Abstract.
EST microsatellite markers were developed in apricot (Prunus armeniaca L.) and grape (Vitis vinifera L.). cDNA libraries from either apricot leaves or grape roots were used in an enrichment procedure for GA and CA repeats. The transferability of EST simple sequence repeat (SSR) markers from apricot and grapevine to other related and unrelated species was examined. Overall, grape primers amplified products in most of the Vitaceae accessions while the apricot primers amplified polymorphic alleles only in closely related species of the Rosaceae. In this taxonomic family, ten EST SSR loci were tested, and one single primer pair, PacB22, was amplified across species and sections in the Prunoideae and Maloideae. Sequencing of EST SSR loci in other species and genera confirmed a higher level of conservation in the microsatellite motif and flanking regions in the Vitaceae compared to the Rosaceae. Two distinct fragments of the PacB22 locus amplified across the Malus and Pyrus genera; however, while the coding region was highly conserved, the microsatellite repeat motif was no longer present. The banding pattern was explained by base substitution and insertion/deletion events in the intronic region of PacB22. This study includes the determination of the degree of polymorphism detected among species and genera in two unrelated taxonomic families and the evaluation of the information provided by the microsatellite repeats and the flanking regions.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Recently, interest has been directed towards the comparative studies of genome organisation in natural and cultivated populations. For this purpose, hypervariable markers such as SSRs (simple sequence repeats, also called microsatellites) have been characterised in a number of crop and tree species (see Powell et al. 1996 for a review). In many cases, microsatellite markers were shown to be transferable across taxa (Rossetto 2001). The conservation of flanking regions and primer binding sites in different genera has been reported, for example, between wheat and barley (Erpelding et al. 1996), soybean and other legume genera (Peakall et al. 1998), Malus and Pyrus (Yamamoto et al. 2001) and among the Vitaceae family (Thomas and Scott 1993; Rossetto et al. 2002). In the Rosaceae, cross-species amplification was tested on Prunus species and between Malus, Fragaria and Prunus genera (Cipriani et al. 1999; Downey and Iezzoni 2000; Sosinski et al. 2000; Dirlewanger et al. 2002). In cases where codominant and hypervariable markers can be transfered over a set of related and unrelated species, microsatellites can be of great interest for the analysis of genetic diversity and for studies on the evolution of species. However, they are usually distinguished by size, which is determined on high-resolution sequencing gels. Compensatory effects within the microsatellite locus such as base substitutions and insertion/deletion events are therefore not detected and the fragments amplified may not be those anticipated. This might lead to errors due to the high chance of independently arising, equally sized alleles (homoplasies) (Grimaldi and Crouau-Roy 1997). Moreover, transferability means not only that amplification but also that the microsatellite primers developed for one species can be used to detect polymorphism at homologous loci in related species. However, successful amplification does not guarantee the presence of the repeat motif within the sequence, leading to a loss in polymorphism across species.
We successfully used an enrichment procedure to isolate microsatellite repeats from genes expressed in grape roots and apricot leaves. We produced a set of microsatellite markers linked to ESTs (expressed sequence tags) and known genes that identify single variable loci. In the current report, the possibility of transferring EST SSR information across species and genera of the Vitaceae and Rosaceae taxa is investigated. The purpose here is not to define phylogenetic relationships across related and unrelated species in the Vitaceae and Rosaceae but rather to determine how informative a set of microsatellites is across two unrelated taxonomic families. Two types of information are compared in this study: the degree of polymorphism detected on high-resolution sequencing gels and the sequences of the microsatellite and its flanking regions.
Materials and methods
Construction of cDNA libraries enriched for microsatellites and primer design (Table 1)
Total RNA was extracted from mature leaves of apricot (Prunus armeniaca L., cultivar Stark Early Orange, SEO) and grape roots (Vitis vinifera L. cv Cabernet Sauvignon). RNA was extracted following the procedure described in Chang et al. (1993) and treated with DNAase RQ1 (Promega). PolyA+ RNA was purified using the Oligotex kit (QIAGEN). Double-stranded cDNA was synthesised with the Promega Riboclone Kit and cDNA larger than 200 bp was selected using spin columns (Pharmacia). The fragment ends were polished and, after purification, 3 ng of cDNA was ligated with the Mlu1 adaptor. Prior to enrichment, 1/20th of the ligation was amplified for 25 cycles with the 21-mer Mlu1 primer (Edwards et al. 1996). The enrichment procedure of Edwards et al. (1996) was used with the modifications described in Butcher et al. (2000). Enrichment for microsatellites was carried out using the total volume of the former amplification which was hybridised on CA- and GA-bound Nylon membranes (Appligene). Following two rounds of enrichment, 8 μl of eluted DNA was amplified by PCR with 0.8 μM of the 21-mer Mlu1 primer in 40 μl of 20 mM Tris, 50 mM KCl, 2 mM MgCl2, 0.2 mM of each dNTP and 1 U of Taq polymerase (Life Technologies) using a Perkin Elmer 9700 thermocycler; 1/20th of the PCR product was cloned in 50 ng of pGEM-T vector (Promega) prior to transformation into DH5α Escherichia coli cells.
Randomly picked clones were amplified using M13 universal primers. Insert DNA was purified on Qiaquick PCR purification columns (QIAGEN) and directly sequenced using an ABI 377 automated DNA sequencer (Applied Biosystems). Oligonucleotide primers complementary to the regions flanking the identified repeat motifs were designed using the program Primer version 0.5 (National Biosciences, Plymouth, Minnesota), setting an annealing temperature of 57.5 °C. Database searches were carried out using the Advanced Blast program at the National Center for Biotechnology Information (Bethesda, Md.).
Cross-species amplification of EST SSR loci
Eight grape microsatellites isolated from an enriched root cDNA library of V. vinifera were tested among 62 individuals out of 46 different species in the Vitaceae family (see Table 2). Classification was as reported by Galet (1967). All accessions were obtained from the national collection of INRA (Institut National de la Recherche Agronomique) based at Montpellier and Bordeaux.
In the Rosaceae family, 21 Prunus accessions including interspecific crosses, one pear (Pyrus communis L.) and six apple (Malus × domestica Bortch.) cultivars and an interspecific cross between Cydonia oblonga (Miller) and P. communis were amplified with each of ten apricot EST SSR primer pairs (see Table 3). These accessions were obtained from the INRA germplasm collection at Bordeaux, Avignon and Angers. Classification was as reported by Rehder (1949).
Genomic DNA from Rosaceae individuals was prepared as described in Lefort and Douglas (1999). In the Vitaceae, DNA was prepared with the QIAGEN Plant DNEASY mini kit. Starting with 10 ng of genomic DNA as a template, PCR conditions were as follows: 2 mM of MgCl2, 0.1 mM of each dNTP, 0.2 μM of each primer, 1× Buffer and 0.5 units of Taq polymerase (Sigma) in 25 μl final volume. Thermocycling conditions were 30 s at 94 °C, followed by 30 cycles of (94 °C for 30 s, 57.5 °C for 30 s and 72 °C for 30 s) and an extension step at 72 °C for 7 min, in a GeneAmp 9700 thermal cycler (Applied Biosystems).
Separation of alleles was performed on a 6% acrylamide sequencing gel in 0.5× TBE buffer at 80 W for 100 to 120 min. One to two microliters of PCR product was mixed with 3 μl of denaturing loading buffer containing 95% formamide, 0.25% bromophenol blue and 0.25% xylen cyanol, and 10 mM of EDTA. This mixture was heated for 5 min at 94 °C to denature the DNA before loading. Gels were stained with silver nitrate following the protocol detailed by Chalhoub et al. (1997) and bands were scored after gel scanning.
PCR cloning of SSR loci and sequence analysis
A dinucleotide (CA/GT)18 microsatellite locus was selected in apricot, PacB22, and screened for sequence variation in the Rosaceae family. Another dinucleotide repeat, vvc34, was screened from Vitaceae genomic DNA. The PacB22 primer pair amplifies a fragment of about 260 bp in P. armeniaca L. and the vvc34 primers amplify a fragment of 200 bp in V. vinifera. PCR conditions were as indicated above. PCR products were purified on QiaQuick PCR purification columns (Qiagen) and cloned in the pGEM-T vector (Promega). Inserts were sequenced in duplicate from various individuals using an automated sequencing system (Genaxis, Nîmes, France). DNA sequences were aligned using the ClustalW software from InfoBiogen (http://www.infobiogen.fr).
Results
Identification of EST SSR loci
Within both enriched cDNA libraries, the insert size ranged from 300 to 1,500 bp with an average of 800 bp. Several clone sequences were discarded prior to primer design for several reasons which included insufficient length of the flanking sequences to design primers, too short sequences between the repeat motif and the poly-A tail or multiple cloned sequences. Details on primers which amplified a single locus in apricot and grape are summarised in Table 2. About 500-bp long sequences from the 5′ and/or 3′ ends of each clone were obtained. TBLAST-X analysis (http://www.ncbi.nlm.nih.gov/BLAST/) identified significant homology to known plant-coding regions for 81% of the apricot and 70% of the grape SSR containing clones (see Table 1). The remaining apricot and grape cDNAs represent novel transcripts. Enrichment in CA and GA motifs was up to 90% but this does not take into account clone redundancy. In apricot, 51.5% of the positive clones were unique based on sequencing data. One clone, PacC3, accounted for up to 19% of the overall library.
SSR length polymorphism
Identical PCR conditions and thermal reactions were used for all EST SSR primers. Optimisation of the PCR conditions especially for relatively distant taxa was not attempted in order to avoid amplification of false positives. Separation of the fragments by standard polyacrylamide sequencing gels and visualisation by silver staining gave, in general, a common "stuttered" banding pattern, but the fragments could easily be distinguished at a resolution of 1 bp. The allele sizes were recorded in base pairs and, for this, the most intense upper band was used. Alleles were scored twice independently by the authors. The degree of polymorphism was counted as the absolute number of different alleles per microsatellite locus.
Of the eight primer pairs that amplified across Vitaceae DNA, all EST SSR loci were variable, two being highly polymorphic, vvc5 and vvc83, with 43 and 42 alleles respectively among 62 accessions tested (Table 2). In the family Vitaceae, most of the EST SSR loci in Cissus and Ampelopsis were homozygous, while most primers failed to amplify DNA from Cissus voinieriana and Parthenocissus semicordata with the exception of two or three loci. However, since only one accession was tested per species, we can not state whether the EST SSR loci are truly homozygous or not in those species and genera. Surprisingly, the vvc34 primer pair amplified more than two fragments in Cissus quadrangularis. This could be explained by gene duplication but cannot be confirmed without segregation analysis in progeny arrays.
During cross-taxa amplification, most of the blanks due to mis-amplification were detected in genera distant from Vitis. These results confirm a negative correlation between the phylogenetic distance and successful amplification. The same tendency was noticeable when considering the level of polymorphism. Interestingly, for the less polymorphic SSR loci (vvc6, vvc7 and vvc19), the low level of allelic variation was not due to a failure in amplification but instead reflects the amplification of predominant alleles.
The most successful cross-species amplification of polymorphic EST SSR loci was within the genus Vitis. There were however a number of exceptions including Vitis rubra accession #10168 which was homozygous at all, but one (vvc34), loci.
All ten apricot EST SSR loci transferred to closely related species such as Prunus domestica, which is a member of the same subgenus as P. armeniaca L., Prunophora (Table 3). In the Prunophora subgenus, the level of ploidy can differ greatly from 2n = 2x to 6x but in most cases, EST SSR loci were polymorphic with two to five alleles per genome. Loci presenting two alleles in the apricot genome were the most polymorphic in related species, except for PacC3 where null alleles were determined by analysis in plum full-sib segregating pedigrees (data not shown). Null alleles can be due to changes in flanking regions, for example at the PacC3 primer binding sites, that prevent primer annealing and result in no amplification.
Across different subgenera, amplification of heterozygote alleles was attained within the subgenus Cerasus, mostly in Prunus cerasus species (2n = 4x), while the Prunus persica accessions were in general homozygous at the respective loci. Table 3 does not show any predominant allele except in intraspecific microsatellite amplification of P. persica DNA, confirming a high level of homozygosity in this species.
Among ten EST SSR loci tested, only the PacB22 primer pairs yielded an unambiguous banding pattern across the Rosaceae genera. Interestingly, in the Maloideae, this locus showed a very similar allelic pattern between the two species, Malus × domestica and Pyrus communis, as well as successful amplification in the Cydonia × Pyrus interspecific accession, Pyronia veitchii. Two clear bands of equal size with no stutters were detected in four of the Malus cultivars and in the pear and P. veitchii accessions.
With the exception of PacB22, cross-genera amplification of EST SSR loci failed or produced a number of bands that could not be interpreted (see PacA10 and PacB35). PacB22 is one of the most-polymorphic EST SSR loci, together with loci PacA18 and PacC13, resulting in 22 to 23 alleles for 29 accessions tested (Table 3). However, this cannot be compared to the most polymorphic vvc loci in the Vitaceae since, in the Rosaceae, we have to account for polyploidy. This can increase considerably the number of alleles observed per locus.
Comparison of nucleotide sequences of the microsatellite locus vvc34 among the Vitaceae species
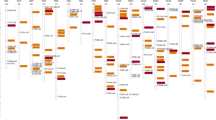
The PCR products amplified across taxa by the vvc34 primer pair were cloned and sequenced from ten species of the genus Vitis. The sequences were analysed to verify the nature of the amplification products compared with one of the two alleles sequenced in V. vinifera, cv Cabernet Sauvignon. The vvc34 microsatellite repeat was systematically present but shorter than in the source cultivar. The alignment by ClustalW analysis revealed a high degree of conservation of vvc34 SSR flanking regions especially across the Vitis genus (Fig. 1). In this genus, allelic variation and length variability within a species and across the Euvitis and Muscadinia subgenus was mainly due to a length variation of the microsatellite alone, combined with point mutation and short insertion/deletion events (2–4-bp long, Fig. 1) within the flanking regions. An interrupted repeat was detected in the related Vitis subgenus, Muscadinia but not in the Euvitis species. Interestingly, a 2-bp indel placed just downstream of the vvc34 forward primer is observed in two geographically distinct species, Vitis mexicana (American) and Vitis davidii (Asian).
Nucleotide comparison of vvc34 EST SSR locus in ten related species and four genera of the Vitaceae family. From 1 to 12, the subgenus Euvitis comprising European (1–2) the V. vinifera accession number in Genbank BQ106729; 3, Vitis sylvestris), american (V. riparia 4–5, labrusca 6 and mexicana 7–8 respectively) and asian (from 9 to 12, V. davidii, Vitis amurensis, Vitis piazeskii, V. romaneti respectively) species; 13 subgenus Muscadinia, species Vitis rotundifolia; 14 P. henryana; 15 and 16 A. chantinii; 17 to 20 C. quadrangularis The microsatellite repeat is underlined. The dot above the sequence indicates interruption in the perfect (CA) repeat in Muscadinia rotundifolia, and arrows the forward and reverse primers
In this sequencing analysis, DNA sequences from Parthenocissus henryana, Ampellocissus chantinii and C. quadrangularis were added as representatives of the more distant genera. Figure 1 shows that, despite point mutations and rather important length variation, the microsatellite repeat is still present. However, it can be either a compound microsatellite repeat as it is in V. vinifera (succession of GC and CA repeats), or a perfect (CA) repeat. It is also interrupted in two distinct positions in P. henryana. Four distinct fragments were amplified and cloned from C. quadrangularis. Surprisingly, one of those presents a deletion downstream to the motif repeat similar to Vitis riparia and Vitis labrusca, while those species are definitely not related. This might be due to the fact that this indel belongs to an extensive hypervariable region, prone to mutation and deletion. It draws attention to potential pitfalls that arise from using flanking sequences too close to the hypervariable microsatellite region for phylogenetic analysis.
Length variation and sequence analysis of the PacB22 flanking regions in the Rosaceae
The PCR products amplified across taxa by the PacB22 primer pairs were cloned and sequenced from three subgenera of the Prunoideae and two Maloideae genera. Along with the microsatellite repeat, 66 nucleotides of the upstream coding region were cloned. They displayed almost perfect conservation at the nucleotide level across species and genera (Fig. 2). One base substitution was observed at position 32 which distinguished members of the Prunoideae from the Maloideae. Single or double consecutive base substitutions were detected in the partial coding region but they were either species-specific (position 19–20) or allele-specific.
Nucleotide comparison of PacB22 EST SSR locus in species of the Rosaceae family. Different alleles in the same accessions are indicated by the name of the accession followed by A or B. 1 P. armeniaca L. SEO (260 bp); 2 Prunus davidiana #1908 (256 bp); 3 P. persica GF305 (250 bp); 4 P. avium Burlat (239 bp); 5 P. cerasus V2327 (239 bp); 6 Prunus japonica × Prunus spinosa cv Jaspi A (244 bp); 7 P. japonica × P. spinosa cv Jaspi B (234 bp); 8 Malus × domestica cv discovery fragment A (220 bp); 9 Malus × domestica cv discovery fragment B (227 bp); 10 Malus × domestica cv MM105 A (220 bp); 11 Malus × domestica cv MM105 B (227 bp); 12 P. communis cv Pine seedlings A (220 bp); 13 P. communis cv Pine seedlings B (213 bp); 14 P. veitchii (220 bp); C consensus sequence. The primary tandem repeat is underlined while gaps are represented by broken lines. Bold characters indicate base substitutions in between Prunoideae and Maloideae, as well as nucleotide similarities across Maloideae alleles. Dots above the sequences show interruption in the apricot microsatellite repeat and uppercase letters above the first 70 bp correspond to the partial 5′ coding region of B22 (accession number BQ134643). Forward and reverse PacB22 primers are displayed by arrows
Across the Prunoideae subfamily, the microsatellite repeat was present but much shorter, down to eight (CA/GT) motifs in P. cerasus and P. avium (Fig. 2). This would explain the low level of polymorphism observed across sections in the Prunoideae. Interestingly, the PacB22 flanking region was perfectly conserved apart from the few base substitutions within the coding region.
Most of the transition events were detected between alleles from the Prunoideae subfamily and species of the Maloideae subfamily. Eleven nucleotide substitutions in the region upstream to the primary tandem repeat clearly distinguish the Prunoideae and Maloideae accessions. Moreover, Fig. 2 shows that electrophoretic size variation detected in the Maloideae was not due to variation in the microsatellite repeat numbers but instead to the occurrence of two deletion events present in the PacB22 flanking region. The first indel and several nucleotide substitutions demonstrate similarities between alleles (named A and B arbitrarily) across two Malus cultivars and the Pyrus and Pyronia veitchii accessions. To determine whether these were from different loci, allele segregation was analysed in Malus × domestica F1 progeny cv Discovery × TN10.8 (data not shown). Twelve individuals were tested along with the parents Discovery and TN10.8 following the above PCR conditions. All 12 progeny and two parents displayed two distinct and equally sized fragments. Therefore, the PacB22 locus in the Maloideae does not follow a single-locus segregation.
Another EST SSR locus, PacA10, was sequenced across the Rosaceae species and genera (data not shown) but the sequence in Malus did not correspond to the initial apricot EST clone. While the PacA10 locus in apricot displayed homology with a chlorophyll a/b binding protein (Table 1), the PCR product amplified with the PacA10 primer pair in Malus × domestica showed similarity with a gene coding for a nonsense-mediated mRNA decay transacting factor in Arabidopsis thaliana (accession number AB017068, id.= 4e-08).
Discussion
In this report, we show that SSRs are retrieved from expressed sequences of grape and apricot species, after little input and without extensive sequencing data. cDNA-SSR markers combine the advantages of microsatellite variability with the information content potentially carried by expressed sequences. This is an efficient and economical technique for accumulating and mapping cDNA sequences from different tissues and developmental stages and, by this means, increasing the density of gene markers on linkage maps of minor crops.
A putative function was assigned by sequence similarities for half of the grape and apricot cDNA-SSR loci; this is in concordance with systematic sequencing and functional annotation of genes in other species where 52% of the sequences were coding for unidentified proteins (Aubourg and Rouzé 2001). Some of them are good candidates for QTL mapping and for the assessment of phenotypic and adaptive variation, e.g. potassium import with the SKOR gene and its relation with fruit quality in grape.
Grape and apricot EST SSR primer pairs were tested for cross-species transferability in the family Vitaceae and Rosaceae respectively. Utility of microsatellite markers between related and less-related species was evaluated not only by the presence or absence of amplification products but also on heterozygous banding patterns and sequence variation in the flanking regions.
In the Vitaceae, high levels of length polymorphism across species and genera, and allele sequence data, confirmed the microsatellite nature of observed variations and the optimal utility of the EST SSR markers. In the Rosaceae, optimal utility of the apricot EST SSR markers was for closely related species belonging to the same subgenus Prunophora. Outside this subgenus, heterozygous banding patterns were rarely attained, except in the tetraploid P. cerasus. Specificity of SSRs for a given taxon can limit the cross-species utility of these markers in other cultivated species or in interspecific crosses. Many authors reported a decline in amplification success with increasing divergence and evolutionary distance between taxa (for example see Whitton et al. 1997). The more-closely related the species are, the better is the amplification and the higher is the allelic diversity. This was confirmed in both the Vitaceae and Rosaceae families. The threshold distance after which no amplification can be expected is shorter in the Rosaceae than in the Vitaceae. This may reflect large genetic distances among Rosaceae taxa and that speciation in the Vitaceae took place rather recently. According to Rossetto (2001), cross transferability within the same genus reaches 76.4%, with 86% displaying polymorphism. Across the Vitaceae genera, Arnold et al. (2002) suggested that cDNA microsatellites were transferred more readily than anonymous SSR markers. In the Rosaceae, such a level of transferability is apparently not the case; polymorphism is particularly low in the Amygdalus subgenus and in other non-source subgenera. More extensive data will be required in the Rosaceae to fully compare anonymous versus transcribed SSR markers.
Our results are consistent with studies performed in the Vitaceae using anonymous SSR markers (Di Gaspero et al. 2000), but in contrast with previous studies in the Rosaceae (Sosinski et al. 2000; Dirlewanger et al. 2002). However, in the two last reports, only amplification and detection of appropriate-sized fragments on Metaphor agarose gels were recorded; polymorphism and verification of true positives by sequencing were not considered. We believe that it is not possible to predict marker transferability into a given species on the basis of its taxonomic classification; it will mainly depend on the evolutionary histories of the respective taxonomic families.
In animals, the potential of interspecific amplification is high (for example see FitzSimmons et al. 1995). In plants, cross-species transferability has been particularly successful in several crop species and forest trees (Westman and Kresovich 1998; Karhu et al. 2000). Conservation of a microsatellite repeat at the waxy locus has been demonstrated in non-related species such as rice, barley and potato (Becker and Heun 1995; Ayres et al. 1997; Milbourne et al. 1998). While in some cases this locus is monomorphic in length (Washington et al. 2000; Domon et al. 2002), such conservation across plant taxa raises the question of the biological role for this microsatellite repeat.
It is important to note that we used unchanged PCR conditions to test the transferability of EST SSRs across taxa. Modifiying the PCR protocol by lowering of annealing temperature may increase transferability. However, false positives can appear and the fragments amplified may not be those anticipated, for example PacA10 in Malus × domestica. Such alleles will be incorrectly scored at an expected EST locus and are a major drawback in genetic comparison and marker-assisted localisation of expressed genes. In many surveys of SSR amplification across plant species and genera, products are selected as useful markers if they amplify consistently and are similar to SSRs in the source taxon in terms of size and polymorphism. This highlights the fact that results are easily misinterpreted when transferring SSR markers across taxa.
Overall, the sequence alignments showed that the microsatellite repeat in the Vitaceae was variable but present in most cases. This study is clearly limited by the number of taxa used and by the number of alleles sequenced. However, other authors recently reported the characterisation of microsatellite flanking regions in other species of the V itaceae family (Di Gaspero et al. 2000; Rossetto et al. 2002). Interestingly, Rossetto et al. (2002) evaluated the potential of EST SSR flanking regions for determining taxonomic relationships within two Vitaceae genera, Cissus and Cayratia. In our case, the purpose was not to perform a phylogenetic study but to compare the potential of information provided by the tandem repeat, as well as the flanking region, in comparative genetics. Clearly, in the Vitaceae, microsatellite and flanking regions are highly conserved. Similar results were reported by Di Gaspero et al. (2000) who showed that there was no difference at the sequence level between European, American and Asian species. However, interruptions in the microsatellite repeat occur in another subgenus, Muscadinia, and enables separation of the two Vitis subgenera, but this needs to be confirmed by more extensive sequencing. More information was obtained by analysis of the flanking regions and the occurrence of insertion/deletion events. We showed sequence similarities, especially a 2-bp deletion event, between two geographically distinct species, V. mexicana (American) and V. davidii (Asian). Along with the former study of Di Gaspero et al. (2000), this clearly demonstrates that classification in the genus Vitis based on the geographical distribution of species may not be reflecting the true phylogenetic relationship.
In view of the low success of cross-genera amplification and size polymorphism in the Rosaceae family, information based on flanking-region sequences makes those EST markers a potentially more useful tool for the study of genome variation and evolution between genera. It is well documented that within a microsatellite locus, length differences between alleles are not only due to a variation in the number of tandem repeat units (Grimaldi and Crouau-Roy 1997; Orti et al. 1997). In the case of the PacB22 EST SSR locus, no variability was found among different species of the Prunoideae subfamily except for microsatellite length shortening and a few base substitutions in the coding region. However, when amplifying across genera in the Rosaceae, base substitutions and deletion events in the flanking region are more informative than the core motif repeat itself. In fact, SSR length variation as normally scored in microsatellite assays is inadequate to assess long-term evolutionary divergence between the Maloideae and the Prunoideae, and in general between relatively distinct taxa. Effectively, we can observe that in distinct species such as Malus × domestica and Pyrus communis, the microsatellite repeat is no longer present. Moreover, allelic similarities between Malus and Pyrus cultivars lead us to propose that those alleles do not belong to the same locus but instead arose either from gene duplication or allopolyploidisation. Segregation analysis in Malus × domestica showed two monomorphic distinct loci. After sequencing, each allelic size class was compared between Malus and Pyrus accessions; one single deletion showed relatedness between one and the other fragment across the subgenera Malus and Pyrus. Previous study in the subfamily Maloideae suggested that the Maloideae genome (x = 17) was derived from allopolyploidy between primitive members of the subfamilies Prunoideae (x = 8) and Spiroideae (x = 9) (Stebbins 1958). Additional loci have frequently been observed for example in wheat, where 29% of primer pairs amplified more than one locus (Bryan et al. 1999), and in apple, 25% (Guilford et al. 1997). This is probably reflecting chromosomal duplication or allopolyploidy in those species. In our case, one distinct locus additional to PacB22 was amplified as confirmed by segregation analysis.
Therefore, the variability recorded within the flanking sequence of the PacB22 locus is sufficient to distinguish between two loci in the same genome, either duplicated or located on two homoeologous chromosomes. This could be particularly useful for differentiating homoeologous chromosomes in allopolyploid species. It also shows that EST-derived markers (RFLP, SNP, SSCP) would represent markers of choice, compared to microsatellite markers, for genome comparisons across genera in the Rosaceae.
References
Arnold C, Rossetto M, McNally J, Henry RJ (2002) The application of SSRs characterized for grape (Vitis vinifera) to conservation studies in Vitaceae. Am J Bot 89:22–28
Aubourg S, Rouzé P (2001) Genome annotation. Plant Physiol Biochem 39:181–193
Ayres NM, McClung AM, Larkin PD, Bligh HFJ, Jones CA, Park WD (1997) Microsatellites and a single-nucleotide polymorphism differentiate apparent amylose classes in an extended pedigree of US rice germ plasm. Theor Appl Genet 94:773–781
Becker J, Heun M (1995) Barley microsatellites: allele variation and mapping. Plant Mol Biol 27:835–845
Bryan GJ, Stephenson P, Collins A, Kirby J, Smith JB, Gale MD (1999) Low levels of DNA sequence variation among adapted genotypes of hexaploid wheat. Theor Appl Genet 99:192–198
Butcher PA, Decroocq S, Gray Y, Moran GF (2000) Development, inheritance and cross-species amplification of microsatellite markers from Acacia mangium. Theor Appl Genet 101:1282–1290
Cipriani G, Lot G, Huang WG, Marrazzo MT, Peterlunger E, Testolin R (1999) AC/GT and AG/CT microsatellite repeats in peach [Prunus persica (L) Batsch]: isolation, characterization and cross-species amplification in Prunus. Theor Appl Genet 99:65–72
Chalhoub BA, Thibault S, Laucou V, Rameau C, Höfte H, Cousin R (1997) Silver staining and recovery of AFLP amplification products on large denaturing polyacrylamide gels. BioTechniques 22:216–220
Chang S, Puryear J, Cairney J (1993) A simple and efficient method for isolating RNA from Pine trees. Plant Mol Biol Rep 11:113–116
Di Gaspero G, Peterlunger E, Testolin R, Edwards KJ, Cipriani G (2000) Conservation of microsatellite loci within the genus Vitis. Theor Appl Genet 101:301–308
Dirlewanger E, Cosson P, Tavaud M, Aranzana MJ, Poizat C, Zanetto A, Arús P, Laigret F (2002) Development of microsatellite markers in peach [Prunus persica (L.) Batsch] and their use in genetic diversity analysis in peach and sweet cherry (Prunus avium L.). Theor Appl Genet 105:127–138
Domon E, Fuijita M, Ishikawa N (2002) The insertion/deletion polymorphisms in the waxy gene of barley genetic resources from East Asia. Theor Appl Genet 104:132–138
Downey SL, Iezzoni AF (2000) Polymorphic DNA Markers in Black Cherry (Prunus serotina) are identified using sequences from Sweet Cherry, Peach and Sour Cherry. J Am Soc Hort Sci 125:76–80
Edwards KJ, Barker JHA, Daly A, Jones C, Karp A (1996) Microsatellite libraries enriched for several microsatellite sequences in plants. BioTechniques 20:758–760
Erpelding JE, Blake NK, Blake TK, Talbert LE (1996) Transfer of sequence tagged site PCR markers between wheat and barley. Genome 39:802–810
FitzSimmons NN, Moritz C, Moore SS (1995) Conservation and dynamics of microsatellite loci over 300 million years of marine turtle evolution. Mol Biol Evol 12:432–440
Galet P (1967) Recherches sur les méthodes d'identification et de classification des Vitacées des zones tempérées. PhD dissertation, Université de Montpellier, Faculté des Sciences, Montpellier, France
Grimaldi MC, Crouau-Roy B (1997) Microsatellite allelic homoplasy due to variable flanking sequences. J Mol Evol 44:336–340
Guilford P, Prakash S, Zhu JM, Rikkerink E, Gardiner S, Bassett H, Forster R (1997) Microsatellites in Malus × domestica (apple): abundance, polymorphism and cultivar identification. Theor Appl Genet 94:249–254
Karhu A, Dieterich JH, Savolainen O (2000) Rapid expansion of microsatellite sequences in pines. Mol Biol Evol 17:259–265
Lefort F, Douglas GC (1999) An efficient micro-method of DNA isolation from mature leaves of four hardwood tree species Acer, Fraxinus, Prunus and Quercus. Ann For Sci 56:259–263
Milbourne D, Meyer RC, Collins AJ, Ramsay LD, Gebhardt C, Waugh R (1998) Isolation, characterisation and mapping of simple sequence repeat loci in potato. Mol Genet Genomics 259:233–245
Orti G, Pearse DE, Avise JC (1997) Phylogenetic assessment of length variation at a microsatellite locus. Proc Natl Acad Sci USA 94:10,745–10,749
Peakall R, Gilmore S, Keys W, Morgante M, Rafalski A (1998) Cross species amplification of Soybean (Glycine max) simple sequence repeats (SSRs) within the genus and other legume genera: Implications for the transferability of SSRs in plants. Mol Biol Evol 15:1275–1287
Powell W, Gordon CM, Provan J (1996) Polymorphism revealed by simple sequence repeats. Trends Plant Sci 1:215–222
Rehder A (1949) Bibliography of cultivated trees and shrubs hardy in the cooler temperate regions of the Northern hemisphere. The Arnold Arboretum of Harvard University, Jamaica Plain, Massachussets, USA
Rossetto M (2001) Sourcing of SSR markers from related plant species. In: Henry R (ed) Plant genotyping – the DNA fingerprinting of plants. CAB International, Wallingford, UK, pp 211–224
Rossetto M, McNally J, Henry RJ (2002) Evaluating the potential of SSR flanking regions for examining taxonomic relationships in the Vitaceae. Theor Appl Genet 104:61–66
Sosinski B, Gannavarapu M, Hager LD, Beck LE, King GJ, Ryder CD, Rajapakse S, Baird WV, Ballard RE, Abbott AG (2000) Characterisation of microsatellite markers in peach [Prunus persica (L) Batsch]. Theor Appl Genet 101:421–428
Stebbins GL (1958) On the hybrid origins of the Angiosperms. Evol 12:267–270
Thomas MR, Scott NS (1993) Microsatellite repeats in grapevine reveal DNA polymorphisms when analysed as sequence-tagged sites (STSs). Theor Appl Genet 86:985–990
Washington JM, Box A, Karakousis A, Barr AR (2000) Developing waxy barley cultivars for food, feed and malt. In: Logue S (ed) Barley genetics VIII. Adelaide University, Adelaide, 3:303–305
Westman AL, Kresovich S (1998) The potential for cross-taxa simple-sequence repeat (SSR) amplification between Arabidopsis thaliana L. and crop brassicas. Theor Appl Genet 96:272–281
Whitton J, Rieseberg LH, Ungerer MC (1997) Microsatellite loci are not conserved across the Asteraceae. Theor Appl Genet 101:1282–1290
Yamamoto T, Kimura T, Sawamura Y, Kotobuki K, Ban Y, Hayashi T, Matsuta N (2001) SSRs isolated from apple can identify polymorphism and genetic diversity in pear. Theor Appl Genet 102:865–870
Acknowledgements.
We thank Drs. C.-E. Durel and M. Tavaud for providing specimens or DNA of Malus × domestica and P. avium respectively, as well as Dr. F. Laigret for performing some of the sequencing reactions. The P. communis and P. veitchii accessions were kindly provided by Dr. Davies from the HRI, East Malling (UK), and maintained in in vitro culture thanks to M. Lansac and J.-P. Eyquard. We acknowledge excellent care of the plants by A. Bonnet, C. Hevin, B. Douens, J.-P. Petit and J.-P. Robert. The authors thank Drs. G. Moran and P. Butcher (CSIRO Forestry and Forest Products, Australia) for critically reading the manuscript. Our study was supported by grants from the Aquitaine region (CCRDT20000307010 and CCRDT20010307007) and from the CIVB (Comité Interprofessionnel des Vins de Bordeaux #23611/23612).
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by C. Möllers
Rights and permissions
About this article
Cite this article
Decroocq, V., Favé, M.G., Hagen, L. et al. Development and transferability of apricot and grape EST microsatellite markers across taxa. Theor Appl Genet 106, 912–922 (2003). https://doi.org/10.1007/s00122-002-1158-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00122-002-1158-z