seeds, using Arabidopsis as a model species. We have used natural variation that has been shown to be present in accessions of Arabidopsis for a large variety of traits (Alonso-Blanco and Koornneef 2000).
Genetic variation for InsP6 and Pi accumulation in seeds and leaves has been analysed, using a novel method for InsP6 detection, for a large number of accessions. Furthermore, quantitative trait locus (QTL) analysis of InsP6 and Pi levels in seeds and leaves has been performed, using an existing recombinant inbred line (RIL) population, in order to identify a gene(s) that is (are) involved in the regulation of InsP6 accumulation.
Materials and methods
Plant material
A set of 101 Arabidopsis accessions was analysed for InsP6 and Pi content in seeds and for Pi content in leaves. The complete list of accessions and the levels of InsP6 and Pi are presented at http://www.natural-eu.org .
A set of 162 RILs derived from crosses between the laboratory strain Ler originated from Northern Europe (Rédei 1992) and the accession Cvi from the Cape Verde Islands (Lobin 1983), were analysed for InsP6 and Pi in seeds and leaves as well. These RILs have been previously characterised for amplified fragment length polymorphism (AFLP) and cleaved amplified polymorphic sequence (CAPS) markers (Alonso-Blanco et al. 1998). Results of InsP6 and Pi analysis of all individual RILs are as well presented at http://www.natural-eu.org .
To construct a near-isogenic line containing a Cvi introgression at the top of chromosome 3 we have used NIL35. NIL35 was constructed by two back crosses of RIL49 with Ler for fine mapping a seed dormancy QTL (Alonso-Blanco et al. 2003), and contained 3 cM of Cvi at the top of chromosome 3, 45 cM on the middle of chromosome 5 and a chromosome segment of 10 cM on the bottom of chromosome 1, in a Ler background, as was determined with AFLP markers. NIL35 has been backcrossed with Ler. In the resulting F2 of this population a plant heterozygous for the top of chromosome 3 and homozygous Ler on chromosome 5 was selected, using molecular markers. The progeny of this plant has been used as a mapping population to refine the map position of the QTL on top of chromosome 3.
A line homozygous for Cvi on the top of chromosome 3 has also been selected from this population and is called NIL26.
Plants were grown in an air-conditioned greenhouse (temperature 22–25 °C), supplemented with additional light (model TDL 58W/84, Philips, Eindhoven The Netherlands) from mid September till early April, providing a day length of 16 h. Plants were grown in 7-cm pots in standard soil.
Extraction and quantification of InsP6 and Pi
Dry seeds (1–4 mg) were boiled for 15–20 min in 0.5 ml of 0.6 M HCl. Leaves (30–60 mg) were extracted by boiling for 15 min in 1 ml of 0.6 M HCL and 10 mM of EDTA. The extracts were centrifuged at 15,000 g for 5 min. The supernatants were diluted 10-times (seeds) or 3-times (leaves) with water, and 20 μl was analysed using a Dionex DX300 HPLC system (Dionex Corporation, Sunnyvale, Calif., USA). Anions were separated on an AS11 (4 × 250 mm) column, preceded by an AG 11 guard column and eluted with NaOH. The elution profile was: 5 min isocratic at 5 mM of NaOH, followed by a 15-min linear gradient with 5–100 mM of NaOH. After each run the column was washed for 5 min with 0.5 M NaOH, followed by a 15-min equilibration at 5 mM. Flow rates were 1 ml·min–1 throughout the run. Contaminating anions in the eluents were removed using an ion trap column (ATC-1), installed between the pump and the sample injection valve. Anions were determined by conductivity detection. Background conductivity was decreased using a ASRS suppressor, with water as a counterflow (5 ml·min–1), operated at 300 mA, controlled by an SRS controller (Dionex Corporation Sunnyvale, Calif., USA). Peaks were identified and quantified by co-elution with known standards. Recoveries of InsP6 were determined by spiking seed samples with Na12-InsP6.
QTL analysis
To map QTLs using the RIL population, a set of 99 markers covering most of the Arabidopsis genetic map was selected from the previously published RIL Ler/Cvi map (Alonso-Blanco et al. 1998). These markers spanned 482 cM, with an average distance between consecutive markers of 5 cM and the largest genetic distance being 12 cM.
The computer program MapQTL version 4.0 (Plant Research International, Wageningen-University and Research Centre, Wageningen, The Netherlands) was used to identify and locate QTLs linked to the molecular markers using both interval mapping and multiple-QTL model mapping (MQM) methods as described in its reference manual (http://www.plant.wageningen-ur.nl/products/mapping/mapqtl/ ). The estimated additive effect and the percentage of variance explained by each QTL as well as the total variance explained by all of the QTLs affecting a trait, were obtained with MapQTL in the final MQM model. For this, different cofactor markers were tested around the putative QTL positions (van Ooijen and Maliepaard 1996), selecting as final cofactors the closest marker to each QTL, i.e. those maximizing the logarithm-of-odds (LOD) score. A LOD score threshold of 2.8 was applied to declare the presence of a QTL, which corresponds to a general genome-wide significance of P = 0.05 for normally distributed data, as was determined by extensive simulation experiments (van Ooijen 1999). We verified this threshold for interval mapping by applying the permutation test to each data set (10,000 repetitions) and found P = 0.05 LOD thresholds between 2.5 and 2.6 for all traits. Two-LOD support intervals were established as approximately 95% confidence intervals (van Ooijen 1992).
For every trait, two-way QTL interactions were analysed by analysis of variance at a significance level of P < 0.005, using the General Linear Model module of the statistical package SPSS version 10.0 (SPSS Incorporated, Chicago). For each analysis, the closest linked markers to the corresponding detected QTL were used as random factors in the ANOVA (the same markers used as cofactors in the MQM mapping with MapQTL).
Fine mapping of the InsP6 and Pi locus
To fine map the major locus for the InsP6 and Pi locus on chromosome 3, DNA of 631 plants derived from the heterozygous NIL26 sister plant was isolated and analysed using the CAPS markers T4P13b and 17D8LE (Table 1). The seeds of recombinants between these two markers were analysed for InsP6 and Pi content, and used for further fine-mapping of the locus. To unambiguously determine the InsP6 phenotype of the recombinants, 12 F3 plants of each recombinant have been analysed subsequentially.
Design of molecular markers
Based on the Columbia genomic sequence (TAIR; http://www.arabidopsis.org ) new cleaved amplified polymorphic sequence (CAPS) (Konieczny and Ausubel 1993) and single sequence length polymorpism (SSLP) (Bell and Ecker 1994) markers were developed. Marker specifications are listed in Table 1.
Results
Extraction and detection of InsP6
A simple and rapid method was needed to extract, detect and quantify InsP6 in a large series of small samples. Anions can easily be separated using exchange chromatography, especially at high pH, and detected by conductivity. Sensitivity of this detection method can largely be increased using post-column removal of the alkali via a suppressor (Weiss 1991). Figure 1 shows chromatograms of a mix of anion standards (A), and an extract of a few mg of Arabidopsis seeds (B), extracted simply by boiling in HCl. Without any further purification, InsP6 (Rt = 15.8) and Pi (Rt = 12.3) could easily be detected. The large peak at 3.2 min in the chromatogram of the seed extract is chloride, added for the extraction. Another major peak in the chromatogram, at 10.9 min, co-eluted with sulphate. Minor peaks, co-eluting with citrate and oxalate were also present, Rt being 12.7 and 11.2 min, respectively.
Recoveries of InsP6 were determined by spiking seed samples, varying in weight from 2 to 7 mg, with authentic Na12 InsP6. Recoveries were found to decrease with increasing sample weight (Fig. 2). The response of the detector was linear with the concentrations of InsP6 and P up to at least 50 mg·l–1 (data not shown). It is concluded that the extraction and detection method is reliable for seed samples smaller than 4 mg of DW. The protocol was sensitive enough to detect InsP6 in sub-milligram samples. To avoid inaccuracy routinely due to weighing errors or a small number of seeds, samples of 2–3 mg were routinely analysed, the smallest sample being 0.8 mg, representing around 50 seeds (Alonso-Blanco et al. 1999).
Natural variation for InsP6 and Pi content among Arabidopsis accessions
For a series of 101 accessions of Arabidopsis, the levels of InsP6 and Pi in mature dry seeds, and in full-grown leaves, collected from vegetative plants, were determined (data presented at http://www.natural-eu.org ). The average InsP6 level in seeds was 16.2 ± 2.3, ranging from 7.0 to 23.1 mg/g of seed. A weak but significant (r = 0.351) positive correlation between InsP6 and Pi levels in seeds was detected. Pi levels in leaves ranged from 4.6 to 30.5 mg/g of fresh weight (FW), averaging at 14.6 ± 5.6 mg/g of FW. The levels of InsP6 in the leaves of these accessions were more than two orders of magnitude lower than in the seeds and were not quantified. In order to test whether InsP6 levels in seeds might represent a overall higher P-status in the plant, seed-InsP6 levels were plotted against leaf-Pi levels (Fig. 3). No significant correlation was observed.
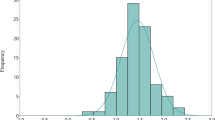
To identify and locate QTLs responsible for the genetic variation in InsP6 and Pi between the accessions Ler and Cvi, we have analysed the RIL population derived from a cross between these accessions for InsP6 and Pi levels in seeds and leaves. Although the analysis of other crosses such as Kyoto-1 × Hey-0 (InsP6 levels 7.0 and 23.1 mg/g of seed, respectively; Fig. 3) might show larger variation, the availability of a permanent mapping population between Ler and Cvi offers unique advantages (Alonso-Blanco and Koornneef 2000). The Ler/Cvi population shows considerable transgression towards both directions, for all traits analysed, as is shown in Fig. 4. This indicates that both accessions carry genes that increase and decrease the content of the different compounds that are analysed. Furthermore, there is a positive correlation between the amounts of InsP6 and Pi in seeds (r = 0.56) and in leaves (r = 0.40) as well as a positive correlation between the amount of InsP6 in seeds compared to the amount in leaves, and for the level of Pi in seeds compared to that of leaves, (r = 0.35 and 0.34, respectively).
Frequency distributions of InsP6 and Pi contents in seeds (A, B) and in leaves (C, D) of plants of the Ler/Cvi RIL population. Arrows correspond to the parental line means and horizontal bars represent their ranges of variation based on the analysis of two different plants. Parental values of InsP6 and Pi in leaves are based on measurements of a single plant
Mapping QTLs for InsP6 and Pi in seeds and leaves
QTL mapping was performed for the quantity of the InsP6 and Pi in seeds and leaves (Fig. 5). In total five genomic regions, affecting InsP6 and Pi levels, were detected, one of them on chromosome 3 affecting all four traits. Additional QTLs were detected for Pi in seeds on the top of chromosome 1, on the bottom of chromosomes 2 and 4, and for InsP6 in leaves on the bottom of chromosomes 1 and 2. The additive effects of these QTLs accounted for 61.8% of the total variance for InsP6 in seeds, 55.3% for Pi in seeds, 35.5% for InsP6 in leaves and 33% for Pi in leaves. The region on the top of chromosome 3 near the AFLP marker DF.77C explained 61.8%, 36.8%, 22.5% and 28.9% of the variation for InsP6 in seeds, Pi in seeds, InsP6 in leaves and Pi in leaves respectively. Therefore, this QTL appears as the major locus responsible for the observed variation in InsP6/Pi content in the Ler/Cvi RIL population, the Ler allele decreasing the content of both InsP6 and Pi in seeds and in leaves. The Ler allele for the QTLs affecting Pi in seeds and InsP6 in leaves (both on chromosome 1) and for Pi in seeds (on chromosome 4) increases the content. The Ler allele for the additional QTLs affecting Pi in seeds and InsP6 in leaves (chromosome 2) decreases the contents. Analysis of QTL interactions detected no significant epistasis between the different loci (P < 0.005).
QTL likelihood maps for InsP6 and Pi contents in seeds and in leaves. Presented are four of the five linkage groups of Arabidopsis. The abscissas correspond to the genetic maps in cM; 1 through 4 indicate the linkage group number. The horizontal dotted lines correspond to the LOD score threshold of 2.8 used to declare the presence of a QTL. Two-LOD support intervals for the significant QTL are shown as black bars along the abscissa. QTL effects are shown in the second panel below the LOD graph. These are given as twice the additive allele effects, i.e. as the mean differences between the two RIL genotypic groups carrying the Ler and Cvi alleles. The effects of Pi in seeds as well as in leaves are multiplied by ten to allow presentation in one figure. A positive QTL effect represents that the Ler allele increases the content. The percentage of phenotypic variance explained by each QTL is reported near the corresponding LOD score peak
Analysis of reciprocal effects and dominance
To analyse the effect of the major QTL on the InsP6 and Pi content in more detail a near-isogenic line (NIL) was constructed. This NIL (NIL26) contained a Cvi introgression on top of chromosome 3 in a Ler background. The analysis of 100 F2 plants of a cross between NIL26 and Ler for seed InsP6 content and the genotype at marker T4P13b, showed a typical monogenic segregation and linkage of the trait to the marker. In addition it revealed the dominance effect of the Cvi allele, heterozygous plants showing the higher InsP6 and Pi levels of NIL26 (Fig. 6). The levels of InsP6 and Pi in seeds of NIL26 are similar to those in Cvi, confirming that indeed the Cvi allele(s) of the introgression on top of chromosome 3 is the major factor responsible for the higher InsP6 and Pi levels of Cvi. Figure 7 shows the levels of InsP6 and Pi in seeds of reciprocal crosses between Ler and NIL26. This data indicates a clear maternal inheritance of InsP6 and Pi levels in seeds, since the InsP6 and Pi levels of the hybrid with Ler as the female parent are similar to that of Ler itself and when NIL26 is the female parent the levels are like that of NIL26 itself.
Fine-mapping of the major InsP6 and Pi locus
Since the region containing the QTL was still approximately 3 cM in size, a further reduction of this region was needed towards the positional or candidate gene cloning. Therefore, a mapping population of 631 plants was generated from a heterozygous NIL26 sister plant. These plants were analysed with two molecular markers (T4P13b and 17D8LE, Table 1) one at the top of chromosome 3 and the other near the end of the Cvi introgression. This revealed 23 recombinants between the two markers showing a recombination frequency of 1.8 cM. Seeds of these recombinants were tested for InsP6 and Pi levels and this confirmed that the InsP6/Pi locus was located in between these two markers. For mapping, nine molecular markers (Table 1) between T4P13b and 17D8LE (which were tested for Ler/Cvi polymorphisms) were generated from the Columbia sequence. All 23 selected recombinants have been analysed with these markers. Hereby the QTL could be mapped to a region of 99 kb, localizing the QTL on BAC T13O15 between markers cT4P13x and cT13O15c, which have respectively 1 and 3 recombinants with the InsP6/Pi locus as is shown in Fig. 8. This region contains 13 open reading frames. Based on the annotation of the Columbia sequence this region includes a putative peptide transporter (gene code: At3g01350), a putative protein transport protein SEC13 (At3g01340), a putative protein kinase (At3g01300), a putative phosphatidylinositolglycan-class N short form (At3g01380), a vacuolar membrane ATPase subunit G1 (At3g01390), a putative RNase H (At3g01410) and seven unknown proteins.
Genetic fine mapping of the major InsP6/Pi QTL. The top of the figure shows the genetic map with AFLP markers (Alonso-Blanco et al. 1998b) of chromosome 3, solid bars indicate Ler DNA, open bars the Cvi alleles. Junctions between open and solid bars indicate crossover breakpoints. The lower part shows the QTL region in detail, essential recombinants which indicate the position of the InsP6/Pi QTL are indicated, with their average InsP6 levels (mg/g seed) Molecular markers with the number of recombinants between the marker and the InsP6/Pi locus are indicated as well. The number of recombinants with the markers nga32 and 17D8LE are underestimated, as the InsP6 levels of heterozygous plants could not be distinguished from Cvi plants. Recombinants with their crossover breakpoint between the other markers have been analysed for InsP6 by determining the seeds of 12 plants derived from these recombinants, to distinguish heterozygous plants from Cvi plants
Discussion
We present the analysis of natural variation for InsP6 and Pi in Arabidopsis thaliana. To be able to analyse a large series of samples we used a simple and rapid detection method, based on anion exchange chromatography and detection through ion-suppressed conductivity. This method has been used before for the analysis of steep water of corn (Hull and Montgomery 1994), but not for plant material. The method is very sensitive, able to detect InsP6 in sub-milligram samples of seeds, with recoveries >90%, provided the sample size does not exceed 4 mg (Fig. 2).
Total levels of P (InsP6 and Pi) in Arabidopsis seeds are higher compared with levels reported for maize, barley and rice (Raboy et al. 2001). However, Coelle et al. (2001) reported around two-times higher levels in seeds of accession Columbia. This might be due to differences in growth conditions, since Lickfett et al. (1999) reported that soil-P levels largely affect InsP6 levels in Brassica seeds. Besides P-fertilization, InsP6 and Pi concentrations in seeds may vary because of many factors including moisture plus climatic factors (Lott et al. 2000). We also found that genetically identical seed batches, harvested at different times, could vary in Pi and Insp6 contents. To avoid these environmental effects we have only compared plants that were grown under identical environmental conditions in the same experiment.
A wide range of variation in InsP6 and Pi levels was observed amongst accessions, varying from Kyoto-1, containing 7.0 mg of InsP6 per gram of seed, to Hey-0 with 23.1 mg of InsP6 per of gram seed. We also observed the typically high and positive correlation of InsP6 with total Pi as has been reported by Raboy (1997) and Raboy et al. (1990), but there are accessions as well that do have altered ratios of InsP6 compared to the Pi levels, indicating that total P accumulation is affected. Altered ratios of InsP6 compared to total P were also described in mutants, isolated in maize (Z. mays L.), barley (H. vulgare L.) and rice (O. sativa L.) (Raboy and Gerbasi 1996; Larson et al. 1998, 2000; Raboy et al. 2000). The observation that total P in mutant seeds remained constant has been explained as an altered biosynthesis of InsP6, rather than differences in uptake from the environment and/or partitioning of P within the plant.
The presence of different types of natural variation among the Arabidopsis accessions provides unique genetic sources to study the genetics and molecular biology of InsP6 levels in plants. Therefore, we performed a QTL mapping procedure to study the natural variation for InsP6 and Pi between the accessions Ler and Cvi. QTL mapping revealed five genomic regions affecting the quantity of InsP6 and Pi in seeds and leaves. One of them, located on top of chromosome 3 is affecting all four traits. This QTL appears as the major locus responsible for the observed variation in InsP6 and Pi contents in the Ler/Cvi RIL population, the Cvi allele increasing the content of both InsP6 and Pi in seeds and in leaves. To Mendelize this major InsP6/Pi QTL we generated a near-isogenic line (NIL26) carrying a 3-cM Cvi genomic region on the top of chromosome 3 in a Ler background. Seeds of this line have higher InsP6 levels than seeds of Ler itself, confirming that indeed the Cvi allele of the QTL is increasing the InsP6/Pi levels. The F2 of a cross between Ler and NIL26 showed a monogenic segregation and revealed the dominance of the Cvi allele at this InsP6/Pi locus. Reciprocal crosses show maternal inheritance. This might indicate that the difference in InsP6 between Ler and Cvi is caused by transport of P from the mother plant into the seed. This is in agreement with the finding that the InsP6 levels compared to the total P levels are not altered, indicating that there is no alteration in the biosynthesis of InsP6 controlled by this locus.
Possible candidate genes for the InsP6/Pi locus are genes which have a function in P translocation. There are four Arabidopsis mutants known to be affected in P accumulation/mobilization. These are pho1, which is exhibiting a reduced P-uptake (Poirier et al. 1991), pho2, a P-accumulator (Delhaize and Randall 1995), pho3, a P deficient mutant probably affected in P uptake by the roots (Zakhleniuk et al. 2001) and ppt1, which is mutated in the phosphate/phosphoenolpyruvate translocator gene (formerly named cue1-1, and allelic to CS3156; Li et al. 1995). However, except for pho3 (which has not been mapped) none of these mutants are candidate genes for the major InsP6/Pi QTL, as they are localised at different genetic positions. PHO1 has recently been cloned and is localised on chromosome 3, but at a lower position than the InsP6/Pi QTL (Hamburger et al. 2002). PHO1 belongs to a gene family but none of the PHO1 homologs map to the same chromosome region as the InsP6/Pi QTL. PHO2 has been mapped to the bottom of chromosome 2, near marker m429 (Delhaize and Randall 1995) and might be a candidate gene for the QTL at the bottom of chromosome 2 that affects InsP6 in leaves and Pi in seeds. PPT1 has been cloned and is located on the middle of chromosome 5 at BAC F19N2 (Streatfield et al. 1999).
Fine mapping of the major QTL using molecular markers localised the InsP6/Pi locus to a 99-kb region containing 13 open reading frames (ORFs). We consider the vacuolar membrane ATPase subunit G, as the most likely candidate gene for the InsP6/Pi locus, among the annotated genes in the 99-kb region of interest. As suggested above, the InsP6/Pi locus might be involved in P-translocation in the plant, and P-transport across membranes depends on the membrane potential, which is generated by ATPases. However, we cannot exclude one of the other genes to play a role in InsP6/Pi accumulation, since only complementation analysis can prove that a candidate gene is responsible for the QTL allelic variation (El-Assal et al. 2001).
Using immortal populations has a great advantage because the same set of plants can be used to analyse different traits that might influence the same processes. Hitz et al. (2002) reported about the biochemical and molecular characterisation of a mutation that confers a decreased raffinose oligosaccharide and InsP6 phenotype on soybean seeds. This can be explained by the fact that InsP6 and raffinose oligosaccharides share at least myo-inositol-1-phosphate and possibly free myo-inositol as a common intermediate (Loewes and Murthy 2000). Since the Ler/Cvi RIL population has also been analysed for oligosaccharide contents and seed storability (Bentsink et al. 2000) we can check whether QTLs affecting these traits co-localize with the QTL identified for InsP6/Pi. This showed that there is no co-localization between the InsP6/Pi loci and the oligosaccharide QTL.
However the InsP6/Pi locus on top of chromosome 3 co-localizes with the QTL for sucrose and seed storability. The Cvi allele of this locus is increasing InsP6 levels, sucrose content and seed storability, indicating that higher InsP6 levels on their own or with higher sucrose levels might result in a better storability. A role of InsP6 in seed quality is suggested by the finding that reducing the level of InsP6 with more than 95% results in loss of viability (Raboy et al. 2001). Except for the major InsP6/Pi QTL we see co-location of the locus affecting Pi in seeds (chromosome 1) with the major seed storability QTL, while decreasing Pi levels correlate with higher storability. Any functional relation between these traits has not been reported and co-location may of course also involve different linked genes.
Another advantage of the Ler/Cvi RIL population is that we can study the link between InsP6 levels and seed size or seed weight. It has been reported that reductions in seed InsP6 have severe impacts on seed and plant growth and function, translating in more severe yield losses (reviewed by Raboy 2001). Alonso-Blanco et al. (1999) studied allelic variation at seed size loci in relation to other life history traits using Ler/Cvi RIL. This analysis revealed 11 loci that affected seed weight and/or seed length. One of them is located on top of chromosome 3 near the locus where the main InsP6/Pi QTL is located; however, this is a QTL with a minor effect and is located below the InsP6/Pi QTL. The effects of these traits are negatively correlated, suggesting that higher InsP6 levels would result in lower seed weight. This is in contrast with results described earlier (Raboy 2001). However, we find a correlation between the major QTL influencing seed size (located on top of chromosome 1) and a QTL affecting Pi levels in seeds. This correlation is much stronger than the one mentioned before, lower Pi levels correlate positively with higher seed weights.
Concluding, there is considerable variation among natural accessions of Arabidopsis that can be used to identify genes that are involved in the regulation of InsP6 accumulation. The use of other accessions may identify additional loci affecting this trait.
References
Alonso-Blanco C, Koornneef M (2000) Naturally occurring variation in Arabidopsis: an underexploited resource for plant genetics. Trends Plant Sci 5:22–29
Alonso-Blanco C, Bentsink L, Hanhart CJ, Blankenstijn-de Vries H, Koornneef M (2003) Analysis of natural allelic variation at seed dormancy loci of Arabidopsis thaliana. Genetics (in press)
Alonso-Blanco C, Peeters AJM, Koornneef M, Lister C, Dean C, van den Bosch N, Pot J, Kuiper MTR (1998) Development of an AFLP based linkage map of Ler, Col and Cvi Arabidopsis thaliana ecotpyes and construction of a Ler/Cvi recombinant inbred line population. Plant J 14:259–271
Alonso-Blanco C, Blankenstijn-de-Vries H, Hanhart CJ, Koornneef M (1999) Natural allelic variation at seed size loci in relation to other life history traits of Arabidopsis thaliana. Proc Natl Acad Sci USA 96:4710–4717
Bell CJ, Ecker JE (1994) Assignment of 30 microsatellite loci to the linkage map of Arabidopsis. Genomics 19:137–144
Bentsink L, Alonso-Blanco C, Vreugdenhil D, Tesnier KJY, Groot SPC, Koornneef M (2000) Genetic analysis of seed-soluble oligosaccharides in relation to seed storability of Arabidopsis. Plant Physiol 124:1595–1604
Coelle P, Maughan JP, Mendoza A, Philip R, Bollinger DW, Veum TL, Vodkin LO, Polacco JC (2001) Generation of low phytic acid Arabidopsis seeds expressing an E. coli phytase during embryo development. Seed Sci Res 11:285–291
Delhaize E, Randall PJ (1995) Characterization of a phosphate-accumulator mutant of Arabidopsis thaliana. Plant Physiol 107:207–231
El-Assal SE-D, Alonso-Blanco C, Peeters AJM, Raz V, Koornneef M (2001) A QTL for flowering time in Arabidopsis reveals a novel allele of CRY2. Nature Genet 29:435–440
Hamburger D, Rezzonico E, Mac-Donald-Comber Petétot J, Somerville C, Poirier Y (2002) Identification and characterization of the Arabidopsis PHO1 gene involved in phosphate loading to the xylem. Plant Cell 14:889–902
Hanakahi LA, Bartlet-Jones M, Chappell C, Pappin D, West SC (2000) Binding of inositol phosphate to DNA-PK and stimulation of double-strand break repair. Cell 102:721–729
Hitz WD, Carlson TJ, Kerr PS, Sebastian SA (2002) Biochemical and molecular characterization of a mutation that confers a decreased raffinosaccharide and phytic acid phenotype. Plant Physiol 128:650–660
Hull SR, Montgomery R (1994) Separation and analysis of 4′-epimeric UDP-sugars, nucleotides, and sugar phosphates by anion-exchange high-performance liquid chromatography with conductimetric detection. Anal Biochem 222:49–54
Konieczny A, Ausubel FM (1993) A procedure for mapping Arabidopsis mutations using co-dominant ecotype-specific PCR-based markers. Plant J 4:403–410
Larson S, Young K, Cook A, Blake T, Raboy V (1998) Linkage mapping of two mutations that reduce the phytic acid content of barley grain. Theor Appl Genet 97:141–146
Larson S, Young K, Blake T, Raboy V (2000) Isolation and genetic mapping of a non-lethal rice low phytic acid I mutation. Crop Sci 40:1397–1405
Lemtiri-Chlieh F, MacRobbie EAC, Brearley CA (2000) Inositol hexaphosphate is a physiological signal regulating the K+-inward rectifying conductance in guard cells. Proc Natl Acad Sci USA 97:8687–8692
Li H-m, Culligan K, Dixon RA, Chory J (1995) CUE1: a mesophyll cell-specific positive regulator of light-controlled gene expression in Arabidopsis. Plant Cell 7:1599–1610
Lickfett T, Matthäus B, Velasco L, Möllers C (1999) Seed yield, oil and phytate concentration in the seeds of two oilseed rape cultivars as affected by a different phosphorus supply. Eur J Agron 11:292–299
Lobin W (1983) The occurrence of Arabidopsis thaliana in the Cape Verde Islands. Arab Inf Serv 20:119–123
Loewus FA, Murthy PPN (2000) Myo-inositol metabolism in plants. Plant Sci 150:1–19
Lott JNA, Ockenden I, Raboy V, Batten GD (2000) Phytic acid and phosphorus in crop seeds and fruits: a global estimate. Seed Sci Res 10:11–33
Lucca P, Hurrell R, Potrykus I (2001) Genetic engeneering approaches to improve the bioavailability and the level of iron in rice grains. Theor Appl Genet 102:392–397
Maugenest S, Martinez I, Lescure AM (1997) Cloning and characterization of a cDNA encoding a maize seedling phytase. Biochem J 322:511–517
Ooijen van JW (1992) Accuracy of mapping quantitative trait loci in autogamous species. Theor Appl Genet 84:803–811
Ooijen van JW (1999) LOD significance thresholds for QTL analysis in experimental populations of diploid species. Heredity 83:613–624
Ooijen van JW, Maliepaard C (1996) MapQTL(tm) version 4.0: software for the calculation of QTL positions on genetic maps. Plant Research International, Wageningen, The Netherlands
Poirier Y, Thoma S, Somerville C, Schiefelbein J (1991) A mutant of Arabidopsis deficient in xylem loading of phosphate. Plant Physiol 97:1087–1093
Raboy V (1997) Accumulation and storage of phosphate and minerals. In: Larkins BA, Vasil IK (eds) Cellular and molecular biology of plant seed development. Kluwer Academic Publishers Dordrecht, pp 441–477
Raboy V (2001) Seeds for a better future: "low phytate" grains help to overcome malnutrition and reduce pollution. Trends Plant Sci 6:458–462
Raboy V, Dickinson D (1987) The timing and rate of phytic acid accumulation in developing soybean seeds. Plant Physiol 85:841–844
Raboy V, Gerbasi P (1996) Genetics of myo-inositol phosphate synthesis and accumulation. In: Biswas B, Biswas S (eds) Myo-inositol phosphates, phosphoinositides and signal transduction. Plenum Press, New York, pp 257–285
Raboy V, Dickinson DB, Neuffer MG (1990) A survey of maize kernel mutants for variation in phytic acid. Maydica 35:383–390
Raboy V, Gerbasi PF, Young KA, Stoneberg SD, Pickett SG, Bauman AT, Murthy PPN, Sheridan WF, Ertl DS (2000) Origin and seed phenotype of maize low phytic acid 1-1 and low phytic acid 2-1. Plant Physiol 124:355–368
Raboy V, Young KA, Dorsch JA, Cook A (2001) Genetics and breeding of seed phosphorus and phytic acid. J Plant Physiol 158:489–497
Rédei GP (1992) A heuristic glance to the past of Arabidopsis genetics. In: Koncz C, Chua N, Schell J (eds) Methods in Arabidopsis research. World Scientific, Singapore, pp 1–15
Safrany ST, Caffrey JJ, Yang X, Shears SB (1999) Diphosphoinositol polyphosphates: the final frontier for inositide research? Biol Chem 380:945–951
Sasakawa N, Sharif M, Hanley MR (1995) Metabolism and biological activities of inositol pentakisphosphate and inositol hexakisphosphate. Biochem Pharmacol 50:137–146
Streatfield SJ, Weber A, Kinsman EA, Hausler RE, Li J, Post-Beittenmiller D, Kaiser WM, Pyke KA, Flugge U-I, Chory J (1999) The phosphoenolpyruvate/phosphate translocator is required for phenolic metabolism, palisade cell development and plastid-dependent nuclear expression. Plant Cell 11:1609–1621
Weiss J (1991) Ionenchromatographie. VCH Verlag, Weinheim New York Basel Cambridge. ISBN 3-527-28236
Wilcox J, Premachandra G, Young K, Raboy V (2000) Isolation of high seed inorganic P, low-phytate soybean mutants. Crop Sci 40:1601–1605
York JD, Odom AR, Murphy R, Ives EB, Wente SR (1999) A phospholipase C-dependent inositol polyphosphate kinase pathway required for efficient messenger RNA export. Science 285:96–100
Zakhleniuk OV, Raines CA, Lloyd JC (2001) pho3: a phosphorus-deficient mutant of Arabidopsis thaliana (L.) Heynh. Planta 212:259–534
Acknowledgements.
The authors thank Diaan Jamar for technical assistance in the InsP6 and Pi analyses. K.Y. received a grant from the China Scholarship Council. L.B. was supported by the Netherlands Organization for Scientific Research and by the European Union (E.U.) program NATURAL (contract QLG2-CT-2001-01097).
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by L. Willmitzer
L. Bentsink and K. Yuan contributed equally to the work
Rights and permissions
About this article
Cite this article
Bentsink, L., Yuan, K., Koornneef, M. et al. The genetics of phytate and phosphate accumulation in seeds and leaves of Arabidopsis thaliana, using natural variation. Theor Appl Genet 106, 1234–1243 (2003). https://doi.org/10.1007/s00122-002-1177-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00122-002-1177-9