Abstract
Mitochondrial glutaminase (GA) plays an essential role in cancer cell metabolism, contributing to biosynthesis, bioenergetics, and redox balance. Humans contain several GA isozymes encoded by the GLS and GLS2 genes, but the specific roles of each in cancer metabolism are still unclear. In this study, glioma SFxL and LN229 cells with silenced isoenzyme glutaminase KGA (encoded by GLS) showed lower survival ratios and a reduced GSH-dependent antioxidant capacity. These GLS-silenced cells also demonstrated induction of apoptosis indicated by enhanced annexin V binding capacity and caspase 3 activity. GLS silencing was associated with decreased mitochondrial membrane potential (ΔΨm) (JC-1 dye test), indicating that apoptosis was mediated by mitochondrial dysfunction. Similar observations were made in T98 glioma cells overexpressing glutaminase isoenzyme GAB, encoded by GLS2, though some characteristics (GSH/GSSG ratio) were different in the differently treated cell lines. Thus, control of GA isoenzyme expression may prove to be a key tool to alter both metabolic and oxidative stress in cancer therapy. Interestingly, reactive oxygen species (ROS) generation by treatment with oxidizing agents: arsenic trioxide or hydrogen peroxide, synergizes with either KGA silencing or GAB overexpression to suppress malignant properties of glioma cells, including the reduction of cellular motility. Of note, negative modulation of GLS isoforms or GAB overexpression evoked lower c-myc and bcl-2 expression, as well as higher pro-apoptotic bid expression. Combination of modulation of GA expression and treatment with oxidizing agents may become a therapeutic strategy for intractable cancers and provides a multi-angle evaluation system for anti-glioma pre-clinical investigations.
Key message
-
Silencing GLS or overexpressing GLS2 induces growth inhibition in glioma cell lines.
-
Inhibition is synergistically enhanced after arsenic trioxide (ATO) or H2O2 treatment.
-
Glutatione levels decrease in GLS-silenced cells but augment if GLS2 is overexpressed.
-
ROS synergistically inhibit cell migration by GLS silencing or GLS2 overexpression.
-
c-myc, bid, and bcl-2 mediate apoptosis resulting from GLS silencing or GLS2 overexpression.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Mammalian glutaminases (GA) catalyze the hydrolytic deamidation of glutamine to glutamate [1], which is further catabolized in the Krebs cycle for the production of ATP, nucleotides, certain amino acids, and lipids [2]. Although there is no doubt about the importance of GA in brain [3, 4], its key role in cancer [5, 6] and oxidative stress has been recently highlighted [7, 8]. In fact, several recent findings support the notion of GA as a multifaceted protein involved in other functions besides glutamate generation from glutamine [1, 5, 9]. Of outstanding interest, the presence of multiple GA transcripts has been demonstrated even in a single cancer cell type [10]. In humans, the GA family consists of two main members: the GLS gene encodes two isoforms, known as kidney (K-type) glutaminase or KGA, and glutaminase C or GAC [4]. These two isoenzymes are usually referred to as GLS [8]. On the other hand, the GLS2 gene codes for liver (L-type) isozymes, named LGA, as well as for a longer isoform named GAB [11, 12]. Both isoenzymes are designated collectively as GLS2 [8]. GLS has been demonstrated to be regulated by oncogenes and to support tumor cell growth [5, 6, 9, 13, 14]. In contrast, GLS2 is a transcriptional target of p53 and has been argued to have tumor suppressor properties, and re-expressing it in p53-deficient cells limits malignancy [7, 15]. It is unclear why these isozymes have such different biological properties despite their ability to catalyse the same enzymatic reaction.
The process of oncogenic transformation places energy demands on cancer cells to support proliferation, expansion, and invasion [16, 17]. Deregulated tumor metabolism is a critical part of oncogenesis and may be targeted for therapeutic benefits [6], including glioma treatment [18]. Both glutamine addiction and GLS activity are critical for c-myc-mediated oncogenesis [9, 14, 19, 20]; they have been linked to glutamine synthetase requirement [17], and proposed as attractive targets for therapeutic intervention [5, 6, 8]. The Warburg effect (increased aerobic glycolysis) [16] and glutaminolysis (increased mitochondrial activity from glutamine catabolism) [5] are well-known hallmarks of cancer [21], and are accompanied by increased lactate production [22] as well as increased generation of reactive oxygen species (ROS) [23–25]. Arsenic trioxide (ATO) and hydrogen peroxide (H2O2) can depolarise mitochondrial membrane potential (ΔΨm), increase intracellular ROS production, and induce apoptosis [24]. Based on these observations, and on the fact that GLS has oncogenic properties [14], and GLS2 isoenzyme can be designed as a tumor suppressor [15] and a ROS regulator [7], we sought to compare and contrast the effects of GLS (KGA and GAC) silencing and GLS2 (GAB) overexpression on malignant properties of tumor cells, alone and when combined with oxidative stress. We used glioma cells lines as a model, because several such cell lines have been shown to be glutamine dependent in culture and to use glutamine as a major substrate for anaplerosis and oxidative metabolism [13]. For GLS silencing experiments, we used the glioblastoma cell lines LN229 and SFxL. Both of these cell lines use glutamine as an anaplerotic precursor for the TCA cycle, and both display significant reductions in ammoniagenesis, cell proliferation, and tumor growth upon GLS silencing [13]. On the other hand, human glioblastoma T98G cell line expresses high amounts of GLS transcripts, while GLS2 transcripts are hardly detectable in these cells [12]. Interestingly, transfection of T98G cells with a GAB cDNA sequence diminished cell proliferation and survival [12].
Methods
Cell lines, culture conditions, stable transfections and RNA interference
All cell lines were tested for mycoplasma contamination. SFxL and LN229 cells were cultured in Dulbecco's modified eagle medium (DMEM) supplemented with 10 % fetal bovine serum (FBS), penicillin/streptomycin, and 6 mM L-glutamine as previously described [13]. All RNA interference (RNAi) experiments used pools of cells. Vectors for RNAi, lentiviral particles, and details have been described previously [13]. Of note, SFxL and LN229 control cells are expressing a non-targeting shRNA. Stably infected pools with adequate silencing were maintained in 1 μg/mL puromycin. In all stable knockdown experiments, very few detached cells were noted in the culture, and these were not included in growth and viability counts. T98G human glioblastoma cells were purchased from American Type Culture Collection and were maintained in minimum essential medium, supplemented with 10 % FBS, 1 % non-essential amino acids, 100 IU/mL penicillin and 100 μg/mL streptomycin, all supplied by Sigma-Aldrich, St. Louis, MO, USA. Cultures were maintained at 37 °C in a humidified atmosphere with 95 % air and 5 % CO2. T98G-GAB and T98G-pcDNA cell lines were obtained by stable transfection of T98G cells with a full cDNA sequence encoding human GAB or empty pcDNA3 vector, respectively, exactly as described previously [12]. The culture medium for the polyclonal populations of T98G-GAB and T98G-pcDNA cells containing the neomycin-resistance gene was supplemented with 0.5 mg/mL geneticin (Sigma-Aldrich). Relative baseline expression of GLS and GLS2 in all assayed cell lines shows that SFxL and LN229 GLS-silenced cell lines significantly diminished GLS expression, and T98G-GAB cell line significantly overexpressed GLS2 (Fig. 1).
Expression of GLS and GLS2 in assayed cell lines. Western blots show that SFxL and LN229 GLS silenced cell lines diminished GLS expression, and T98G-GAB cell line effectively overexpressed GLS2 isoform. Transacted controls were equivalent to non-transfected cells for all type of cells. Data are from one of three representative experiments. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as loading control
Cell viability assays
For viability assays, 5 × 104 cells in 100 μL of medium were seeded in a 96-well culture plate. Next, the cells were exposed to increasing (0–300 μM) concentrations of ATO (Sigma-Aldrich) or H2O2 (Sigma-Aldrich) for 15 min, 1, 6, 24, 48 and 72 h. After treatments, the medium was removed, the cells were washed with phosphate-buffered saline (PBS), and cell number was evaluated. In brief, 10 μL of 3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium (MTS) (Promega, Southampton, UK) was added to each well (0.5 mg/mL), and then the plates were incubated at 37 °C for 3 h. The absorbance at 570 nm was measured using an Elisa BioRad Microplate Reader (BioRad, Hercules, CA, USA).
Annexin V and caspase 3 activity assays
Apoptosis was quantified by flow cytometry after staining cells with R-phycoerythrin (R-PE)-labelled annexin V (Invitrogen, Grand Island, NY, USA) and propidium iodide (PI). After 48 h of ATO treatment (5 μM for SFxL and LN229 pairs, and 50 μM for T98G derivative cells), 1 × 106 cells/mL were harvested and centrifuged at 900×g for 5 min; the pellets were washed twice with PBS and resuspended in 100 μl of annexin V binding buffer (0.14 M NaCl, 2.5 mM CaCl2, 0.01 M HEPES pH 7.4). Annexin V (5 μL) was added to the samples and incubated in the dark for 30 min. For PI assay (Sigma-Aldrich), the same amount of cells was washed with PBS and then resuspended in 500 μl of PBS containing 20 μg/mL RNase A, 0.1 % Triton X-100 and PI (20 μg/mL). Untreated cells were used as control for double staining. Samples were kept on ice after incubation until fluorescence-activated cell sorting (FACS) analysis was performed, and apoptosis evaluated based on the percentage of annexin V-positive cells. Annexin V can be detected in both the early and late stages of apoptosis. PI enters the cell in late apoptosis or necrosis. Early apoptosis is defined by annexin V+/PI− staining, late apoptosis is defined by annexin V+/PI+ staining and necrosis is defined by annexin V−/PI+ staining. Fluorimetric assay of caspase-3 activity was performed according to the instructions of the manufacturer without modifications (Promega, Madison, Wisconsin, USA).
Evaluation of mitochondrial membrane potential
ΔΨm was assessed using a sensitive fluorescent probe 5,50,6,60-tetrachloro-1,10,3,30-tetraethyl-benzamidazolocarbocyanin iodide (JC-1, Invitrogen), calculated as the ratio of fluorescence intensities at 640 ± 30 nm and 525 ± 20 nm (640/525), with excitation at 485 nm and a beamsplitter at 505 nm. Red emission from the dye is attributed to a potential-dependent aggregation of JC-1 in the mitochondria. Green fluorescence reflects the monomeric form of JC-1, appearing in the cytoplasm after mitochondrial membrane depolarization. 0.25–0.5 × 106 cells were grown on 25-cm2 flasks, followed by ATO (2.5 μM, 48 h) or H2O2 (200 μM, 1 h) treatment. Cells were then incubated with 1.5 mM JC-1 dye diluted in culture medium at 37 °C for 30 min. Cells were washed with PBS and analyzed immediately using a FACScalibur (Becton and Dickinson, Franklin Lakes, NJ Franklin Lakes, NJ, USA).
Intracellular antioxidant levels
Total glutathione (GSH + GSSG), oxidized glutathione (GSSG) and reduced glutathione (GSH) were measured spectrophotometrically by the glutathione reductase (GR) recycling assay at 412 nm in the presence of 5,5′-dithiobis(2-nitrobenzoic) acid (DTNB) (Sigma-Aldrich). 0.75 × 106 cells were grown on 25-cm2 flasks suspensions and left untreated or were treated with different concentrations of ATO or H2O2. For SFxL, SFxL-GLS(−), LN229, LN229-GLS(−) cells, 2.5 μM ATO (24 h), and 200 μM H2O2 (1 h) were used. For T98G, T98G-GAB, T98G-pcDNA cells, 5 μM ATO (24 h) and 300 μM H2O2 (1 h) were used. For sample preparation, cells were trypsinized and washed twice in cold PBS, and then resuspended to the same volume in PBS, lysed in PBS supplemented with 0.1 % Triton X-100, 0.6 % sulfosalicylic acid, vortexed, and then incubated 30 min at 4 °C. After centrifugation for 4 min at 3,000×g, supernatant was recovered and divided in two aliquots: one was conserved at 4 °C for GSH + GSSG quantification, the other one was treated with 2-vinylpyridine (C f = 5 %) during 1 h at room temperature (RT), and triethanolamine (C f = 1 %) during 10 min at RT to mask the GSH present in the extract and prevent its measurement [26]. Using a 96-well culture plate, 20 μL of the supernatant was added to 120 μL of DTNB reagent (0.040 g of DTNB dissolved in 10 mL of PBS). This was mixed and incubated for 30 s at RT. Then, 60 μL of 1 mM NADPH was added, and the absorbance was read at 412 nm, using an Elisa BioRad Microplate Reader (BioRad, Hercules, CA, USA). The amount of GSH was obtained by subtracting the amount of GSSG from that of total glutathione.
Transwell migration assay
Migration assays were performed using a 24-well Boyden chamber plate with 8-μm pore size polycarbonate membrane filters (BD- Biosciences San Jose, CA, USA). SFxL, SFxL-GLS(−), LN229, LN229-GLS(−) cells (2× 104/well), and T98G, T98G-GAB, T98G-pcDNA cells (4 × 104/well) were placed in the upper part of the Boyden chamber containing DMEM and 10 % FBS, the lower chamber also contained DMEM and 10 % FBS. When treated cells were assayed, after a period of cell adhesion (12 h) the medium was changed to introduce the oxidizing agents (2.5 μM ATO for SFxL and LN229 cells, and 5 μM ATO for T98G cells, 24 or 48 h; and 1 μM H2O2 for SFxL and LN229 cells, and 2 μM H2O2 for T98G cells, 24 h) to both compartments of the chamber. The culture media was changed daily (ATO) or every 12 h (H2O2). After incubation, the cells on the membrane filter were fixed with methanol and stained with 0.05 % crystal violet for 30 min. The cells on the upper surface of the filter were removed with a cotton swab. The membranes were then rinsed in PBS until excess stain was removed. The membranes were then air-dried for 20 min. The migratory cells were determined by counting the cells that migrated to the lower side of the filter by bright field microscopy at ×200 magnification. Leica model SP5 II and Nikon model Optiphot-II microscopes, with ×20 objectives, were used to examine the cells, with Leica LAS AF and Nikon Image Pro plus image acquisition software, respectively. Ten random fields were counted for each filter, and each sample was assayed in triplicate.
Protein expression
The cells were harvested and washed in PBS, keep in ice and lysed in RIPA buffer supplemented with a Complete Protease Inhibitor kit (Roche, Indianapolis, IN, USA). Lysates were centrifuged at 10,000×g for 10 min at 4 °C, and the supernatants were collected and stored at −80 °C. Protein concentration was determined using the Bradford Protein Assay Kit (BioRad, Hercules, CA, USA). After heating for 5 min in boiling water, 25 μg of protein was resolved on 12 % polyacrylamide-SDS gels and then electrotransferred to nitrocellulose membranes. Membranes were washed in TBST (20 mm Tris–HCl, pH 7.5, and 500 mm NaCl, containing 0.1 % Tween 20) and blocked for 1 h in TBST containing 0.5 % Tween 20 and 5 % dried milk, or 5 % bovine serum albumin (BSA). The following antibodies were purchased: c-myc (Abcam, Cambridge, UK), bcl-2 (Cell Signaling, Danvers, MA, USA), bid (Cell Signaling), and used as recommended by suppliers. After incubation with respective primary antibodies, membranes were incubated with a horseradish peroxidase-conjugated secondary antibody (Sigma-Aldrich), for 1 h at RT with gentle rocking, and visualized on Kodak X-ray film using an enhanced chemiluminiscence (ECL) detection system (SuperSignal West Pico, Pierce, Rockford, IL, USA). The membranes were stripped for 30 min in Stripping Buffer (60 mM Tris–HCl pH 6.8, 0.7 % β-mercaptoethanol, 2 % SDS) and re-probed with anti-glyceraldehyde-3-phosphate dehydrogenase (GAPDH) antibody as a loading control (Sigma-Aldrich). Independent experiments validated that this stripping procedure did not lead to loss of signal.
Statistical analysis
Data were expressed as the mean ± SD from independent experiments. Statistical analysis was performed using SPSS 13.0 for Windows. Comparisons between two groups were performed by unpaired-test. Multiple comparisons between more than two groups were performed by one-way analysis of variance (ANOVA). Significance was considered to be P < 0.05. Experiments were performed at least three times.
Results
Silencing GLS or overexpressing GLS2 induces growth inhibition in different glioma cell lines and inhibition is synergistically augmented after oxidative stress
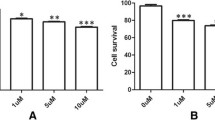
Cell proliferation (MTS) assays were performed to characterize growth of control and glutaminase-modified glioma cell lines (Fig. 2). In all inhibitory studies, non-transfected controls are equivalent to transfected controls. To avoid repetition, only non-silencing controls are shown for SFxL and Ln229 cells. White bars for SFxL and LN229 cells represent cells transfected with a non-silencing shRNA (Fig. 2). A rapid (24 h) effect was found in proliferation ratio when GLS was silenced in SFxL (41 %) and LN229 cells (38 %). Minor, but significant (P < 0.01), effect was depicted in T98G cells (63 %) in cells overexpressing GLS2 (Fig. 2). No significant differences were found at 48 or 72 h (results not shown). Then, we used these human glioma cancer cell lines to evaluate the effects of ATO and H2O2 in cell survival. To assess the effect of these agents, each viability value was standardized setting 100 % to the respective value without treatment. All tested cell lines showed dose- and time- dependent sensitivity to oxidative stress by both oxidative agents. Thus, these cell lines provide us an excellent system to evaluate how silencing of GLS or overexpression of GLS2 alters survival of glioma cell lines when an oxidative stress occurs. First, MTS assays were carried out to study the responsiveness of SFxL versus SFxL-GLS(−) to ATO (Fig. 3a). GLS-silenced cells were more sensitive to this oxidative agent than control cells. For example, a significant (P < 0.01) synergistic inhibition was shown (50 %) after 24 h of 20 μM ATO incubation. When incubation time was 48 h, more than 20 % inhibition was found at 2.5 μM ATO, but the highest effect was depicted at 5, 10, and 20 μM (more than 50 % inhibition). Finally, after 72-h treatment, the effect was stronger, 35 % at 2.5 μM and 50 % at 5, 10, and 20 (Fig. 3a). Regarding H2O2 effect, in SFxL pair, after 1 h (at 50, 100, and 200 μM) and 6 h (at 50 and 100 μM) treatments, the highest differences were observed (Fig. 3b). Moreover, significant differences were found at 200 μM (6 h). In LN229 system, similar survival rates were found (Fig. 3c,d). After treatment for 72 h, more than 75 % inhibition was noticed at 10 and 20 μM ATO (Fig. 3c). Large differences (P < 0.01) were also obtained for H2O2 treatment (Fig. 3d). Specifically, inhibition was > 25 % at 100 μM (6 h) and 200 μM (15 min), and > 35 % at 200 μM (1 and 6 h). For 300 μM H2O2 50, 60 and 80 % inhibition were found respectively (15 min, 1 and 6 h). When overexpression of GLS2 was studied, opposite results were obtained at 48 h for lower (10 and 20 μM) and higher (50 μM) ATO (Fig. 3e). When ATO concentration was 50 μM T98G-GAB cells showed higher survival ratio for 48-h treatment but no differences were observed after 24 and 72 h. However, clear inhibition values were found at 50 μM (24 and 72 h) and even at lower concentrations (5 and 10 μM) for 48 h. For H2O2 treatment, a similar heterogeneous pattern of survival was obtained, including a clear higher survival rate at 100 μM (6 h) (Fig. 3f). However, significant lower values were obtained at 200 μM (15 min and 1 h) as well as at 300 μM (1 and 6 h). For T98G and T98G-pcDNA control cell lines significant differences were not found (results not shown) neither for ATO or H2O2 treatment.
Effect of GLS silencing or GLS2 overexpression on the proliferation of glioma cells after 24 h without treatment. Proliferation of SFxL-GLS(−) and LN229-GLS(−) versus SFxL and LN229 cells, and T98G-GAB versus T98G cells, respectively, were assayed by MTS method. White bars for SFxL and LN229 cells represent cells transfected with a non-silencing shRNA. For T98G, white bar represent value for wild-type cells. Dark grey bars represent values for GLS-silenced cells, or cells overexpressing GLS2. Light gray bars represent values for transacted control (T98G-pcDNA cell line). Data are presented as the mean ± S.E. of triplicate experiments. Value for each control cell type was set at 100 %. Differences were always significant (n = 3, *P < 0.01) between SFxL-GLS(−), LN229-GLS(−) or T98G-GAB cells and their respective control cells. Transacted controls were equivalent to non-transfected cells for all type of cells. Three asterisks indicated significantly different values between T98G-GAB cells and T98G-pcDNA cells (***P < 0.01)
Dose-dependent decrease in cell viability of the assayed glioma cell lines by MTS method after treatment with ATO or H2O2. a Bar chart showing percentages of survival in SFxL (white bars) and SFxL-GLS(−) (gray bars) cells when treated with 1.25–20 μM ATO for 24, 48, and 72 h. b Bar chart showing percentages of survival in SFxL (white bars) and SFxL-GLS(−) (gray bars) cells when treated with 50–300 μM H2O2 for 15 min, 1 h, and 6 h. c Bar chart showing percentages of survival in LN229 (white bars) and LN229-GLS(−) (gray bars) cells when treated with 1.25–20 μM ATO for 24, 48, and 72 h. d Bar chart showing percentages of survival in LN229 (white bars) and LN229-GLS(−) (gray bars) cells when treated with 50–300 μM H2O2 for 15 min, 1 h, and 6 h. e Bar chart showing percentages of survival in T98G (white bars) and T98G-GAB (gray bars) cells when treated with 10–50 μM ATO for 24, 48, and 72 h. f Bar chart showing percentages of survival in T98G (white bars) and T98G-GAB (gray bars) cells when treated with 50–300 μM H2O2 for 15 min, 1 h, and 6 h. Each value is mean from three independent experiments. Error bars are not shown in this figure for clarity. Transfected controls were equivalent to non-transfected cells for all type of cells. Asterisks indicated significantly decreased values between SFxL-GLS(−), LN229-GLS(−), or T98G-GAB cells and their respective control cells (*P < 0.05, **P < 0.01). Three asterisks indicated significantly increased values between T98G-GAB cells and their respective control cells (***P < 0.05)
GLS silencing synergizes with induction of intrinsic apoptosis but GLS2 overexpression can reduce apoptosis after oxidative stress
When assaying SFxL and LN229 pairs, after treatment for 48 h with 5 μM of ATO, the percentage of early apoptotic cells was 69 and 70 %, respectively (both P < 0.01 versus control), compared to untreated control cells (23 and 28 %) (Fig. 4a). Then, ATO clearly induced higher apoptotic activity in these glioma cells when GLS was silenced. Conversely, after treatment with 50 μM of ATO (48 h) T98G and T98G-GAB cells showed 60 and 30 % of apoptosis, respectively (Fig. 4a). Additional fluorimetric assays were then performed to verify whether GLS- and GLS2-mediated apoptosis was associated with caspase 3 activity and, consequently, through the intrinsic mechanisms. A large increase of caspase-3 activity was observed in both SFxL and LN229 pair cells after 48 h of exposure to 5 μM of ATO (Fig. 4b). Simultaneously, a clear decrease in caspase 3 activity was found in the pair of T98G and T98G-GAB cells when exposed to 50 μM of ATO (48 h).
Assessment of apoptosis. Color code for bars as depicted in Fig. 2. a Apoptosis assay using cytometry after staining with annexin V-PE/propidium iodide (PI). b Caspase 3 activity using a fluorimetric assay as described in the materials and methods. Results are expressed as percentage of relative fluorescence units (RFU) per minute per milligram of protein detected in untreated cells, which are arbitrarily expressed as 100 %. SFxL and LN229 cells were treated with 5 μM ATO (48 h), and T98G cells were treated with 50 μM ATO (48 h). For SFxL and LN229 pair cells caspase activity of GLS-silenced cells were set at 100 %. Conversely, for T98G cell type cells caspase activity was set at 100 % for wild-type T98G. Data are presented as the mean ± S.E. of triplicate experiments. Differences were always significant (n = 3, *P < 0.01) between SFxL-GLS(−), LN229-GLS(−), or T98G-GAB cells and their respective control cells. Transacted controls were equivalent to non-transfected cells for all type of cells. Three asterisks indicated significantly different values between treated T98G-GAB cells and treated T98G-pcDNA cells (***P < 0.01)
GLS silencing and GLS2 overexpression synergize with oxidative stress evoking formation of dysfunctional mitochondria
To prove whether the reduction of viability produced by ATO and H2O2 was the result of accumulation of dysfunctional mitochondria, the ΔΨm dissipation as a consequence of a 48 h-long treatment with ATO, or a 1 h-short treatment with H2O2, was analyzed in detail. As expected, control tumor cells showed a high mean red fluorescence (set to 100 %) after staining with JC-1 (Fig. 5). Cells silencing GLS or overexpressing GLS2 show levels of fluorescence 10–20 % lower than controls. After treatment with oxidant agents (ATO and H2O2), the mean red fluorescence of the mitochondria dropped (40–55 %) in cells with knocked-down expression of GLS, indicating that the ΔΨm had collapsed, and that GLS silencing and ROS have a synergistic effect against functional mitochondria. As shown in Fig. 5 GLS silencing was able to produce sustained ΔΨm dissipation that was highly increased after a ROS insult (P < 0.01). SFxL and SFxL-GLS(−) cells, with or without treatment of 2.5 μM ATO (48 h), showed a significant reduction (P < 0.01) in the mean red fluorescence intensity (Fig. 5a), which, in repeated experiments, was similar to that seen for LN229 and LN229-GLS(−) cells (Fig. 5a). When experiments were carried out using the T98G glioma model, including cells overexpressing GLS2, similar results were found for 5 μM ATO (48 h) (Fig. 5a). On the other hand, treatment of glioma cells with cytotoxic doses of H2O2 (200 μM) causes a more rapid (1 h) loss of ΔΨm (Fig. 5b). Also in this case, such effect was shown for SFxL and LN229 models. Equally, significant lower ΔΨm were found for the T98G glioma model that overexpresses GLS2 after 200 μM H2O2 (1 h) treatment (Fig. 5b).
Flow cytometry analysis showing alterations in the ΔΨm of SFxL, LN229, and T98G cells. Upon completion of treatments, the cells were stained with JC-1 and the alterations in ΔΨm were analyzed as mentioned in methods. White bars represent non-silencing controls for SFxL and LN229 cells, as well as non-transfected controls for T98G cell line. Transfected controls were equivalent to non-transfected cells for all type of cells. Dark grey bars represent values for GLS-silenced cells, or cells overexpressing GLS2. Light gray bars represent values for control T98G-pcDNA cell line. Non-hatched bars represent untreated cells. Hatched bars represent treated cells with ATO (a) or H2O2 (b). a Bar chart showing percentages of relative fluorescence intensity of ΔΨm in SFxL versus SFxL-GLS(−) cells, LN229 versus LN229-GLS(−) cells, and T98G and T98G-pcDNA versus T98G-GAB cells for untreated (unfilled pattern) and 2.5 μM (SFxL and LN229 derivative cells) or 5 μM (T98G derivative cells) ATO (48 h) treated cells (angular stripes pattern). b Bar chart showing percentages of relative fluorescence intensity of ΔΨm in SFxL versus SFxL-GLS(−) cells, LN229 versus LN229-GLS(−) cells, and T98G and T98G-pcDNA versus T98G-GAB cells for untreated (unfilled pattern) and 200 μM H2O2 (1 h) treated cells (angular stripes pattern). Each value is mean ± S.E. from at least three independent experiments. Asterisk indicated significantly different values between treated and control (untreated) cells (*P < 0.01). Two asterisks indicated significantly different values between treated silenced/overexpressed cells and treated control cells (**P < 0.01). Three asterisks indicated significantly different values between treated T98G-GAB cells and treated T98G-pcDNA cells (***P < 0.01)
Antioxidant glutathione levels were decreased in GLS-silenced glioma cells but augmented in glioma cells overexpressing GLS2. Both effects were augmented after an oxidative insult
Levels of GSH, the major cellular antioxidant, were affected by both GLS silencing and GLS2 overexpression (Fig. 6a). However, the GSH/GSSG ratio was not significantly modified in any assayed cell type (Fig. 6b). Nevertheless, when cells were treated with 2.5 μM ATO (24 h) or 200 μM H2O2 (1 h), differences were much larger between control cells and their counterparts (Fig. 6c,e). Curiously, after treatment with oxidative agents, a clear difference appeared in GSH/GSSG ratio (P < 0.01); specifically lower GSH/GSSG ratios for SFxL-GLS(−) and LN229-GLS(−), and higher GSH/GSSG ratios for T98G-GAB cells (Fig. 6d, f).
Glutathione levels (nmol/mg protein) were measured in pooled control or GLS-silenced/GLS2-transfected cells. Color code for bars as depicted in Fig. 2. a GSH levels in SFxL-GLS(−), LN229-GLS(−), and T98G-GAB cells versus SFxL, LN229, and T98G/T98G-pcDNA cells, respectively. b GSH/GSSG ratios in SFxL-GLS(−), LN229-GLS(−), and T98G-GAB cells versus SFxL, LN229, and T98G/T98G-pcDNA cells, respectively. c GSH levels in SFxL-GLS(−), LN229-GLS(−), and T98G-GAB cells versus SFxL, LN229, and T98G/T98G-pcDNA cells, respectively, after 24-h ATO treatment (2.5 μM for SFxL and LN229 pairs, and 5 μM for T98G derivative cells). d GSH/GSSG ratios in SFxL-GLS(−), LN229-GLS(−) and T98G-GAB cells versus SFxL, LN229 and T98G/T98G-pcDNA cells, respectively, after 24-h ATO treatment (2.5 μM for SFxL and LN229 pairs, and 5 μM for T98G derivative cells). e GSH levels in SFxL-GLS(−), LN229-GLS(−), and T98G-GAB cells versus SFxL, LN229, and T98G/T98G-pcDNA cells, respectively, after 1-h treatment with H2O2 (200 μM for SFxL and LN229 pairs, and 300 μM for T98G derivative cells). f GSH/GSSG ratios in SFxL-GLS(−), LN229-GLS(−), and T98G-GAB cells versus SFxL, LN229, and T98G/T98G-pcDNA cells, respectively, after 1-h treatment with H2O2 (200 μM for SFxL and LN229 pairs, and 300 μM for T98G derivative cells). Differences were always significant (n = 3, *P < 0.01) between SFxL-GLS(−), LN229-GLS(−), or T98G-GAB cells and their respective control cells. Trasfected controls were equivalent to non-transfected cells for all type of cells. Two asterisks (**P < 0.01) indicated significantly different values between treated silenced/overexpressed cells and untreated control cells (panels c and e versus a, and panels d and f versus b). Three asterisks indicated significantly different values between treated T98G-GAB cells and treated T98G-pcDNA cells (***P < 0.01)
Inhibition of cell migration by GLS silencing or by GLS2 overexpression, and synergistic effect after a ROS insult
For ATO treatment, the number of cells migrating across the upper well was decreased by 25 % and 20 % (P < 0.05), respectively, when GLS was silenced in SFxL and LN229 cells, after oxidative insult for 24 h (Fig. 7a). Treatment was also assayed after 48 h and similar results were found. The rate of migrated cells was equally lower in T98G-GAB cells than the control cells. In fact, we observed that the migration rate of T98G-GAB cells decreased approximately 30 % (P < 0.05) compared to both transacted and non-transfected control cells (Fig. 7a). Treatment was also assayed after 48 h showing significant differences that were somewhat reduced in scope (Fig. 7a). When oxidative stress was induced with 1 μM of H2O2 (24 h), the number of SFxL and LN229 cells migrating across the upper well also decreased significantly (20 and 22 % respectively) when compared with the respective non-silencing control cells (P < 0.05) (Fig. 7b). The same experiment was performed with 2 μM of H2O2 (24 h) for T98G-GAB cells and they showed a similar 30 % decrease (P < 0.05), compared to both transfected and non-transfected control cells (Fig. 7b). Representative images of migrated cells are shown for non-silencing control and silenced SFxL, LN229 pair cells, as well as for non-transfected control T98G and T98G-GAB cells, after ATO (Fig. 8a) and H2O2 treatment (Fig.8b), respectively. A representative synergistic effect between GLS inhibition and ATO treatment (5 μM, 48 h) is shown for LN229 pair of cells (Fig. 9a). As higher rates of migration inhibition were obtained after ATO treatment (Fig. 9b), panels for non-treated cells were omitted.
GLS silencing and GLS2 overexpression reduces glioma cells migration after an oxidative insult. Color code for bars as depicted in Fig. 2. Unfilled pattern means 24-h treatment. Dotted pattern means 48-h treatment. a Cell migration of SFxL-GLS(−) cells versus SFxL, LN229-GLS(−) cells versus LN229, and T98G-GAB cells versus T98G/T98G-pcDNA after ATO treatment. b Cell migration of SFxL-GLS(−) cells versus SFxL, LN229-GLS(−) cells versus LN229, and T98G-GAB cells versus T98G/T98G-pcDNA after H2O2 treatment. Data are the mean of three independent experiments. Transacted controls were equivalent to non-transfected cells for all type of cells. Asterisk indicated significantly different values between treated silenced/overexpressed cells and treated control cells (*P < 0.01). Two asterisks indicated significantly different values between 24-h treated silenced/overexpressed cells and 48-h treated control cells (**P < 0.01). Three asterisks indicated significantly different values between treated T98G-GAB cells and treated T98G-pcDNA cells (***P < 0.01)
Photomicrographs of crystal violet-stained cells are shown for treated SFxL, LN229, and T98G derivative cells. The number of transmigrated glioma cells was counted at ×200 magnification in ten microscopic fields randomly selected and calculated as mean cell number per field. Trasfected controls did not show differences with non-transfected cells. Because space reasons only one type of control cells is shown for each assayed glioma. a Representative photomicrographs are shown after ATO treatment (24 h). b Representative photomicrographs are shown after H2O2 treatment (24 h)
Photomicrographs of crystal violet-stained cells are shown for untreated and ATO-treated LN229 and LN229-GLS(−) cells. After culture for 12 h, more untreated cells migrated to the centre of the wound field, whereas less ATO-treated cells and even fewer ATO-treated plus GLS-silencing cells did so. The number of transmigrated glioma cells was counted at ×200 magnification in ten microscopic fields randomly selected and calculated as mean cell number per field. a Representative photomicrograph of LN229 and LN229-GLS(−) cells is shown with or without 5 μM ATO treatment (48 h). b Relative number of migrated cells are represented (value for untreated LN229 cells was set at 100 %). White bars represent cells transfected with a non-silencing shRNA. Grey bars represent values for GLS-silenced cells. Asterisk indicated significantly different values between GLS silenced LN229 and non-silencing control for untreated or treated cells (*P < 0.01). Two asterisks indicated significantly different values between untreated and treated non-silencing cells (**P < 0.01). Three asterisks indicated significantly different values between untreated and treated GLS silenced LN229 cells (***P < 0.05)
GLS silencing or GLS2 overexpression reduced the expression of c-myc, as well as changed the expression of bid and bcl-2
To verify the role of GLS silencing in the expression of the oncoprotein c-myc, as well as pro-apoptotic bid and anti-apoptotic bcl-2 proteins, Western blot analysis of these key proteins was performed with lysates of SFxL, SFxL-GLS(−), LN229, and LN229-GLS(−) glioma cells after 48-h treatment with 2.5 μM ATO. Indeed c-myc, bid, and bcl-2 were present in all samples, although at different levels (Fig. 10a). Our find that protein levels vary between control and their corresponding GLS-silenced cell line is in agreement with our earlier findings where GLS-silenced cells showed a less tumorigenic phenotype. Note that levels of c-myc in GLS-silenced cells are much lower (ten times lower for SFxL and two times lower for LN229, based on densitometry than those for controls (Fig. 10b). These data suggest that c-myc plays a more important role in SFxL cells than in LN229 cells. Conversely, the abundance of bid band was augmented by twofold for both SFxL and LN229 cells, in comparison to GLS-silenced cells, respectively (Fig. 10b). This finding is consistent with an essential role of GLS in the effect of this pro-apoptotic factor in those cells. Additional support to the hypothesis that apoptosis is a pivotal process related to GLS expression came from bcl-2 protein expression levels: anti-apoptotic bcl-2 was less than 2.5- and 6-fold, respectively, expressed in both SFxL and LN229 cells, versus to their corresponding GLS-silenced cells (Fig. 10b). GLS2 is a key isoenzyme linked to pro-apoptotic and tumor suppressor properties [7, 8, 15]. Consistently, when GLS2 was overexpressed in T98G cells, c-myc and bcl-2 were less than 4- and 6-fold respectively expressed. In contrast, pro-apoptotic bid was 2.3-fold overexpressed (Fig. 11).
Results of expression of c-myc, bid and bcl-2 proteins in SFxL and LN229 cells silenced cells. a Western blot analysis of glioma cell shows significant differences in the expression of some key proteins when simultaneously GLS was silenced and cells were treated during 48 h with 2.5 μM ATO. GAPDH was used as loading control. Data are from one of three representative experiments. b Relative levels of proteins as assessed by grey values. White bars represent cells transfected with a non-silencing shRNA. Grey bars represent values for GLS-silenced cells. Quantifications were normalized to value obtained for GAPDH protein expression and represented as integrated optical density (IOD). Asterisk indicated significantly different values between GLS silenced SFxL or GLS silenced LN229 cells and their respective non-silencing control cells (*P < 0.01). Two asterisks indicated significantly different values between GLS silenced LN229 cells and non-silencing control cells (**P < 0.05)
Results of expression of c-myc, bid and bcl-2 proteins in T98G cells overexpressing GAB versus cells transfected with the empty vector. a Western blot analysis of glioma cell shows significant differences in the expression of some apoptotic key proteins when GAB was overexpressed. Non-transfected controls were equivalent to transacted controls. To avoid repetition, only transacted controls are shown. GAPDH was used as loading control. Data are from one of three representative experiments. b Relative levels of proteins as assessed by grey values. White bars represent value for T98G-pcDNA (transfected control). Grey bars represent values for cells overexpressing GAB isoform. Quantifications were normalized to value obtained for GAPDH protein expression and represented as integrated optical density (IOD). Asterisks indicated significantly different values between T98G-GAB cells and transacted control T98G-pcDNA cells (*P < 0.01; **P < 0.05)
Discussion
Expression of glutaminase isoenzymes is a key switch in oncogenic signaling pathways
Glutaminase expression has been shown to be increased in several tumor types, such as lymphoma, prostate, brain (glioblastoma), and kidney cancers [9, 18]. Elevated glutaminase activity is under regulatory control of the oncogenic transcription factor c-myc, which itself is amplified in many cancers [5, 9, 20]. Glutaminase is also upregulated by other oncogenic signaling pathways [27], and by the small GTPase Rho in a NF-κB-dependent manner [6]. Defective signaling through tightly regulated apoptotic pathways that involve oxidative stress can contribute to both unbalance in apoptosis [25] and development of cancer [23]. Glutamine (via glutamate) is one of the precursor amino acid for glutathione, which is present within the cell as GSH and GSSG forms [8]. The ratio of GSH/GSSG is the most important regulator of the cellular redox potential [25]. Total GSH is used by cells for many purposes, but the GSH/GSSG ratio may indicate that the cells can utilize both GSH and GSSG, and both forms of glutathione could be used to bind free sulfydryl groups of cysteine residues in proteins [8]. In our experiments, T98G cells transfected with GLS2 showed augmentation of both GSH and GSSG, while SFxL and LN229 cells which are deplete of GLS exhibited lower levels of both GSH and GSSG, consistent with GLS2's prominent role in controlling overall redox status [8, 15]. In addition, silencing of GLS kills cells that overexpress c-myc and catabolize glutamine [20]. On the other hand, while lacking GLS expression, breast cell lines have shown a higher level of GLS2 [17], according with the higher p53 levels given the ability of p53 to regulate GLS2 [7, 15]. Nonetheless, an isoenzymatic balance exists between GLS and GLS2 in most human cancers [8], and simultaneous control of the expression of both isoenzymes might be used as a valuable tool for some types of cancer.
Lower c-myc and bcl-2 expression, as well as higher pro-apoptotic bid expression were found as potential mediators of glioma cell apoptosis resulting from the combination of GLS-silencing and ATO treatment. To highlight, GA isoenzymes can regulate ROS generation [7], generate pro-apoptotic signals [8], and disturb reprogrammed metabolism (glutaminolysis) [14]. One of the biochemical hallmarks of tumor cells involves the reprogramming of energy metabolism from primarily oxidative phosphorylation (OXPHOS) towards an enhanced rate of anaerobic glycolysis, a phenomenon termed the Warburg effect [16]. Interestingly, inhibition of GLS also provoked a significant decrease in the ratio of O-glycosylated to total proteins [28]. Accordingly, posttranslational modification of proteins by O-linked β-N-acetylglucosamine (O-GlcNAcylation) is a key metabolic regulator of glucose metabolism in cancer cells. In fact, blocking glycosylation of phosphofructokinase 1 (PFK1) reduced cancer cell proliferation in vitro and impaired tumor formation in vivo [29].
A synergistic effect by GLS silencing or GLS2 overexpression plus an oxidative insult decreased apoptosis, antioxidant status and cellular motility
In this study, silencing GLS or overexpressing GLS2 genes decreased glioma cell survival. This effect was increased by an oxidative insult. Overall, the survival experiments firstly suggest that longer periods of oxidative stress were required to achieve similar antiproliferative effects with ATO versus H2O2. Secondly, GLS silencing had a stronger synergistic inhibitory effect in cell survival than GLS2 overexpression. Third, only higher concentrations and longer periods of both ATO and H2O2 evoke decreased cell viability in the T98G pair. A kind of balance exists and, in some other conditions, an opposite effect was observed. To test whether the decrease in SFxL and LN229 cell number observed after oxidative treatment was the result of apoptosis, ATO-treated cells were assayed for annexin V/PI staining and caspase 3 activity. We also tested apoptosis and caspase 3 activity for critical conditions that evokes higher survival ratios for T98G-GAB cells. Altogether, these results suggest that both the antiproliferative effect of GLS silencing and the higher survival in GLS2 overexpressing cells may be caused by opposite ratios of intrinsic apoptosis. Additionally, after GLS2 overexpression T98G glioma cells became more resistant to oxidative stress, reinforcing the idea of GLS2 having a key role in GSH antioxidant function [7, 8, 15]. Both GLS silencing and GLS2 overexpression showed diminished ΔΨm and cell viability, but the decrease was substantially augmented after inducing oxidative stress with either agent, ATO or H2O2. Apoptosis and mitochondrial swelling is often associated with the loss of ΔΨm [25]. In our research, these results mirror the data obtained for annexin V positive cells, as well as for caspase 3 activity, indicating that loss of ΔΨm is linked to GLS and ROS-mediated cell killing. Loss of ΔΨm is an early event of the apoptosis processes, which could be regulated by other pro-survival effectors, including higher glutathione levels [8]. Significantly, after oxidative treatments, GLS silencing disturbs redox balance severely disrupting ROS scavenging by GSH, as evidenced by a prominent lower GSH/GSG ratio in GLS silenced glioma cells. On the contrary, T98G-GAB cells showed the highest GSH/GSSG ratio after both ATO and H2O2 treatment. Evidently, this increased GSH/GSSG appeared insufficient to effectively counteract the pro-apoptotic pathway. These striking results obtained for GSH/GSSG ratios could be explained by the key role of GLS isoenzymes regulating oxidative stress [7, 15].
In addition, the lack of sensitivity of non-cancerous cell lines to the clinically relevant concentrations of ATO also suggests that this treatment is an adequate strategy to study the effect of GLS inhibition and/or GLS2 overexpression on both oxidative stress and cancer growth. Certainly, non-transformed cells are less sensitive to oxidative injury [25], and tumor cells tend to display higher dependence on glutamine than non-malignant cells [5]. Moreover, H2O2 sensitivity of tumor cells can be modified by GSH-dependent mechanisms [25]. On the other hand, invasive ability and aberrant motility are key hallmarks of cancer [8]. To address whether GLS inhibition or GLS2 overexpression plays a role in regulating the migration of SFxL, LN229, and T98G cells, we investigated the influence of an oxidative insult on cells migration by a transwell assay, using controls and GLS/GLS2 modified cells. The results point out an important relation between GA isoenzymes expression and migration rate of these glioma cell lines after both ATO and H2O2 treatment, and suggest that the decrease in migration rate might be synergistically related to the downregulation of GLS expression and/or overexpression of GLS2.
Future prospect of combination therapy as an effective strategy to combat glioma
Recently, selective pharmacological inhibition of bcl-2 has shown antitumor activity inducing apoptosis in bcl-2-dependent hematological cancers [30]. Efforts against glioma malignancies have focused on therapies which target key intracellular apoptotic pathways which may confer tumor resistance, such as p53 family, and more recently miRNAs [31]. This research by Liu et al. proposes a combination therapy which incorporates chemotherapy, radiotherapy, and selective apoptotic inhibitors and miRNA-based therapy, which is likely to be the future direction for the treatment of glioblastoma. Otherwise, caspase 3 and bcl-2 family proteins have been also involved in therapies targeting apoptosis signaling pathways in T98G cells [31]. In a very recent research, the expression of KGA and GAC isoforms was knocked down in T98G and T98G-GAB; the GLS silencing reduced proliferation and viability of glioblastoma cells and strengthen the antiproliferative effect evoked by GLS2 overexpression. Therefore, combination of negative modulation of GA isoforms arising from GLS gene with the introduction of GLS2 gene product, GAB, may in the future provide a useful means to curb glioblastoma growth in situ [32].
As previously proposed, GA modification will not be effective as a single-arm therapy but will be a part of a more complex strategy that may involve other metabolic targets, including inhibition of glycolysis [18]. Combination and sequential treatments to improve therapeutic efficacy are recommended [33], including synergistic antitumor effects of several natural antioxidants with current anticancer drugs [34]. In other study, an allosteric inhibitor of glutamine metabolism, the small molecule bis-2-(5-phenylacetamido-1,2,4-thiadiazol-2-yl)ethyl sulfide (BPTES) has been found to slow growth of glioblastoma cells targeting GLS but not GLS2 isoenzymes [18]. The small molecule 968, a dibenzophenanthridine, was recently shown to inhibit GAC, blocking the proliferation and inhibiting tumor formation in mouse xenograft models. Of outstanding interest, an additive effect was observed when cancer cells are dosed with 968 and BPTES simultaneously [35]. As a main conclusion of our research, GLS silencing and overexpression of GLS2 while simultaneously disrupting glutathione-dependent antioxidant capacity can synergistically contribute to an important effect against tumor cell growth caused by ROS (ATO or H2O2 insult). Consistently, blocking GLS and/or overexpressing GLS2 could help to reset cellular metabolism toward normal cell growth, providing a new strategy to combat some types of cancer, as those having mutant isocitrate dehydrogenase 1 (IDH1) cancer cells [18]. In another investigation, stable transfection with GLS2 inhibited cell growth of T98G glioblastoma through downregulation of O6-methylguanine-DNA methyltransferase, and sensitized these cells to alkylating agents as temozolomide and carmustine. Consequently, enrichment with GLS2, in addition to inhibiting glioma growth, may facilitate chemotherapeutic intervention [36].
Reprogramming glutamine metabolism is a promising tool against cancer
Cancer often involves perturbations of normal signal transduction, including glutamine metabolism [2, 5, 13, 16]. Although metabolic profiles of tumors depend on both the genotype and tissue of origin [20], cancer cells have a very different metabolic profile compared to normal cells [21]. Of note, targeting metabolism of breast cancer cells with ATO has been a very useful strategy. Interestingly, lower sensitivity of normal cells to the concentrations of ATO tested in this study states that this agent as a promising treatment against solid tumors in vivo [24]. Given the importance of glutamine metabolism in maintaining the redox state in cancer [8] future therapeutic approaches could synergize with ATO or H2O2 treatment. In fact, when combined with H2O2, the growth-suppressive effect on pancreatic cancer cells was notably augmented, indicating that cancer cells are markedly more sensitive to ROS when glutamine metabolism is impaired and glutathione levels are reduced [37]. Hence, these authors revealed that inhibition of glutaminase in cancer cells can potentially synergize with therapies that increase intracellular ROS, for example chemotherapy and radiation [37].
Multiple nuclear proteins may regulate the transcription of the human GLS2 gene promoter [38], suggesting its possible involvement in transcription regulation [3]. Interestingly, there is a growing body of evidence pointing to glutamine as a signaling molecule involved in gene expression [8]. The significance of the nuclear GLS2 localization could be as simple as being an enzyme controlling in situ the glutamine levels in the nucleoplasm and therefore being indirectly involved in the expression of glutamine-regulated genes [3]. It is becoming clear that metabolic reprogramming network is induced by many proteins, such as IDH1, c-myc, p53, GLS, GLS2, and other molecules that act both independently and in concert with each other [39]. Therefore, RNAi-mediated downregulation of GLS and overexpression of GLS2 could be valuable tools for defining gene expression and functional programs downstream of oncogenic GLS as well as tumor suppressor GLS2 signals, which, in turn, may be helpful to isolate novel diagnostic markers, and for designing tumor-specific intervention at downstream targets of GLS and GLS2.
References
Márquez J, de la Oliva AR, Matés JM, Segura JA, Alonso FJ (2006) Glutaminase: a multifaceted protein not only involved in generating glutamate. Neurochem Int 48:465–471
DeBerardinis RJ, Lum JJ, Hatzivassiliou G, Thompson CB (2008) The biology of cancer: metabolic reprogramming fuels cell growth and proliferation. Cell Metab 7:11–20
Olalla L, Gutiérrez A, Campos JA, Khan ZU, Alonso FJ, Segura JA, Márquez J, Aledo JC (2002) Nuclear localization of L-type glutaminase in mammalian brain. J Biol Chem 277:38939–38944
Márquez J, Tosina M, de la Rosa V, Segura JA, Alonso FJ, Matés JM, Campos-Sandoval JA (2009) New insights into brain glutaminases: beyond their role on glutamatergic transmission. Neurochem Int 55:64–70
Dang CV (2009) MYC, microRNAs and glutamine addiction in cancers. Cell Cycle 8:3243–3245
Wang JB, Erickson JW, Fuji R, Ramachandran S, Gao P, Dinavahi R, Wilson KF, Ambrosio AL, Dias SM, Dang CV et al (2010) Targeting mitochondrial glutaminase activity inhibits oncogenic transformation. Cancer Cell 18:207–219
Suzuki S, Tanaka T, Poyurovsky MV, Nagano H, Mayama T, Ohkubo S, Lokshin M, Hosokawa H, Nakayama T, Suzuki Y et al (2010) Phosphate-activated glutaminase (GLS2), a p53-inducible regulator of glutamine metabolism and reactive oxygen species. Proc Natl Acad Sci U S A 107:7461–7466
Matés JM, Segura JA, Martín-Rufián M, Campos-Sandoval JA, Alonso FJ, Márquez J (2013) Glutaminase isoenzymes as key regulators in metabolic and oxidative stress against cancer. Curr Mol Med 13:514–534
Gao P, Tchernyshyov I, Chang TC, Lee YS, Kita K, Ochi T, Zeller KI, De Marzo AM, Van Eyk JE, Mendell JT et al (2009) c-Myc suppression of miR-23a/b enhances mitochondrial glutaminase expression and glutamine metabolism. Nature 458:762–765
Martín-Rufián M, Tosina M, Campos-Sandoval JA, Manzanares E, Lobo C, Segura JA, Alonso FJ, Matés JM, Márquez J (2012) Mammalian glutaminase Gls2 gene encodes two functional alternative transcripts by a surrogate promoter usage mechanism. PLoS One 7:e38380
de la Rosa V, Campos-Sandoval JA, Martín-Rufián M, Cardona C, Matés JM, Segura JA, Alonso FJ, Márquez J (2009) A novel glutaminase isoform in mammalian tissues. Neurochem Int 55:76–84
Szeliga M, Obara-Michlewska M, Matyja E, Łazarczyk M, Lobo C, Hilgier W, Alonso FJ, Marquez J, Albrecht J (2009) Transfection with liver-type glutaminase cDNA alters gene expression and reduces survival, migration and proliferation of T98G glioma cells. Glia 57:1014–1023
Cheng T, Sudderth J, Yang C, Mullen AR, Jin ES, Matés JM, DeBerardinis RJ (2011) Pyruvate carboxylase is required for glutamine-independent growth of tumor cells. Proc Natl Acad Sci U S A 108:8674–8679
Wise DR, DeBerardinis RJ, Mancuso A, Sayed N, Zhang XY, Pfeiffer HK, Nissim I, Daikhin E, Yudkoff M, McMahon SB et al (2008) Myc regulates a transcriptional program that stimulates mitochondrial glutaminolysis and leads to glutamine addiction. Proc Natl Acad Sci U S A 105:18782–18787
Hu W, Zhang C, Wu R, Sun Y, Levine A, Feng Z (2010) Glutaminase 2, a novel p53 target gene regulating energy metabolism and antioxidant function. Proc Natl Acad Sci U S A 107:7455–7460
Vander Heiden MG, Cantley LC, Thompson CB (2009) Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science 324:1029–1033
Kung HN, Marks JR, Chi JT (2011) Glutamine synthetase is a genetic determinant of cell type-specific glutamine independence in breast epithelia. PLoS Genet 7:e1002229
Seltzer MJ, Bennett BD, Joshi AD, Gao P, Thomas AG, Ferraris DV, Tsukamoto T, Rojas CJ, Slusher BS, Rabinowitz JD et al (2010) Inhibition of glutaminase preferentially slows growth of glioma cells with mutant IDH1. Cancer Res 70:8981–8987
Yuneva M, Zamboni N, Oefner P, Sachidanandam R, Lazebnik Y (2007) Deficiency in glutamine but not glucose induces MYC-dependent apoptosis in human cells. J Cell Biol 178:93–105
Yuneva MO, Fan TW, Allen TD, Higashi RM, Ferraris DV, Tsukamoto T, Matés JM, Alonso FJ, Wang C, Seo Y et al (2012) The metabolic profile of tumors depends on both the responsible genetic lesion and tissue type. Cell Metab 15:157–170
Marin-Valencia I, Yang C, Mashimo T, Cho S, Baek H, Yang XL, Rajagopalan KN, Maddie M, Vemireddy V, Zhao Z et al (2012) Analysis of tumor metabolism reveals mitochondrial glucose oxidation in genetically diverse human glioblastomas in the mouse brain in vivo. Cell Metab 15:827–837. Erratum in. Cell Metab 16:686
Maher EA, Marin-Valencia I, Bachoo RM, Mashimo T, Raisanen J, Hatanpaa KJ, Jindal A, Jeffrey FM, Choi C, Madden C et al (2012) Metabolism of [U-13 C]glucose in human brain tumors in vivo. NMR Biomed 25:1234–1244
Weinberg F, Hamanaka R, Wheaton WW, Weinberg S, Joseph J, Lopez M, Kalyanaraman B, Mutlu GM, Budinger GR, Chandel NS (2010) Mitochondrial metabolism and ROS generation are essential for Kras-mediated tumorigenicity. Proc Natl Acad Sci U S A 107:8788–8793
Sun RC, Board PG, Blackburn AC (2011) Targeting metabolism with arsenic trioxide and dichloroacetate in breast cancer cells. Mol Cancer 10:142
Matés JM, Segura JA, Alonso FJ, Márquez J (2012) Oxidative stress in apoptosis and cancer: an update. Arch Toxicol 86:1649–1665
Rahman I, Kode A, Biswas SK (2006) Assay for quantitative determination of glutathione and glutathione disulfide levels using enzymatic recycling method. Nat Protoc 1:3159–3165
Thangavelu K, Pan CQ, Karlberg T, Balaji G, Uttamchandani M, Suresh V, Schüler H, Low BC, Sivaraman J (2012) Structural basis for the allosteric inhibitory mechanism of human kidney-type glutaminase (KGA) and its regulation by Raf-Mek-Erk signaling in cancer cell metabolism. Proc Natl Acad Sci U S A 109:7705–7710
Donadio AC, Lobo C, Tosina M, de la Rosa V, Martín-Rufián M, Campos-Sandoval JA, Matés JM, Márquez J, Alonso FJ, Segura JA (2008) Antisense glutaminase inhibition modifies the O-GlcNAc pattern and flux through the hexosamine pathway in breast cancer cells. J Cell Biochem 103:800–811
Yi W, Clark PM, Mason DE, Keenan MC, Hill C, Goddard WA 3rd, Peters EC, Driggers EM, Hsieh-Wilson LC (2012) Phosphofructokinase 1 glycosylation regulates cell growth and metabolism. Science 337:975–980
Souers AJ, Leverson JD, Boghaert ER, Ackler SL, Catron ND, Chen J, Dayton BD, Ding H, Enschede SH, Fairbrother WJ et al (2013) ABT-199, a potent and selective BCL-2 inhibitor, achieves antitumor activity while sparing platelets. Nat Med 19:202–208
Liu J, Albrecht AM, Ni X, Yang J, Li M (2013) Glioblastoma tumor initiating cells: therapeutic strategies targeting apoptosis and MicroRNA pathways. Curr Mol Med 13:352–357
Szeliga M, Bogacińska-Karaś M, Różycka A, Hilgier W, Marquez J, Albrecht J (2013) Silencing of GLS and overexpression of GLS2 genes cooperate in decreasing the proliferation and viability of glioma cells. Tumor Biol. doi:10.1007/s13277-013-1247-4
Lee MJ, Ye AS, Gardino AK, Heijink AM, Sorger PK, MacBeath G, Yaffe MB (2012) Sequential application of anticancer drugs enhances cell death by rewiring apoptotic signaling networks. Cell 149:780–794
Matés JM, Segura JA, Alonso FJ, Márquez J (2011) Anticancer antioxidant regulatory functions of phytochemicals. Curr Med Chem 18:2315–2338
Katt WP, Ramachandran S, Erickson JW, Cerione RA (2012) Dibenzophenanthridines as inhibitors of glutaminase C and cancer cell proliferation. Mol Cancer Ther 11:1269–1278
Szeliga M, Zgrzywa A, Obara-Michlewska M, Albrecht J (2012) Transfection of a human glioblastoma cell line with liver-type glutaminase (LGA) down-regulates the expression of DNA-repair gene MGMT and sensitizes the cells to alkylating agents. J Neurochem 123:428–436
Son J, Lyssiotis CA, Ying H, Wang X, Hua S, Ligorio M, Perera RM, Ferrone CR, Mullarky E, Shyh-Chang N et al (2013) Glutamine supports pancreatic cancer growth through a KRAS-regulated metabolic pathway. Nature 496:101–105
Pérez-Gómez C, Matés JM, Gómez-Fabre PM, del Castillo-Olivares A, Alonso FJ, Márquez J (2003) Genomic organization and transcriptional analysis of the human L-glutaminase gene. Biochem J 370:771–784
Soga T (2013) Cancer metabolism: key players in metabolic reprogramming. Cancer Sci 104:275–281
Acknowledgements
We apologize for omissions of original references because of space limitations. Thanks are due to Rosa M. Moreno and Juan I. Rodríguez-Arranz for their valuable help in experimental tasks. This work was supported by Ministerio de Educación of Spain, PHB2010-0014-PC; Ministerio de Ciencia y Tecnología of Spain, SAF2010-17573; Junta de Andalucía, CVI-6656, Spain; PI-0825-2010, Junta de Andalucía, Spain; grant RD06/1012 of the RTA RETICS network from the Spanish Health Institute Carlos III, Spain. R.J.D. is supported by grants from the NIH (R01 CA157996), the Cancer Prevention and Research Institute of Texas (HIRP100437 and RP101243), the Robert A. Welch Foundation (I-1733), and the Damon-Runyon Cancer Research Foundation. M.S. and J.A. received support from the Ministry of Science and Higher Education (National Science Centre), grant NN401 039238. Thanks are also due to CAPES/DGU 250/11, Brazil.
Conflict of interest
The authors declare no conflict of interests related to this study.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Martín-Rufián, M., Nascimento-Gomes, R., Higuero, A. et al. Both GLS silencing and GLS2 overexpression synergize with oxidative stress against proliferation of glioma cells. J Mol Med 92, 277–290 (2014). https://doi.org/10.1007/s00109-013-1105-2
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00109-013-1105-2