Abstract
The androgen receptor (AR) is a transcriptional factor that has a pivotal role in the development of normal and also cancerous prostate. Therefore, analyzing AR signaling is essential to understand cancerogensis and proliferation of prostate cancer (PCa). Inhibitor of growth 1 (ING1) and ING2 are tumor suppressors with reduced expression in many cancer types. There are also indications of misregulation of ING1 and ING2 in PCa. However, the roles of ING1 and ING2 in PCa and AR signaling are poorly understood. Here, we show that surprisingly the ING1b knockdown (KD) represses AR-mediated transactivation on AR key target genes in the human LNCaP PCa cells. This is associated with growth reduction of LNCaP cells by ING1 KD. In line with this, using Ing1 knockout (KO) mice, we provide further evidence that ING1 deficiency downregulates prostate-specific AR target genes in vivo. Further analyses suggest that KD of ING1b results in induction of both cellular senescence and the cell cycle inhibitor p16 INK4a. The unexpected finding that the ING1 KD results in growth inhibition was further analyzed and can be explained by a compensatory mechanism through enhanced levels of ING2 protein in ING1-deficient condition. Accordingly, the data suggest that ING2 interacts with AR and hampers the AR transcriptional activation, causes growth arrest, and induces cellular senescence. The data further suggest that ING2 upregulates p16 INK4a, which is a novel target for ING2. Taken together, our data suggest that ING2 is a novel corepressor for AR. ING2 levels are increased upon downregulation of ING1 expression indicating a compensatory mechanism and suggests a novel crosstalk between ING1 and ING2 tumor suppressors to inhibit AR signaling and induce cellular senescence in PCa cells.
Key message
• The tumor suppressors ING1 and 2 are dysregulated in human prostate cancer.
• ING1 deficiency reduces AR-mediated gene expression in vitro and in vivo.
• ING2, like ING1, inhibits AR-mediated transactivation and prostate cancer cell growth.
• ING1 regulates ING2.
• ING1 and ING2 crosstalk with each other to inhibit AR signaling in prostate cancer.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Prostate cancer (PCa) is the most common diagnosed cancer and the second leading cause of cancer mortality for males in many western countries [1]. While age is a major risk factor for this disease, hormone levels might also contribute to growth of the tumor. Androgens play an important role by regulating the growth of both normal prostate and PCa via the ligand-controlled transcription factor androgen receptor (AR) [2], which is a member of the nuclear hormone receptor superfamily. Therefore, AR represents a major drug target in the treatment of PCa.
The AR signaling is activated by the binding of its ligands either androgens (AR agonists) or antiandrogens (AR antagonists) to the receptor, leading to AR translocation into the nucleus where it binds to androgen response element (ARE) on the promoter of its target genes [3]. Upon binding to ARE, the liganded AR recruits coregulators which as a complex regulate the transcription of AR target genes [4]. The recruitment of coactivators leads to transactivation, while the recruitment of corepressors results in inhibition of AR target gene expression. The genes regulated by AR are important for the development and physiological function of the normal prostate as well as for the proliferation of cancerous prostate tissue. The AR signaling does not only play a role in transcriptional regulation but can also direct the fate of PCa. The effect of androgens and the AR on the invasion and proliferation of PCa cells have been well established [5, 6]. On the other hand, AR antagonists and the AR have also been reported to be involved in the induction of apoptosis and cellular senescence of PCa cells [7, 8]. However, the possibility to inhibit the androgen-activated AR by cellular tumor suppressors that act as AR corepressors is not fully understood.
The inhibitor of growth (ING) family of proteins represents a tumor suppressor family. Among the family members, ING1 and ING2 have been studied as a subgroup due to their highly conserved amino acid sequences and similarity in tumor suppressive functions [9]. ING1b is the most widely expressed isoform of ING1, while ING2a is the major isoform of ING2. Their tumor suppressor functions have been concerned with many biological processes including gene transcription, cell growth control, apoptosis, and cellular senescence [10, 11]. Their role as tumor suppressors is supported by the fact that their expression is decreased or lost in many human tumors [12]. Moreover, both Ing1 and Ing2 knockout (KO) mice were reported to promote tumor development [13–15]. However, analyzing the available microarray gene expression data from Oncomine database [16] has revealed that ING1 or ING2 messenger RNA (mRNA) is controversially either upregulated or downregulated in PCa compared to normal prostate gland [17–21]. One study has revealed an inverse expression of ING1 and ING2 in human PCa specimens, correlating a reduced ING1 expression with an increase of ING2 level [18].
A recent study of our research group reports that ING1b acts as a novel corepressor for AR [22]. ING1b was found to physically interact with AR both in vitro and in vivo. The overexpression of ING1b resulted in an inhibition of AR-mediated transactivation and the growth of PCa cells, while the primary mouse embryonic fibroblasts (MEFs) from ING1b KO mouse [13] exhibited enhanced AR activity [22].
Surprisingly, knockdown (KD) of ING1b also resulted in reduction of AR signaling and inhibition of PCa cell growth. Similar results were also detected for prostate-specific AR target genes in Ing1 KO mice. Therefore, we were wondering whether the ING1b-related factor, ING2, might compensate for ING1 and also interferes with AR signaling in ING1-deficient condition. The data suggest that ING2 expression is increased in both the complete Ing1 KO mice prostate and the ING1b KD LNCaP cells. Also, we show that ING2 represses AR-mediated transactivation, which could be the underlying molecular mechanism of repression of AR-mediated transactivation in the genetic Ing1 KO or ING1b KD backgrounds. It further indicates that ING2 is also a corepressor for the AR. Accordingly, the expression of ING2a downregulates ING1b, which indicates a crosstalk between the two tumor suppressors. Thus, we provide a novel crosstalk between ING1 and ING2 and a compensatory mechanism in regulating AR-mediated transactivation.
Materials and methods
Cell culture and retroviral transduction
MEFs [14] isolated from 15.5-day embryos were cultured in DMEM supplemented with 10 % fetal bovine serum (FBS), 1 % penicillin/streptomycin, and 25 mM HEPES (pH 7.8). The androgen-dependent human PCa cell line LNCaP [23] was cultured in RPMI1640 supplemented with 10 % FBS, 1 % penicillin/streptomycin, 25 mM HEPES (pH 7.8), and 1 % sodium pyruvate. Castration-resistant prostate cancer (CRPCa) cells C4-2, PC3, and PC3-AR were cultured as described earlier [22]. Phoenix-Eco, which is a retroviral packaging cell line for the generation of helper-free ecotropic retroviruses, was cultured in the same medium as MEFs, except that FBS was 10 % heat inactivated. All cells were cultivated in a humidified atmosphere with 5 % CO2 at 37 °C. The cells were cultured in charcoal-stripped serum (CSS) containing media when hormone depletion was needed.
Retroviral gene transfer into LNCaP cells was performed essentially as described earlier [22]. In brief, for virus production, Phoenix-Eco cells are transiently transfected with the retroviral vector of interest using the CaPO4 method. After 48 h, the medium containing viral particles was filtered, diluted with fresh medium in the presence of polybrene, and added to the human PCa cells, which were expressing the ecotropic receptor for the ecotropic virus entry. Infected cells were selected with puromycin for 10–15 days for successful transduction. Selected cells were then pooled for the assays.
Growth assays
LNCaP cells were seeded on the 6-well tissue culture plates in appropriate medium. Then, the retroviral transduction was applied to the cells. After 48 h, the medium was replaced with the ones containing 5 % FBS and different AR ligands. After a further cultivation of 48 h, antibiotic selection of the transduced cells in the presence or absence of AR ligands was performed for 10–15 days. The resistant cells were fixed with 1 % glutaraldehyde, stained with 0.1 % crystal violet as an indirect measure of cell number, and the cell-associated crystal violet in dried plates was solubilized with Sørenson’s solution as described [24]. Absorbance was measured at 590 nm. Growth curves were plotted as previously explained [22].
Scratch assays
The scratch assays were performed according to Jansson et al. [24]. The images acquired were further analyzed quantitatively by using TScratch computing software [25]. Cell migration is presented as percent gap closure, calculated using the following formula:
Senescence-associated beta-galactosidase staining
The staining was performed as described previously [26, 27] directly on the transduced cells survived from antibiotic selection. The percentage of stained cells was determined following counting at least 400 cells per well under light microscopy.
Dichlorofluorescein assays
For detection of reactive oxygen species (ROS) levels, dichlorofluorescein (DCF) assays were used according to Lorenz et al. [28]. The fluorescence was measured in Fluoroskan Ascent (Labsystems, Helsinki, Finland) at excitation wavelength 485 nm and emission filter 538 nm. The values were normalized to the protein concentrations. The normalized values were depicted as fold ROS level after setting the values obtained for the control cells arbitrarily to 1.
Reporter gene assay
The reporter gene assays were performed as described earlier [22].
Quantitative reverse transcription PCR
RNA was isolated from the cells and mice organs using peqGOLD TriFast (Peqlab, Erlangen, Germany) according to the manufacturer’s protocol. Two-step quantitative reverse transcription PCR (qRT-PCR) was conducted using High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems, Foster City, CA, USA) and the SsoFast™ EvaGreen® Supermix (Bio‐Rad, München, Germany), gene-specific primers, and Bio-Rad CFX96™ Real-Time PCR detection system. qRT-PCR results were analyzed via ΔΔCt method [29] using CFX manager software from Bio-Rad. The primer sequences are listed in Supplementary Tables S1 and S2.
Antibodies and Western blot analyses
Preparation of whole-cell lysates and Western blotting were performed as described elsewhere [22]. Protein extraction from mice organs was performed on the organic phase remaining after RNA isolation with TriFast according to the manufacturer’s protocol. Protein pellet was dissolved in 1 % SDS by incubating it at 50 °C. Insoluble material was then removed by centrifugation (12,000 rpm, 10 min, 4 °C). The protein supernatant was transferred to a fresh tube for further analysis. The primary antibodies used for immunodetection were AR (Biogenex, 256M), ING1b (BD Biosciences, 550455), ING1 all isoforms (Upstate, 05-720 and Santa Cruz, sc-374295), ING2a (Proteintech, 11560-1-AP), ING3 (ABclonal, A5832), ING4 (ABclonal, A5833), ING5 (ABclonal, A7288), p21 (Cell Signaling, 2946), p27 (Santa Cruz, sc-1641), PARP (Cell Signaling, 9546), and α-tubulin (Abcam, ab15246). As secondary antibody, horseradish peroxidase-conjugated anti-mouse IgG (Santa Cruz, sc‐2005) or anti-rabbit IgG (Santa Cruz, sc‐2370) were used. LabImage 1D software (Kapelan Bio Imaging Solutions, Leipzig, Germany) was applied for quantification of protein of interest relative to the loading control (α-tubulin). In order to dephosphorylate the whole-cell lysate, 50 μg of protein extract was incubated with 2 U of the FastAP alkaline phosphatase (Thermo Fisher Scientific, MA, USA) at 37 °C for 3 h.
Co-immunoprecipitation
The co-immunoprecipitation (Co-IP) experiments were carried out as described previously [22]. Briefly, a specific antibody against ING2a (Santa Cruz, sc-67646) or normal goat IgG (negative control) was bound on protein G magnetic beads. Whole-cell extract from LNCaP-ING2a cells was incubated with the antibody-loaded beads for 2 h at 4 °C. After washing, the beads were resuspended in SDS buffer, boiled at 99 °C. Then, SDS buffer containing precipitated protein complex was subjected to 10 % SDS-PAGE for detection by Western blotting.
Statistical analysis
Two-tailed unpaired Student’s t test was performed for differential comparison between two groups using GraphPad Prism software. A value of p < 0.05 was considered as statistically significant.
Results
Ing1 KO and ING1b KD downregulate endogenous AR target genes in vivo and in cell culture
Our previous findings have revealed that ING1b acts as a novel corepressor for AR and represses the AR-mediated transcription [22]. To investigate how ING1 affect the AR activity in vivo, male Ing1 KO mice were used [14]. This mouse strain, which shares quite similar phenotype with the isoform-specific ING1b KO mice [13], is characterized by reduced size, hypersensitivity to radiation, and elevated incidence of lymphomas. However, there are no other obvious morphological, physiological, or behavioral abnormalities in Ing1 KO mouse, indicating that Ing1 function is dispensable for the viability of mice under normal physiological conditions [14].
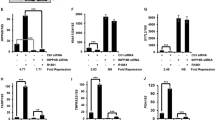
qRT-PCR analysis of various organ-specific AR target genes [30–37] indicates unexpectedly that mRNA expression of Pbsn, Nkx3.1, and Fkbp5 are downregulated in the prostate and seminal vesicles of male Ing1 KO mice (Fig. 1a). Interestingly, this downregulation has not been detected for key AR target gene expression in testis, liver, or kidney (Fig. 1b–d). In line with the data of prostate, Ing1 KO MEFs show also downregulation of key AR target genes in response to androgens (Fig. 1e).
Ing1 KO reduces AR-induced gene expression in vivo. a–d Different organs from wild type and Ing1 KO male mice (n = 3 mice per group, 6 months old) were analyzed for organ-specific AR target genes by qRT-PCR. Rpl13a and Hprt were used as housekeeping genes for normalization. The normalized mRNA levels are shown as fold levels after setting the wild-type values arbitrarily at 1. Error bars indicate SEM. e MEFs from wild type and Ing1 KO male mice were cultured in 10 % CSS medium and treated 48 h with solvent control or the synthetic androgen methyltrienolone (R1881, 0.1 nM) in hormone-depleted condition. Thereafter, RNA was isolated, and qRT-PCR analysis was conducted using specific primers for mouse AR target genes Tnf-α, Nkx3.1, and Sox9. Rpl13a was used for normalization. The normalized mRNA levels are shown as fold changes after setting the solvent values arbitrarily at 1. *p < 0.05
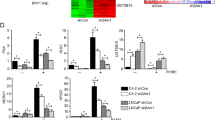
Since our previous data have indicated that the ING1b isoform-specific KO MEFs exhibit enhanced expression of AR target genes compared to wild-type MEFs, we knocked down ING1b in AR-expressing prostate cancer cell line LNCaP using retroviral transduction with the vectors expressing short hairpin RNA against ING1b (sh-ING1b). Functional ING1b KD was confirmed by Western blotting and also by downregulation of the ING1b target gene BCL2 [22] (Fig. 2a, b). In agreement with the data obtained for prostate and seminal vesicles of Ing1 KO mice, ING1b KD reduces androgen-induced gene expression of endogenous AR target genes in LNCaP cells (Fig. 2c). This inhibitory effect was confirmed by mRNA expression analysis of prostate-specific antigen (PSA) gene in response to different concentration of androgens (Fig. 2d). Similar results were obtained for PC3-AR cells (Fig. 2e, f).
ING1b KD inhibits AR-mediated gene expression. a Western blot analyses of the ING1b and α-tubulin proteins in LNCaP cells transduced with vector expressing short hairpin RNA either against firefly luciferase (sh-control) or against ING1b (sh-ING1b). b qRT-PCR analysis of ING1b target gene BCL2 for the cells explained in a. c qRT-PCR analysis of retrovirally transduced LNCaP cells. After 10-day antibiotic selection, the transduced cells were starved for 2 days in 10 % CSS medium; then, the cells treated with the solvent control or R1881 (1 nM) or the natural agonist dihydrotestosterone (DHT, 10 nM) in CCS medium. d qRT-PCR analysis of PSA in a similar condition explained in c treated with the indicated concentration of R1881. Error bars represent SEM. e Western blot analyses of the ING1b and α-tubulin proteins in PC3-AR cells transduced with the indicated vectors. f qRT-PCR analysis with PC3-AR cells in the same experimental setup as explained in c. *p < 0.05
Taken together, these findings indicate that ING1 deficiency downregulates AR target genes in vivo and in vitro. The data further suggest that ING1 has a role in regulating AR transcriptional activity in prostate and seminal vesicles in mice.
ING1b KD inhibits growth and migration and induces cellular senescence in LNCaP cells
Given the fact that the suppression of ING1b promotes neoplastic transformation [38] and ING1b-deficient mice develop B cell lymphoma [13], it was asked whether ING1b KD promotes growth in LNCaP cells. Unexpectedly, the results from growth assays of LNCaP cells under treatment of different AR ligands suggest that ING1b KD inhibits growth in these cells, although no synergistic effect was seen between ING1b KD and AR ligands (Fig. 3a, b).
ING1b KD results in inhibition of growth and migration and induces cellular senescence. a Growth assays for the LNCaP cells under treatment with solvent, low (L) and high (H) concentrations of R1881 (10 pM and 1 nM, respectively), DHT (10 nM), Casodex (Cas 1 μM), or atraric acid (AA 30 μM). The values obtained with the empty vector treated with solvent were set arbitrarily at 1. b Growth curves for the stably transduced LNCaP cells under treatment with the indicated AR ligands at different time points during cell proliferation assays. c Scratch assays to analyze migration of the stably transduced LNCaP cells. The same scratched regions were captured under the brightfield microscope at indicated time points. The scale bars indicate 300 μm. d Quantification of the gap closure of the scratch assays described in c. e SA-β-Gal staining was used as a marker of cellular senescence for transduced LNCaP cells. f mRNA expression of p14, p16, and E2F1 in the transduced LNCaP cells. The mean ± SEM values from at least two independent experiments are shown. *p < 0.05
To investigate the effect of ING1b KD on migratory behavior of LNCaP cells, scratch assays were conducted by using the stably transduced cells. Compared to the results seen with the control cells, ING1b KD reduces migration rate of LNCaP cells to heal the wounds created in the cell monolayers (Fig. 3c, d).
ING1b is involved in apoptosis through mitochondria [39] which are an important source of ROS within most mammalian cells [40]. Therefore, to disclose the underlying mechanisms for growth reduction upon ING1b KD, we initially analyzed the ROS production and apoptosis. The data exhibited neither an increase of ROS level nor the induction of apoptosis marker cleaved PARP in the ING1b KD LNCaP cells (Supplementary Fig. S1A, B). In line with this, the microscopic observations do not show any signs of increased apoptotic cells or cell detachment (data not shown).
Excluding ROS induction and apoptosis, ING1b KD cells were examined for induction of cellular senescence by conducting senescence-associated beta-galactosidase (SA-β-Gal) staining. Intriguingly, the data indicate the induction of cellular senescence in ING1b KD cells compared to the control ones (Fig. 3e). To get insights into the molecular pathway by which ING1b KD induces growth arrest and cellular senescence in LNCaP cells, the expression of cell cycle inhibitors p14 ARF, p16 INK4a, p21 CIP1/WAF1, and p27 KIP1 along with cell cycle mediator E2F1 was examined. qRT-PCR results indicated that ING1b KD upregulates p16 mRNA expression level in LNCaP cells, while no change for p14 mRNA level was detected (Fig. 3f). In line with this, E2F1 mRNA expression is downregulated in ING1b KD cells (Fig. 3f). However, the p21 expression was not influenced by ING1b KD in LNCaP cells (Supplementary Fig. S2A, B). Although ING1b KD does not affect the expression of p27, it abrogates androgen-induced stabilization of p27 protein in LNCaP cells (Supplementary Fig. S2C, D). This result underlines the role of ING1b as a mediator of androgen-induced p27 stabilization in LNCaP cells.
Taken together, these results suggest that ING1b KD inhibits LNCaP cell proliferation and migration. The data further suggest that ING1b KD induces cellular senescence and p16 and reduces the E2F1 mRNA level.
ING1b regulates the stability of ING2a protein
Contrary to our expectation, ING1b KD reduces the expression of key AR target genes and induces growth arrest and p16 expression. Since the AR protein level is not affected by ING1b KD in LNCaP cells (data not shown), one hypothesis could be that knockdown of ING1b leads to enhanced expression of a putative protein, which is responsible for suppressing AR target genes in ING1b KD cells. To examine this hypothesis, inhibition of the protein synthesis was employed. Intriguingly, the protein synthesis inhibitor cycloheximide reduced the ING1b KD repressive effect on TMPRSS2 mRNA level in response to androgens (data not shown) indicating that a putative factor is synthesized upon ING1 KD.
Since ING1 isoforms are differentially expressed during cellular senescence [41] and the ING1b KO mouse has displayed upregulation of ING1a and/or ING1c isoforms [13], it was asked whether ING1b deficiency in LNCaP cells is compensated by upregulation of other ING1 variants. Nevertheless, the results of Western blot analysis and protein quantification suggest no upregulation of other ING1 isoforms in LNCaP-ING1b KD cells (Fig. 4a).
ING1b regulates ING2a in LNCaP cells and mouse prostate and seminal vesicles. a, b Western blot analysis of ING1 isoforms and ING2a in LNCaP cells that were transduced with the indicated vectors and treated with solvent, R1881 (1 nM), or Casodex (Cas 1 μM). α-Tubulin served as a loading control. ING1a, ING1c, ING1d, and ING2a protein levels were normalized to α-tubulin and indicated. c Western blot analysis of ING2a in prostate and seminal vesicles of wild type and Ing1 KO mice (n = 3). The normalized ING2a protein values are indicated. The values underneath the brackets are shown as mean ± SEM of each mice group. LNCaP cells transduced with either ING2a expressing vector or empty vector were loaded as positive controls at left lanes. d Western blot analyses of the ING1b and ING2a proteins for the LNCaP cells in a similar condition explained in a, b with the indicated vectors: empty or ING1b expression vectors. e Western blot analyses of the ING1b and ING2a proteins for the PC3-AR cells using the same experimental setup as explained in a, b, and d
Since there are structural and functional overlaps among ING family members [42], it was reasoned to analyze the expression of ING2 with the highest amino acid sequence similarity to ING1 in ING1 KD cells to find compensatory feedback loop. Human ING2 has two known mRNA isoforms; however, only ING2a protein isoform with a molecular weight of 33 kDa has been experimentally detected at protein level. The amino acid sequence of human ING2a displays more than 70 % homology with ING1b, and it shares many of the functional characteristics of ING1b [10]. For analyzing ING2 protein and mRNA expression levels, both Western blot and qRT-PCR were performed in ING1b KD expressing LNCaP cells and compared with those of the corresponding control cells. Interestingly, ING1b KD in LNCaP cells upregulates ING2a protein level in a ligand-dependent manner (Fig. 4b).
To validate this result in vivo, ING2a protein expression was investigated in prostate and seminal vesicles of Ing1 KO mice as these organs showed the downregulation of AR target genes (Fig. 1a). The results display the upregulation of ING2a protein level in Ing1 KO mice (Fig. 4c). In line with this, ectopic expression of ING1b downregulates strongly ING2a protein in LNCaP cells (Fig. 4d). Very similarly, ING1b regulates ING2a protein level in PC3-AR cells (Fig. 4e). Moreover, an increased protein expression of ING2a was detected in CRPCa C4-2 and PC3 cells compared to androgen-dependent LNCaP cells (Supplementary Fig. S3), while ING1b protein expression is decreased in these CRPCa cells [22]. Accordingly, elevated mRNA level of ING2a in PC3 cells compared to LNCaP cells has been reported [43].
Notably, the protein levels of ING3, ING4, and ING5 are not altered by ING1b in LNCaP and PC3-AR cells (Supplementary Fig. S4A, B).
Thus, ING1b negatively regulates ING2a protein level suggesting a novel crosstalk between ING1 and ING2.
ING2a represses the AR-mediated transactivation and proliferation, and it induces cellular senescence and p16
To confirm whether the upregulation of ING2a is responsible for the reduced AR target gene expression in ING1 KD cells, the role of ING2a was investigated in LNCaP cells. To this end, LNCaP cells were stably transduced with ING2a or empty vector (Fig. 5a), and then, the mRNA expression of PSA, TEMPRSS2, and NKX3.1 was analyzed. qRT-PCR results suggest that ING2a represses the androgen-induced expression of endogenous AR-regulated genes (Fig. 5b).
ING2a inhibits both AR-mediated gene expression and LNCaP proliferation and induces cellular senescence. a Western blot analysis of ING2a in transduced LNCaP cells. b qRT-PCR analysis with the experimental conditions similar to Fig. 2c using the empty or ING2a expression vectors. c Crystal violet staining of LNCaP cell colonies after stable transduction. d Growth assays were performed similar to Fig. 3a with the indicated vectors. e Growth curves were plotted for the stably transduced LNCaP cells similar to Fig. 3b. f SA-β-Gal staining for transduced LNCaP cells. The scale bars indicate 100 μm. g Percentage of the SA-β-Gal-positive cells by counting minimum of 400 cells per cell type. h qRT-PCR results for p16 and E2F1 mRNA expression in transduced LNCaP cells. *p < 0.05
To explain the second upper band detected by Western blot analysis of ING2a protein, the phosphorylation of ING2a has been reported [44]. To test this post-translational modification of ING2a protein, the protein extracts from control and ING2a-overexpressed cells were incubated with or without the phosphatase. The results indicate that not only the overexpressed ING2a but also the endogenous ING2a proteins were phosphorylated in LNCaP cells (Supplementary Fig. S5).
To explain the inhibitory role of ING1b KD on the growth of LNCaP cells, proliferation assays were performed upon ectopic expression of ING2a. The results suggest that ING2a prevents the growth of LNCaP cells, although the fold reduction in growth is rather independent of the AR ligands (Fig. 5c–e). Interestingly, this growth arrest could be explained by induction of cellular senescence in LNCaP-ING2a cells (Fig. 5f, g), as it is observed for the ING1b KD cells (Fig. 3e). In line with this, apoptosis marker cleaved PARP is not induced by ING2a in LNCaP cells (Supplementary Fig. S6).
It has been previously reported that ING2a is able to induce p21 [45]. However, it seems that the p21 induction by ING2a is dependent on the cellular context because LNCaP-ING1 KD cells showed no detectable change for p21 expression, but for p16 (Fig. 3f and Supplementary Fig. S2A, B). Therefore, it was concluded that ING2a can induce p16 in LNCaP cells. Expectedly, qRT-PCR result of increased p16 levels in LNCaP-ING2a cells confirmed this hypothesis, and accordingly, the expression of the p16 downstream target E2F1 was downregulated (Fig. 5h).
To further investigate the crosstalk between ING1 and ING2, Western blot results suggest that ING2a overexpression indeed results in remarkable downregulation of ING1b protein level as expected (Fig. 6). These results further indicate that AR enhances ING2a protein level in response to androgen when compared to the solvent control treatment (Fig. 6).
ING2a downregulates ING1b protein level. Western blot analysis of ING1b and ING2a proteins as described similar to Fig. 4b
Thus, the data indicate that ING2a represses AR-mediated transactivation and it inhibits growth in LNCaP cells. Furthermore, the data suggest that ING2a induces cellular senescence and p16 in the same cellular context.
ING2a interacts with AR and inhibits AR-responsive promoters
Given the fact that ING2a represses the AR transcriptional activity (Fig. 5b), it was asked whether ING2a interacts with the AR and plays a corepressor role for AR. To answer this question, Co-IP assays were performed using the whole-cell lysate of LNCaP-ING2a cells (Fig. 7a). Intriguingly, AR protein was detected in the immunoprecipitates with anti-ING2 antibody. Of note, this interaction was induced in the presence of androgen. In the negative control, no AR was detected using a nonspecific antibody (Fig. 7a). To analyze functional role of ING2 on AR-responsive promoters, PC3-AR cells were transiently cotransfected with either empty vector or a vector expressing ING2a along with AR-responsive reporter constructs MMTV-luc or PSA-luc. The data suggest that ectopic expression of ING2a inhibits AR-mediated transactivation of both reporters in response to androgen treatment (Fig. 7b, c).
ING2a interacts with AR and inhibits androgen-induced promoters. a Co-IP experiments to analyze interaction between ING2a and AR. Whole-cell lysates of LNCaP-ING2a cells treated with solvent or R1881 (1 nM) for 2 h were immunoprecipitated with anti-ING2a (α-ING2a) and blotted for AR and ING2a. Goat IgG was used as an antibody control. b Reporter gene assays were performed with PC3-AR cells. The cells were seeded out in 10 % CSS medium and transiently cotransfected with 1 μg of either empty vector or vector expressing ING2a along with 1 μg reporter construct b MMTV-luc or c PSA-luc and pCMV‐lacZ (0.2 μg) which is used as an internal control for normalization. Sixteen hours later, the cells were treated with solvent control or R1881 (1 nM) for 3 days in 10 % CSS medium. Cells were then lysed, and the measured luciferase values were normalized to β-galactosidase activity. The normalized luciferase values are shown as fold induction after setting the solvent values arbitrarily at 1. *p < 0.05
Thus, the results suggest ING2 as an interacting partner of AR and an inhibitor of AR-responsive promoters.
In sum, ING1b KD inhibits growth, migration, and AR-mediated transcriptional activation on key target genes, which is in accordance with the in vivo results from Ing1 KO mice. Since ING1b is known as a tumor suppressor and AR corepressor [10, 22], the obtained results were unexpected. Analyzing potential mechanisms suggests that these unexpected findings might be due to a compensatory mechanism through upregulation of ING2a protein. The data indicate that ING2a can also inhibit growth and key target genes of AR and induces cellular senescence and p16. This compensatory mechanism indicates that ING1 expression regulates ING2 expression suggesting a novel crosstalk among ING1 and ING2 in regulating various AR functions. Thus, the obtained findings provide evidence for functional interactions among AR, ING1, and ING2.
Discussion
Since AR is the main drug target for PCa therapy, understanding AR signaling will shed light into PCa carcinogenesis. So far known, the AR exerts its biological functions mainly by regulating the transcription of its target genes through interaction with and recruitment of various factors including coactivators or corepressors [4, 46]. Our previous findings indicate that the tumor suppressor ING1b is a novel androgen-activated AR corepressor [22]. As ING1b downregulates the AR-mediated transcription in response to androgens, our prediction was that the inhibition of endogenous ING1b should upregulate the AR transactivation. In contrast, the data suggest unexpectedly that agonist-stimulated gene expression was repressed upon knockdown of ING1b in LNCaP cells. In line with this, the prostate-specific AR target genes have been also revealed to be downregulated in Ing1 KO mice. Moreover, LNCaP-ING1 KD cells grow and migrate less than control LNCaP cells. Interestingly, Ing1 KO mice have reduced size in comparison to the wild-type one [14]. Of note, this reduction in body weight is two times more in males than females. Although analyzing young (6 weeks old) Ing1 KO mice has revealed no abnormalities with fertility and prostate phenotype [14], based on the obtained results at 6-month-old Ing1 KO mice, the fertility and prostate phenotype need to be rechecked in detail with older Ing1 KO mice. Induction of cellular senescence and p16 could explain the growth reduction in LNCaP-ING1b KD cells.
By analyzing possible underlying mechanisms for these unexpected results, the data suggest that ING2 is negatively regulated by ING1 in vitro and in vivo. Very interestingly, PCa specimens with reduced ING1 expression indicate higher level of ING2 [18]. Our data might explain this phenomenon by that ING1 regulates the expression of ING2. A downregulation of ING1 can lead to an upregulation of ING2. ING2, like ING1b, is a tumor suppressor and shares high amino acid sequence homology with ING1b. Further, we show that ING2 has tumor suppressive and AR corepressor function in PCa cells, which compensates for the deficiency of ING1. Nevertheless, they are exclusive components of the SIN3A/HDAC1-2/SAP30 complex in repressing the gene transcription [10]. Our findings indicate that ING2 is the possible mediator by which AR-induced transactivation is inhibited in ING1b KD or Ing1 KO contexts. ING2, like ING1b, has been found to associate with corepressor alien and enhance alien-induced gene silencing mediated by selected members of nuclear hormone receptors and E2F1 [47].
In addition, ING2 induces p16 in LNCaP cells, which explain the enhanced level of p16 in LNCaP-ING1 KD cells with elevated ING2 expression. This finding along with the indicated ING2-mediated cellular senescence in human PCa cells might be the molecular reasons for the increase of SA-β-Gal activity in LNCaP-ING1b KD cells. The regulatory function of ING2 to induce cellular senescence has been described in human primary cells [48]. Our finding is the first report regarding the induction of cellular senescence by ING2 in PCa cells. It is of very interest that recently the relevance of cellular senescence as a physiological barrier against tumor initiation and progression is well established [49–51]. However, the mechanism by which ING2 causes cell growth inhibition by either cellular senescence or apoptosis seems dependent on the cellular context as discussed for ING1 [22, 52, 53].
Apart from reduced size and higher incidence of lymphomas, Ing1 KO mice exhibit no other obvious morphological, physiological, or behavioral abnormalities, indicating that Ing1 function is dispensable for the viability under normal physiological conditions [14] and Ing2 might play a compensatory role in Ing1-depleted condition. In spite of high homology between ING1 and ING2, a different phenotype has been observed in their corresponding KO mice models [13–15]. Ing2 KO mice are characterized by defective spermatogenesis in males and higher incidence of soft tissue sarcomas [15]. Interestingly, the major tumor type observed in Ing2 KO mice was histiocytic sarcoma which showed increased incidence preferentially in males. Moreover, Ing2 KO mice have displayed a decrease in acinar dilation in prostate. Also, analysis of the publicly available dataset (GSE18610 at http://www.ncbi.nlm.nih.gov/geo/) from Ing2 KO mice (2–3 months old) [15] has shown the significantly increased expression of testis-specific AR target genes Rhox5 and Sox9 in testes of Ing2 KO mice compared to wild-type ones. All these evidences support a putative role for ING2 in crosstalk with AR in male mice.
Homologs of ING1 and ING2 are found also in insects, nematodes, and plants suggesting a very important biological role. Therefore, the relatively mild phenotype in Ing1 KO mice was surprising. One possibility of the mild phenotype might be that the reduction of ING1 is compensated by an increased expression of the ING2 tumor suppressor family member.
Interestingly, overexpression of ING1b strongly downregulates ING2a protein level; however, ING2a mRNA level remains unchanged (data not shown) suggesting that ING1 does not affect the ING2 promoter activity. In addition, the administration of the proteasome inhibitor MG132 does not lead to an increase of ING2a protein level in ING1b-overexpressed cells (data not shown) suggesting that proteasomal degradation is not involved to reduce ING2 protein levels. Therefore, we hypothesize that ING1-ING2 crosstalk is mediated at post-transcriptional level. Several possibilities might exist, e.g., directly through inhibiting ING2 translation by ING1 or alternatively indirectly through induction of miRNAs by ING1 that inhibit ING2 translation. Although overexpression of ING2a strongly downregulates the ING1b protein level, ING2a KD decreases unexpectedly ING1b protein as well, but interestingly not the ING1b mRNA level (data not shown) suggesting that ING1-ING2 crosstalk is not a simple vice versa mechanism rather involving other factors. Thus, we speculate that an optimum level of ING2a is required to maintain the levels of ING1b and vice versa. In case of an imbalance, the translation machinery but not the transcriptional machinery might be used to regulate the levels of the other ING factor.
Analyzing the expression of ING1 or ING2 in human PCa samples for use as possible tumor or prognostic markers indicated contrary results showing either upregulation or downregulation of their expression levels [17–21]. Considering the here identified crosstalk between ING1 and ING2, we suggest that it might be useful to detect the expression levels of both ING factors within the same PCa specimen to obtain a better correlation with PCa.
Taken together, our findings provide evidence for a novel crosstalk and a crossregulation between ING1 and ING2 in regulating AR-mediated transactivation and suggest that ING2 acts as a novel corepressor that inhibits AR signaling, PCa cell growth, and migration. The data suggest that cellular senescence is the possible underlying mechanism for ING2-mediated growth repression in PCa cells.
References
Siegel RL, Miller KD, Jemal A (2015) Cancer statistics, 2015. CA Cancer J Clin 65(1):5–29
Dehm SM, Tindall DJ (2007) Androgen receptor structural and functional elements: role and regulation in prostate cancer. Mol Endocrinol 21(12):2855–2863
Claessens F, Verrijdt G, Schoenmakers E, Haelens A, Peeters B, Verhoeven G, Rombauts W (2001) Selective DNA binding by the androgen receptor as a mechanism for hormone-specific gene regulation. J Steroid Biochem Mol Biol 76(1-5):23–30
Baniahmad A (2005) Nuclear hormone receptor co-repressors. J Steroid Biochem Mol Biol 93(2-5):89–97
Jenster G (1999) The role of the androgen receptor in the development and progression of prostate cancer. Semin Oncol 26(4):407–421
Xu Y, Chen SY, Ross KN, Balk SP (2006) Androgens induce prostate cancer cell proliferation through mammalian target of rapamycin activation and post-transcriptional increases in cyclin D proteins. Cancer Res 66(15):7783–7792
Hessenkemper W, Roediger J, Bartsch S, Houtsmuller AB, van Royen ME, Petersen I, Grimm MO, Baniahmad A (2014) A natural androgen receptor antagonist induces cellular senescence in prostate cancer cells. Mol Endocrinol 28(11):1831–1840
Lee EC, Zhan P, Schallhom R, Packman K, Tenniswood M (2003) Antiandrogen-induced cell death in LNCaP human prostate cancer cells. Cell Death Differ 10(7):761–771
He GH, Helbing CC, Wagner MJ, Sensen CW, Riabowol K (2005) Phylogenetic analysis of the ING family of PHD finger proteins. Mol Biol Evol 22(1):104–116
Guerillon C, Larrieu D, Pedeux R (2013) ING1 and ING2: multifaceted tumor suppressor genes. Cell Mol Life Sci 70(20):3753–3772
Schafer A, Karaulanov E, Stapf U, Doderlein G, Niehrs C (2013) Ing1 functions in DNA demethylation by directing Gadd45a to H3K4me3. Genes Dev 27(3):261–273
Walzak AA, Veldhoen N, Feng X, Riabowol K, Helbing CC (2008) Expression profiles of mRNA transcript variants encoding the human inhibitor of growth tumor suppressor gene family in normal and neoplastic tissues. Exp Cell Res 314(2):273–285
Coles AH, Liang H, Zhu Z, Marfella CG, Kang J, Imbalzano AN, Jones SN (2007) Deletion of p37Ing1 in mice reveals a p53-independent role for Ing1 in the suppression of cell proliferation, apoptosis, and tumorigenesis. Cancer Res 67(5):2054–2061
Kichina JV, Zeremski M, Aris L, Gurova KV, Walker E, Franks R, Nikitin AY, Kiyokawa H, Gudkov AV (2006) Targeted disruption of the mouse ing1 locus results in reduced body size, hypersensitivity to radiation and elevated incidence of lymphomas. Oncogene 25(6):857–866
Saito M, Kumamoto K, Robles AI, Horikawa I, Furusato B, Okamura S, Goto A, Yamashita T, Nagashima M, Lee TL et al (2010) Targeted disruption of Ing2 results in defective spermatogenesis and development of soft-tissue sarcomas. PLoS One 5(11):e15541
Rhodes DR, Yu J, Shanker K, Deshpande N, Varambally R, Ghosh D, Barrette T, Pandey A, Chinnaiyan AM (2004) ONCOMINE: a cancer microarray database and integrated data-mining platform. Neoplasia 6(1):1–6
Lapointe J, Li C, Higgins JP, van de Rijn M, Bair E, Montgomery K, Ferrari M, Egevad L, Rayford W, Bergerheim U et al (2004) Gene expression profiling identifies clinically relevant subtypes of prostate cancer. Proc Natl Acad Sci U S A 101(3):811–816
Singh D, Febbo PG, Ross K, Jackson DG, Manola J, Ladd C, Tamayo P, Renshaw AA, D'Amico AV, Richie JP et al (2002) Gene expression correlates of clinical prostate cancer behavior. Cancer Cell 1(2):203–209
Tomlins SA, Mehra R, Rhodes DR, Cao X, Wang L, Dhanasekaran SM, Kalyana-Sundaram S, Wei JT, Rubin MA, Pienta KJ et al (2007) Integrative molecular concept modeling of prostate cancer progression. Nat Genet 39(1):41–51
Vanaja DK, Cheville JC, Iturria SJ, Young CY (2003) Transcriptional silencing of zinc finger protein 185 identified by expression profiling is associated with prostate cancer progression. Cancer Res 63(14):3877–3882
Varambally S, Yu J, Laxman B, Rhodes DR, Mehra R, Tomlins SA, Shah RB, Chandran U, Monzon FA, Becich MJ et al (2005) Integrative genomic and proteomic analysis of prostate cancer reveals signatures of metastatic progression. Cancer Cell 8(5):393–406
Esmaeili M, Jennek S, Ludwig S, Klitzsch A, Kraft F, Melle C, Baniahmad A (2016) The tumor suppressor ING1b is a novel corepressor for the androgen receptor and induces cellular senescence in prostate cancer cells. J Mol Cell Biol. doi:10.1093/jmcb/mjw007
Protopopov AI, Li J, Winberg G, Gizatullin RZ, Kashuba VI, Klein G, Zabarovsky ER (2002) Human cell lines engineered for tetracycline-regulated expression of tumor suppressor candidate genes from a frequently affected chromosomal region, 3p21. J Gene Med 4(4):397–406
Jansson KH, Lynch JE, Lepori-Bui N, Czymmek KJ, Duncan RL, Sikes RA (2012) Overexpression of the VSSC-associated CAM, beta-2, enhances LNCaP cell metastasis associated behavior. Prostate 72(10):1080–1092
Geback T, Schulz MM, Koumoutsakos P, Detmar M (2009) TScratch: a novel and simple software tool for automated analysis of monolayer wound healing assays. Biotechniques 46(4):265–274
Dimri GP, Lee X, Basile G, Acosta M, Scott G, Roskelley C, Medrano EE, Linskens M, Rubelj I, Pereira-Smith O et al (1995) A biomarker that identifies senescent human cells in culture and in aging skin in vivo. Proc Natl Acad Sci U S A 92(20):9363–9367
Roediger J, Hessenkemper W, Bartsch S, Manvelyan M, Huettner SS, Liehr T, Esmaeili M, Foller S, Petersen I, Grimm MO et al (2014) Supraphysiological androgen levels induce cellular senescence in human prostate cancer cells through the Src-Akt pathway. Mol Cancer 13:214
Lorenz V, Hessenkemper W, Rodiger J, Kyrylenko S, Kraft F, Baniahmad A (2011) Sodium butyrate induces cellular senescence in neuroblastoma and prostate cancer cells. Horm Mol Biol Clin Invest 7(1):265–272
Pfaffl MW (2001) A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res 29(9):e45
Felder MR, Watson G, Huff MO, Ceci JD (1988) Mechanism of induction of mouse kidney alcohol dehydrogenase by androgen. Androgen-induced stimulation of transcription of the Adh-1 gene. J Biol Chem 263(28):14531–14537
Ikeda H, Serria MS, Kakizaki I, Hatayama I, Satoh K, Tsuchida S, Muramatsu M, Nishi S, Sakai M (2002) Activation of mouse Pi-class glutathione S-transferase gene by Nrf2 (NF-E2-related factor 2) and androgen. Biochem J 364(Pt 2):563–570
Lund SD, Miller D, Chapman V, Ganschow RE (1988) Androgen regulation of murine beta-glucuronidase expression: identification and characterization of a nonresponse variant. Genetics 119(1):151–156
Magee JA, Chang LW, Stormo GD, Milbrandt J (2006) Direct, androgen receptor-mediated regulation of the FKBP5 gene via a distal enhancer element. Endocrinology 147(1):590–598
Melia MJ, Bofill N, Hubank M, Meseguer A (1998) Identification of androgen-regulated genes in mouse kidney by representational difference analysis and random arbitrarily primed polymerase chain reaction. Endocrinology 139(2):688–695
Rennie PS, Bruchovsky N, Leco KJ, Sheppard PC, McQueen SA, Cheng H, Snoek R, Hamel A, Bock ME, MacDonald BS et al (1993) Characterization of two cis-acting DNA elements involved in the androgen regulation of the probasin gene. Mol Endocrinol 7(1):23–36
Xu Q, Lin HY, Yeh SD, Yu IC, Wang RS, Chen YT, Zhang C, Altuwaijri S, Chen LM, Chuang KH et al (2007) Infertility with defective spermatogenesis and steroidogenesis in male mice lacking androgen receptor in Leydig cells. Endocrine 32(1):96–106
Zhou W, Wang G, Small CL, Liu Z, Weng CC, Yang L, Griswold MD, Meistrich ML (2011) Gene expression alterations by conditional knockout of androgen receptor in adult Sertoli cells of Utp14b jsd/jsd (jsd) mice. Biol Reprod 84(2):400–408
Garkavtsev I, Kazarov A, Gudkov A, Riabowol K (1996) Suppression of the novel growth inhibitor p33ING1 promotes neoplastic transformation. Nat Genet 14(4):415–420
Bose P, Thakur S, Thalappilly S, Ahn BY, Satpathy S, Feng X, Suzuki K, Kim SW, Riabowol K (2013) ING1 induces apoptosis through direct effects at the mitochondria. Cell Death Dis 4:e788
Adam-Vizi V, Chinopoulos C (2006) Bioenergetics and the formation of mitochondrial reactive oxygen species. Trends Pharmacol Sci 27(12):639–645
Soliman MA, Berardi P, Pastyryeva S, Bonnefin P, Feng X, Colina A, Young D, Riabowol K (2008) ING1a expression increases during replicative senescence and induces a senescent phenotype. Aging Cell 7(6):783–794
Coles AH, Jones SN (2009) The ING gene family in the regulation of cell growth and tumorigenesis. J Cell Physiol 218(1):45–57
Unoki M, Kumamoto K, Robles AI, Shen JC, Zheng ZM, Harris CC (2008) A novel ING2 isoform, ING2b, synergizes with ING2a to prevent cell cycle arrest and apoptosis. FEBS Lett 582(28):3868–3874
Ythier D, Larrieu D, Binet R, Binda O, Brambilla C, Gazzeri S, Pedeux R (2010) Sumoylation of ING2 regulates the transcription mediated by Sin3A. Oncogene 29(44):5946–5956
Larrieu D, Ythier D, Brambilla C, Pedeux R (2010) ING2 controls the G1 to S-phase transition by regulating p21 expression. Cell Cycle 9(19):3984–3990
Heinlein CA, Chang C (2001) Role of chaperones in nuclear translocation and transactivation of steroid receptors. Endocrine 14(2):143–149
Fegers I, Kob R, Eckey M, Schmidt O, Goeman F, Papaioannou M, Escher N, von Eggeling F, Melle C, Baniahmad A (2007) The tumor suppressors p33ING1 and p33ING2 interact with alien in vivo and enhance alien-mediated gene silencing. J Proteome Res 6(11):4182–4188
Pedeux R, Sengupta S, Shen JC, Demidov ON, Saito S, Onogi H, Kumamoto K, Wincovitch S, Garfield SH, McMenamin M et al (2005) ING2 regulates the onset of replicative senescence by induction of p300-dependent p53 acetylation. Mol Cell Biol 25(15):6639–6648
Collado M, Serrano M (2010) Senescence in tumours: evidence from mice and humans. Nat Rev Cancer 10(1):51–57
Nardella C, Clohessy JG, Alimonti A, Pandolfi PP (2011) Pro-senescence therapy for cancer treatment. Nat Rev Cancer 11(7):503–511
Prieur A, Peeper DS (2008) Cellular senescence in vivo: a barrier to tumorigenesis. Curr Opin Cell Biol 20(2):150–155
Han XR, Bai XZ, Sun Y, Yang Y (2014) Nuclear ING2 expression is reduced in osteosarcoma. Oncol Rep 32(5):1967–1972
Wang Y, Wang J, Li G (2006) Leucine zipper-like domain is required for tumor suppressor ING2-mediated nucleotide excision repair and apoptosis. FEBS Lett 580(16):3787–3793
Acknowledgments
We are grateful to Dr. Ignacio Palmero for providing the knockdown vector for ING1b. This work was supported by the German Academic Exchange Service (DAAD) to ME and German Cancer Aid to AB.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Electronic supplementary material
Below is the link to the electronic supplementary material.
ESM 1
(PDF 300 kb)
Rights and permissions
About this article
Cite this article
Esmaeili, M., Pungsrinont, T., Schaefer, A. et al. A novel crosstalk between the tumor suppressors ING1 and ING2 regulates androgen receptor signaling. J Mol Med 94, 1167–1179 (2016). https://doi.org/10.1007/s00109-016-1440-1
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00109-016-1440-1