Abstract
The serine/threonine kinase homeodomain-interacting protein kinase (HIPK2) is a tumor suppressor and functions as an evolutionary conserved regulator of signaling and gene expression. This kinase regulates a surprisingly vast array of biological processes that range from the DNA damage response and apoptosis to hypoxia signaling and cell proliferation. Recent studies show the tight control of HIPK2 by hierarchically occurring posttranslational modifications such as phosphorylation, small ubiquitin-like modifier modification, acetylation, and ubiquitination. The physiological function of HIPK2 as a regulator of cell proliferation and survival has a downside: proliferative diseases. Dysregulation of HIPK2 can result in increased proliferation of cell populations as it occurs in cancer or fibrosis. We discuss various models that could explain how inappropriate expression, modification, or localization of HIPK2 can be a driver for these proliferative diseases.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Architecture and functions of HIPK2
The activities of enzymes mediating posttranslational modifications (PTMs) such as protein kinases are themselves often regulated by PTMs, thus creating highly wired and complex signaling networks. The systematic analysis of cancer genomes for mutated genes revealed a strong overrepresentation of kinases [1]. One of the kinases found to be dysregulated in cancer is the homeodomain-interacting protein kinase (HIPK2) [2].
Already, the identification of HIPK2 as an interactor of the homeodomain transcription factor NKx-1.2, suggested its role in transcriptional regulation [3]. Meanwhile, more HIPK2 interaction partners have been found which can be roughly divided in two groups. Group 1 includes a bewildering long and continuously growing list of transcription factors and accessory proteins of the transcriptional machinery. Group 2 consists a heterogeneous group of signaling proteins that have enzymatic functions (e.g., as kinases, acetyl transferases, or ubiquitin E3 ligases) or function as scaffolding proteins (e.g., Han11 and Axin) [4, 5]. Accordingly, HIPK2 serves to augment or to repress gene expression. HIPK2 has been implicated in a perplexingly large group of different signaling pathways. These include the Salvador–Warts–Hippo pathway, BMP, Wnt/Wingless and Notch signaling, Redox regulation, p53 activation, as well as TGFβ and hypoxia signaling [4–6]. All studies share the finding that HIPK2 does not function as a highly connected and essential canonical core component of the respective signaling pathway, but rather as an accessory regulator. The function of HIPK2 could be interpreted as an auxiliary protein that serves to shape the signal output from the signaling cascade and/or as a signal integrator that acts to connect different signaling pathways. The multitude of HIPK2 interactors is also reflected by a complex domain architecture, raising the possibility that this kinase may also function as a scaffold protein. HIPK2 and the other two HIPK family members HIPK1 and HIPK3 share an N-terminal kinase domain with >90 % sequence homology [3]. The C-terminal part contains an interaction domain for the homeodomain transcription factors and a PEST motif that overlap with a region containing a small ubiquitin-like modifier (SUMO)-interacting motif (SIM). These regions are followed by an autoinhibitory domain and a C-terminal region that is rich in short repeats of S, Q, or A.
Physiological functions of HIPK2
Cell proliferation and survival
Mice lacking the Hipk2 gene are not born in Mendelian ratios, but approximately 40 % die in the first 3 days after birth. The surviving animals show a reduced weight gain and remain abnormally small [7–9]. These data suggest a role of HIPK2 for cell proliferation, but imply also the occurrence of compensatory mechanisms that allow survival of a fraction of HIPK2−/− mice. These compensatory events could also account for the inconsistent results obtained from the proliferation analysis of HIPK2−/− mouse embryonic fibroblasts (MEFs). While one study revealed an increased proliferation rate of HIPK2−/− MEFs [10], a second study showed a decreased proliferation of HIPK2-deficient MEFs [9]. Since cultured cells can cope with the effects of gene deletions by employing alternative signaling pathways, inducible knockout systems and the use of non-immortalized primary cells will help to clarify this issue. An important function of HIPK2 in cell proliferation is corroborated by a study where shRNA-mediated knockdown of HIPK2 in erythroid precursor cells was shown to inhibit terminal erythropoiesis [11]. In any case, the overexpression of HIPK2 frequently leads to a cell cycle stop and to the induction of cell death [12, 13], showing that the amount of this kinase is of critical importance. A recent study revealed fascinating insights into the mechanisms employed by HIPK2 to control cell division [8]. This study shows that HIPK2 controls cytokinesis and prevents tetraploidization by binding and phosphorylating histone H2B at the midbody, a transient structure connecting two daughter cells at the end of cytokinesis. Depletion of HIPK2 by gene knockout or RNA interference results in absent phosphorylation of H2B at Ser14, a modification that is required for efficient cell cleavage [8].
The cell death-regulating function of HIPK2 is mediated by several mechanisms. Classical p53-induced apoptosis is enhanced by the HIPK2-mediated phosphorylation of p53 at Ser46 which allows the subsequent CBP-mediated p53 acetylation and results in induced expression of proapoptotic genes [12, 13]. P53-independent pathways include HIPK2-mediated phosphorylation and degradation of the transcriptional corepressor CtBP which in turn allows upregulation of proapoptotic genes and cell death [14]. The in vivo role of HIPK2 in the control of cell death was also revealed by the characterization of knockout mice. HIPK2-deficient animals have increased expression of antiapoptotic Brn3a target genes and show increased survival of neurons in the trigeminal ganglion [15]. While these studies collectively show a proapoptotic function of HIPK2, the kinase has also opposing functions in specialized cell types. HIPK2-deficient mice exhibit a progressive loss of enteric neurons during postnatal development, which does not involve apoptosis but rather depends on autophagy [7].
Differentiation
The function of HIPK2 as a regulator of cell proliferation and survival is closely linked to its contribution to cell differentiation. In Drosophila, HIPK2 (dHIPK2) regulates eye development in the fly by targeting the corepressor Groucho. DHIPK2-mediated phosphorylation of Groucho has been shown to attenuate its contact to DNA-bound transcription factors and thereby altering gene expression [16, 17]. In this regard, dHIPK2 promotes the Notch pathway which acts at multiple points in eye development by antagonizing the gene repressing activity of Groucho and inactivation of the Dhipk2 gene results in small, rough eyes and pupal lethality with rare escaper adults [18]. The role of HIPK2 for eye development is also seen in mammals, as HIPK2−/− HIPK1+/− mice often show small eyes with lens deficiency and abnormally thickened and laminated retinas [19]. HIPK2 knockout mice have been also thoroughly characterized for defects in neuronal development. HIPK2 is required for postnatal development of enteric dopaminergic neurons via TGFβ-induced BMP signaling [20]. The analysis of HIPK1/2 double-deficient mouse embryos revealed the occurrence of homoeotic transformations at the axial skeleton and defects in the closure of the dorsal neural tube [19, 21].
The multitude of biological processes employing HIPK2 raises the question how a single kinase can participate in so many different signaling pathways. One widely used principle is the modification of signaling proteins by PTMs, which control the formation of protein/protein interactions and influence the stability, enzymatic activity or localization of a protein.
Regulation of HIPK2 by PTMs
Since PTMs act as reversible molecular switchboards, the function of HIPK2 can only be understood by deciphering these protein-encoded mechanisms and needs mapping of PTM patterns which provide a “protein code” to control numerous functions. Two recent studies describe the modification of HIPK2 by multiple phosphorylations throughout the entire kinase [22, 23]. While mass spectrometric analysis of GST-tagged wild-type HIPK2 revealed 23 phosphorylation sites [23], the analysis of immunoprecipitated Flag-tagged HIPK2 allowed the identification of 17 modification sites [22]. A comparison of these published sites with data retrieved from the public PhosphoSitePlus© database is given in Fig. 1 and shows that 10 of these sites are found in at least two different and independent experimental settings. Both studies show that the vast majority of phosphorylation sites that were identified in unstimulated cells are mediated by autophosphorylation. What is the function of these modification sites? Detailed information are now available for the activation loop which is contained within the kinase domain and which needs to be phosphorylated at key amino acids to allow correct positioning of catalytic groups [24]. HIPK2 phosphorylation in the activation loop occurs at Tyr354 and Ser357 [22, 23]. Tyr354 is well conserved between HIPK2 and related kinases (Fig. 2) and functional experiments show that this phosphorylation proceeds via an intramolecular mechanism. In contrast to DYRK2 where tyrosine cis-autophosphorylation is essential for its kinase activity [25], the tyrosine autophosphorylation of HIPK2 is not an absolute prerequisite for its enzymatic activity. Mutation of Tyr354 rather results in out-of-target phosphorylation activity and cytoplasmic relocalization of the kinase [23]. Only mutation of both phosphorylated activation loop residues at Tyr354 and Ser357 precluded HIPK2 kinase activity and its effects on gene expression [22]. These results imply that the kinase is already produced in a constitutively active form that is independent from upstream activating kinases. This also raises the need to control the activity of HIPK2 by other mechanisms that are discussed below. The possible functions of the phosphorylation sites outside from the activation loop are not known. GST pulldown experiments showed the necessity of HIPK2 phosphorylation for binding to the prolyl isomerase Pin1 [22]. Since Pin1 is also important for HIPK2-mediated phosphorylation of p53 at Ser46 [26], these data suggest that one or several of these phosphorylation sites can control Pin1-dependent regulation of HIPK2 conformation.
Identification of HIPK2 phosphorylation sites. The structure of HIPK2 is schematically shown at the left; the location of the autoinhibitory domain (AID) and the speckle retention signal (SRS) are indicated. The various basal phosphorylation sites identified in unstimulated cells by published papers [22, 23] and retrieved from the public PhosphoSitePlus© database are displayed
Sequence alignment of activation loop segments of HIPK2 and related kinases. Amino acid sequence alignment of the indicated kinases was performed using the ClustalW2 program. Identical amino acids are marked with an asterisk and similar residues with a colon. The conserved catalytic aspartate residue in the catalytic loop is marked. The DFG and APE motifs of the activation loop are shown in red
Differential conformation of HIPK2 could also explain the observation that HIPK2 phosphorylation is a prerequisite for modification of HIPK2 by SUMOylation [27] and acetylation (M.L.S., unpublished). Attachment of SUMO1 to Lys25 of HIPK2 is promoted by the SUMO E3 ligase Pc2. HIPK2-mediated inducible phosphorylation of Pc2 is required for its ability to increase HIPK2 SUMOylation in response to DNA damage and thus establishes an autoregulatory feedback loop between the kinase and its cognate E3 ligase [27]. SUMO modification of HIPK2 is neither required for its localization in nuclear bodies nor for its recruitment to promyelocytic leukemia nuclear bodies (PML-NBs) [28]. But SUMO proteins cannot only be conjugated covalently to target lysines as they also have the ability to bind proteins noncovalently to SIMs. Also HIPK2 contains a SIM in its C-terminus [29] and mutation or deletion of the SIM precludes localization of the kinase in nuclear speckles and recruitment to PML-NBs [30]. A further study demonstrated the importance of the SIM for the ability of overexpressed HIPK2 to suppress cell proliferation [31].
While phosphorylation of HIPK2 is required for its SUMOylation, attachment of SUMO itself controls the acetylation status [32], thus providing a demonstrative example how PTMs can occur in a hierarchical fashion. SUMOylated HIPK2 is found in constitutive association with the histone deacetylase (HDAC)3, which ensures the deacetylation of HIPK2 in unstressed cells. Elevated levels of reactive oxygen species (ROS) lead to the non-enzymatic deconjugation of SUMO due to reversible and direct inhibition of SUMO-conjugating enzymes [33]. Accordingly, elevated ROS levels lead to the de-SUMOylation of HIPK2 which in turn reduce its association with HDAC3 and allow CBP/p300-mediated acetylation of HIPK2 at 10 lysines. Functional experiments in reconstituted knockout cells showed that cells lacking HIPK2 or harboring the acetylation mimicking HIPK2-10KQ are largely protected from H2O2-triggered cell death, thus revealing that HIPK2 acetylation protects from ROS-triggered apoptosis. These data also imply that the threshold leading to ROS-triggered cell demise is controlled by HIPK2 acetylation. Since tumor cells frequently show elevated ROS levels, HIPK2 acetylation may ensure their survival even under these adverse conditions as discussed in more detail below. Two acetylated residues are contained in the nuclear localization signal (NLS)1. Accordingly, a modification-specific antibody specifically recognizing the acetylated NLS1 showed that the acetylated kinase does not localize in nuclear speckles but rather in the nucleoplasm [32].
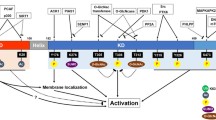
The recent years have provided mounting evidence that the relative abundance of HIPK2 is tightly controlled by degradative ubiquitination. Four different ubiquitin E3 ligases (MDM2, Siah, WSB-1, and Fbx3-formed SCF) ensure the tight limitation of HIPK2 amounts [34–37]. These different E3 ligases can be activated to degrade HIPK2 in different physiological settings. Under conditions of nonsevere DNA damage (where p53 is not phosphorylated at Ser46), activation of p53 allows expression of MDM2 which in turn mediates ubiquitin/proteasome-dependent degradation of HIPK2 [36]. Continuous HIPK2 ubiquitination in unstressed cells is also mediated by Siah1 and WSB-1, thus ensuring efficient restriction of HIPK2 levels under these conditions. Under conditions of genotoxic stress, WSB-1-mediated HIPK2 ubiquitination is blocked by an unknown pathway, while inducible phosphorylation of Siah1 by ATM/ATR leads to the disruption of the HIPK2/Siah1 complex. This in turn results in HIPK2 stabilization, p53 Ser46 phosphorylation, and the induction of cell death [38], as schematically depicted in Fig. 3. Another physiologically relevant setting resulting in HIPK2 degradation is hypoxia. Low oxygen availability leads to the induction of the ubiquitin E3 ligase Siah2 which binds to HIPK2 and causes its efficient polyubiquitination and proteasomal degradation. As HIPK2 is contained in gene-repressing complexes that prevent the expression of specific genes under normoxic conditions, hypoxia-induced proteasomal elimination of the kinase allows full induction of hypoxia-triggered genes [34]. MDM2-mediated HIPK2 ubiquitination has been mapped to Lys1182 in mouse HIPK2 [36], but the lysines modified by the other ubiquitin E3 ligases are not known and should be identified in the future. Another important future task is the identification of PTMs occurring in primary cells and under conditions known to regulate HIPK2 activity.
Regulation of HIPK2 stability. In unstressed cells HIPK2 is constantly degraded through complex formation with the ubiquitin E3 ligases Siah1, WSB-1, and Fbx3 which target HIPK2 for proteasomal degradation. In response to genotoxic stress, the checkpoint kinases ATM and ATR are activated and mediate the phosphorylation of Siah1, resulting in its dissociation from HIPK2. Nonsevere DNA damage leads to cell cycle arrest and recovery, whereas HIPK2-mediated phosphorylation of p53 at Ser46 promotes apoptosis
Pathophysiological functions of HIPK2 in proliferative diseases
Dysregulation of HIPK2 frequently results in increased proliferation as it is typical in cancer or fibrosis, a process that is characterized by the excessive formation of fibrous tissue [39, 40]. A recently published systematic approach identified HIPK2 as a key regulator of kidney fibrosis [41]. Three different animal models for this disease allowed the identification of this kinase as a driver of kidney fibrosis: HIV-associated nephropathy, unilateral ureteral obstruction, and folic acid-induced renal fibrosis. The starting point for this study was the HIV transgenic mouse model (Tg26), where the proviral transgene recapitulates the renal pathological changes observed in the human disease. The determination of differentially regulated genes was followed by computational analysis of involved transcription factors and protein–protein interaction subnetworks, thus leading to the identification of HIPK2 as a dysregulated candidate kinase. Evidence for a causative role of HIPK2 in kidney fibrosis was revealed by the analysis of knockout transgenic hybrid mice that express the HIV transgene and lack the HIPK2 genes (KO-Tg26 mice). While Tg26 mice develop tubulointerstitial injury and fibrosis and are characterized by proteinuria and lower concentrations of serum urea nitrogen, the KO-Tg26 animals are largely protected from these symptoms [41]. The kidneys of Tg26 mice show upregulated levels of HIPK2 proteins which inversely correlates with the levels of Siah1, one of the ubiquitin E3 ligases, leading to the degradation of HIPK2. As kidneys of Tg26 mice show elevated levels of ROS, it would be interesting to determine whether also the acetylation status of HIPK2 is changed in this disease model. Another recent study implicated dysregulated HIPK2 levels in idiopathic pulmonary fibrosis (IPF), a disease that is characterized by aberrant proliferation of lung fibroblasts, tissue remodeling, and extracellular matrix deposition [42]. The analysis of HIPK2 expression in IPF samples and in primary fibroblast cell cultures showed the frequent occurrence of loss of heterozygosity (LOH) for the HIPK2 locus at 7q32-34 and concomitantly lower levels of mRNA and protein [42]. While kidney fibrosis was triggered by overexpression of HIPK2, IPF was accompanied by a diminished expression of the kinase.
The role of HIPK2 as a tumor-suppressing kinase was initially revealed by the analysis of HIPK2−/− mice. Although these animals do not develop spontaneous tumors, treatment with a skin carcinogenesis protocol results in elevated formation of skin tumors [10]. Increased tumor formation already appeared in heterozygous mice, suggesting that HIPK2 functions as a haploinsufficient tumor suppressor. The tumor-suppressing function of HIPK2 was also recapitulated in cell culture models, since expression of a constitutively active form of H-ras induced enlarged colonies in HIPK2−/− MEFs [10]. In addition, an unbiased genome-wide screen for genetic alterations in radiation-induced thymic lymphomas generated from p53+/− and p53−/− mice corroborated HIPK2s tumor-suppressing effect and revealed a frequent LOH for the Hipk2 gene [43]. Of note, point mutations of HIPK2 are only rarely observed [44] and might not even be the cause for tumor development but rather be the consequence of high mutation rates and genomic instabilities as they are typical for cancer cells [45]. A number of studies describe decreased expression levels of HIPK2 in various tumors including thyroid and breast carcinomas [46], papillary thyroid carcinomas, and follicular thyroid carcinomas [47]. These finding are in line with the idea that LOH or impaired expression facilitates tumor growth and accordingly, colon cancers with increased HIPK2 expression have a better outcome than tumors with low expression [48]. In marked contrast, a number of studies show elevated expression of HIPK2 in tumors such as pilocytic astrocytoma [49, 50] and cervical cancer [51].
Mechanisms mediating pathophysiological inactivation of HIPK2
How can overexpression of a tumor suppressor favor cell proliferation? This situation resembles the situation occurring in fibrosis where both over- and underexpression of HIPK2 were reported. These seemingly contradictory findings might be explained by various models which are discussed here. Although the limited quality of commercially available HIPK2-detecting antibodies may handicap the interpretation of protein expression data in tumor samples, the frequent observation of dysregulated protein amounts could mean that only optimal amounts of HIPK2 are functional. This “optimum model” is schematically depicted in Fig. 4a and implies that higher concentrations of HIPK2 are less active, possibly due to the formation of inactive protein aggregates or misfolded HIPK2. Another important aspect was provided by several studies that revealed the importance of intracellular HIPK2 localization. This “localization model” is schematically shown in Fig. 4b and reflects results from a number of studies showing the delocalization of HIPK2 in cancer. Overexpression of the high-mobility group A1 protein as it frequently happens in tumor cells causes the relocalization of HIPK2 from nucleus speckles to the cytosol [52]. Another example for an aberrant HIPK2 localization comes from AML1(RUNX1)-associated leukemic cells expressing the oncogenic fusion protein PEBP2β-SMMHC. Expression of this fusion protein sequesters HIPK2 to filamentous structures in the cytosol and thus interferes with its functions in the nucleus [53]. The recent progress in the identification and functional characterization of PTMs regulating HIPK2 function enable the development of tools that allow the convenient determination of HIPK2 activity. The “PTM model” (Fig. 4c) reflects the recent findings that modifications of HIPK2 regulate its numerous activities. It will be therefore interesting in future studies to determine the modification status of HIPK2 in patient material. One particular aspect of HIPK2 regulation might be of special relevance in solid tumors such as glioblastomas which are characterized by hypoxic areas. Insufficient vascularization and blood supply is accompanied by low oxygen availability and induction of the ubiquitin E3 ligase Siah2 which binds to HIPK2 and causes its efficient polyubiquitination and proteasomal degradation. HIPK2 downregulation under hypoxic conditions also impairs DNA damage-induced p53 Ser46 phosphorylation [34]. While these findings suggests a molecular mechanism by which hypoxic cancer cells can escape chemotherapeutic drug treatment [34], a confirmatory follow-up study revealed that HIPK2 expression and p53 Ser46 phosphorylation in hypoxic cells can be rescued by inhibition of the proteasome [54]. Whether also HIPK2 is a suitable target for drugs causing its specific inhibition or activation remains to be studied in the future.
References
Bignell GR, Greenman CD, Davies H, Butler AP, Edkins S, Andrews JM, Buck G, Chen L, Beare D, Latimer C et al (2010) Signatures of mutation and selection in the cancer genome. Nature 463:893–898
Puca R, Nardinocchi L, Givol D, D'Orazi G (2010) Regulation of p53 activity by HIPK2: molecular mechanisms and therapeutical implications in human cancer cells. Oncogene 29:4378–4387
Kim YH, Choi CY, Lee SJ, Conti MA, Kim Y (1998) Homeodomain-interacting protein kinases, a novel family of co-repressors for homeodomain transcription factors. J Biol Chem 273:25875–25879
Rinaldo C, Siepi F, Prodosmo A, Soddu S (2008) HIPKs: jack of all trades in basic nuclear activities. Biochim Biophys Acta 1783:2124–2129
Calzado MA, Renner F, Roscic A, Schmitz ML (2007) HIPK2: a versatile switchboard regulating the transcription machinery and cell death. Cell Cycle 6:139–143
Poon CL, Zhang X, Lin JI, Manning SA, Harvey KF (2012) Homeodomain-interacting protein kinase regulates hippo pathway-dependent tissue growth. Curr Biol. doi:10.1016/j.cub.2012.06.075
Chalazonitis A, Tang AA, Shang Y, Pham TD, Hsieh I, Setlik W, Gershon MD, Huang EJ (2011) Homeodomain interacting protein kinase 2 regulates postnatal development of enteric dopaminergic neurons and glia via BMP signaling. J Neurosci 31:13746–13757
Rinaldo C, Moncada A, Gradi A, Ciuffini L, D'Eliseo D, Siepi F, Prodosmo A, Giorgi A, Pierantoni GM, Trapasso F et al (2012) HIPK2 controls cytokinesis and prevents tetraploidization by phosphorylating histone H2B at the midbody. Mol Cell 47:87–98
Trapasso F, Aqeilan RI, Iuliano R, Visone R, Gaudio E, Ciuffini L, Alder H, Paduano F, Pierantoni GM, Soddu S et al (2009) Targeted disruption of the murine homeodomain-interacting protein kinase-2 causes growth deficiency in vivo and cell cycle arrest in vitro. DNA Cell Biol 28:161–167
Wei G, Ku S, Ma GK, Saito S, Tang AA, Zhang J, Mao JH, Appella E, Balmain A, Huang EJ (2007) HIPK2 represses beta-catenin-mediated transcription, epidermal stem cell expansion, and skin tumorigenesis. Proc Natl Acad Sci U S A 104:13040–13045
Hattangadi SM, Burke KA, Lodish HF (2010) Homeodomain-interacting protein kinase 2 plays an important role in normal terminal erythroid differentiation. Blood 115:4853–4861
D'Orazi G, Cecchinelli B, Bruno T, Manni I, Higashimoto Y, Saito S, Gostissa M, Coen S, Marchetti A, Del Sal G et al (2002) Homeodomain-interacting protein kinase-2 phosphorylates p53 at Ser 46 and mediates apoptosis. Nat Cell Biol 4:11–19
Hofmann TG, Moller A, Sirma H, Zentgraf H, Taya Y, Droge W, Will H, Schmitz ML (2002) Regulation of p53 activity by its interaction with homeodomain-interacting protein kinase-2. Nat Cell Biol 4:1–10
Zhang Q, Yoshimatsu Y, Hildebrand J, Frisch SM, Goodman RH (2003) Homeodomain interacting protein kinase 2 promotes apoptosis by downregulating the transcriptional corepressor CtBP. Cell 115:177–186
Wiggins AK, Wei G, Doxakis E, Wong C, Tang AA, Zang K, Luo EJ, Neve RL, Reichardt LF, Huang EJ (2004) Interaction of Brn3a and HIPK2 mediates transcriptional repression of sensory neuron survival. J Cell Biol 167:257–267
Choi CY, Kim YH, Kwon HJ, Kim Y (1999) The homeodomain protein NK-3 recruits Groucho and a histone deacetylase complex to repress transcription. J Biol Chem 274:33194–33197
Choi CY, Kim YH, Kim YO, Park SJ, Kim EA, Riemenschneider W, Gajewski K, Schulz RA, Kim Y (2005) Phosphorylation by the DHIPK2 protein kinase modulates the corepressor activity of Groucho. J Biol Chem 280:21427–21436
Lee W, Andrews BC, Faust M, Walldorf U, Verheyen EM (2009) Hipk is an essential protein that promotes Notch signal transduction in the Drosophila eye by inhibition of the global co-repressor Groucho. Dev Biol 325:263–272
Inoue T, Kagawa T, Inoue-Mochita M, Isono K, Ohtsu N, Nobuhisa I, Fukushima M, Tanihara H, Taga T (2010) Involvement of the Hipk family in regulation of eyeball size, lens formation and retinal morphogenesis. FEBS Lett 584:3233–3238
Zhang Q, Wang Y (2007) Homeodomain-interacting protein kinase-2 (HIPK2) phosphorylates HMGA1a at Ser-35, Thr-52, and Thr-77 and modulates its DNA binding affinity. J Proteome Res 6:4711–4719
Aikawa Y, Nguyen LA, Isono K, Takakura N, Tagata Y, Schmitz ML, Koseki H, Kitabayashi I (2006) Roles of HIPK1 and HIPK2 in AML1- and p300-dependent transcription, hematopoiesis and blood vessel formation. EMBO J 25:3955–3965
Saul VV, de la Vega L, Milanovic M, Krüger M, Braun T, Fritz-Wolf K, Becker K, Schmitz ML (2013) HIPK2 kinase activity depends on cis-autophosphorylation of its activation loop. J Mol Cell Biol 5:27–38
Siepi F, Gatti V, Camerini S, Crescenzi M, Soddu S (2013) Homeodomain-interacting protein kinase 2 (HIPK2) catalytic activity and specificity are regulated by activation-loop Y354 autophosphorylation. Biochim Biophys Acta 1833:1443–1453
Johnson LN, Noble ME, Owen DJ (1996) Active and inactive protein kinases: structural basis for regulation. Cell 85:149–158
Lochhead PA, Sibbet G, Morrice N, Cleghon V (2005) Activation-loop autophosphorylation is mediated by a novel transitional intermediate form of DYRKs. Cell 121:925–936
Grison A, Mantovani F, Comel A, Agostoni E, Gustincich S, Persichetti F, Del Sal G (2011) Ser46 phosphorylation and prolyl-isomerase Pin1-mediated isomerization of p53 are key events in p53-dependent apoptosis induced by mutant huntingtin. Proc Natl Acad Sci U S A 108:17979–17984
Roscic A, Moller A, Calzado MA, Renner F, Wimmer VC, Gresko E, Ludi KS, Schmitz ML (2006) Phosphorylation-dependent control of Pc2 SUMO E3 ligase activity by its substrate protein HIPK2. Mol Cell 24:77–89
Hofmann TG, Jaffray E, Stollberg N, Hay RT, Will H (2005) Regulation of homeodomain-interacting protein kinase 2 (HIPK2) effector function through dynamic small ubiquitin-related modifier-1 (SUMO-1) modification. J Biol Chem 280:29224–29232
Engelhardt OG, Boutell C, Orr A, Ullrich E, Haller O, Everett RD (2003) The homeodomain-interacting kinase PKM (HIPK-2) modifies ND10 through both its kinase domain and a SUMO-1 interaction motif and alters the posttranslational modification of PML. Exp Cell Res 283:36–50
de la Vega L, Frobius K, Moreno R, Calzado MA, Geng H, Schmitz ML (2011) Control of nuclear HIPK2 localization and function by a SUMO interaction motif. Biochim Biophys Acta 1813:283–297
Sung KS, Lee YA, Kim ET, Lee SR, Ahn JH, Choi CY (2011) Role of the SUMO-interacting motif in HIPK2 targeting to the PML nuclear bodies and regulation of p53. Exp Cell Res 317:1060–1070
de la Vega L, Grishina I, Moreno R, Kruger M, Braun T, Schmitz ML (2012) A redox-regulated SUMO/acetylation switch of HIPK2 controls the survival threshold to oxidative stress. Mol Cell 46:472–483
Bossis G, Melchior F (2006) Regulation of SUMOylation by reversible oxidation of SUMO conjugating enzymes. Mol Cell 21:349–357
Calzado MA, de la Vega L, Moller A, Bowtell DD, Schmitz ML (2009) An inducible autoregulatory loop between HIPK2 and Siah2 at the apex of the hypoxic response. Nat Cell Biol 11:85–91
Choi DW, Seo YM, Kim EA, Sung KS, Ahn JW, Park SJ, Lee SR, Choi CY (2008) Ubiquitination and degradation of homeodomain-interacting protein kinase 2 by WD40 repeat/SOCS box protein WSB-1. J Biol Chem 283:4682–4689
Rinaldo C, Prodosmo A, Mancini F, Iacovelli S, Sacchi A, Moretti F, Soddu S (2007) MDM2-regulated degradation of HIPK2 prevents p53Ser46 phosphorylation and DNA damage-induced apoptosis. Mol Cell 25:739–750
Shima Y, Shima T, Chiba T, Irimura T, Pandolfi PP, Kitabayashi I (2008) PML activates transcription by protecting HIPK2 and p300 from SCFFbx3-mediated degradation. Mol Cell Biol 28:7126–7138
Winter M, Sombroek D, Dauth I, Moehlenbrink J, Scheuermann K, Crone J, Hofmann TG (2008) Control of HIPK2 stability by ubiquitin ligase Siah-1 and checkpoint kinases ATM and ATR. Nat Cell Biol 10:812–824
Wynn TA (2011) Integrating mechanisms of pulmonary fibrosis. J Exp Med 208:1339–1350
Wallace K, Burt AD, Wright MC (2008) Liver fibrosis. Biochem J 411:1–18
Jin Y, Ratnam K, Chuang PY, Fan Y, Zhong Y, Dai Y, Mazloom AR, Chen EY, D'Agati V, Xiong H et al (2012) A systems approach identifies HIPK2 as a key regulator of kidney fibrosis. Nat Med 18:580–588
Ricci A, Cherubini E, Ulivieri A, Lavra L, Sciacchitano S, Scozzi D, Mancini R, Ciliberto G, Bartolazzi A, Bruno P et al (2012) Homeodomain-interacting protein kinase2 in human idiopathic pulmonary fibrosis. J Cell Physiol. doi:10.1002/jcp.24129
Mao JH, Wu D, Kim IJ, Kang HC, Wei G, Climent J, Kumar A, Pelorosso FG, DelRosario R, Huang EJ et al (2012) Hipk2 cooperates with p53 to suppress gamma-ray radiation-induced mouse thymic lymphoma. Oncogene 31:1176–1180
Li XL, Arai Y, Harada H, Shima Y, Yoshida H, Rokudai S, Aikawa Y, Kimura A, Kitabayashi I (2007) Mutations of the HIPK2 gene in acute myeloid leukemia and myelodysplastic syndrome impair AML1- and p53-mediated transcription. Oncogene 26:7231–7239
Bakhoum SF, Compton DA (2012) Chromosomal instability and cancer: a complex relationship with therapeutic potential. J Clin Invest 122:1138–1143
Pierantoni GM, Bulfone A, Pentimalli F, Fedele M, Iuliano R, Santoro M, Chiariotti L, Ballabio A, Fusco A (2002) The homeodomain-interacting protein kinase 2 gene is expressed late in embryogenesis and preferentially in retina, muscle, and neural tissues. Biochem Biophys Res Commun 290:942–947
Lavra L, Rinaldo C, Ulivieri A, Luciani E, Fidanza P, Giacomelli L, Bellotti C, Ricci A, Trovato M, Soddu S et al (2011) The loss of the p53 activator HIPK2 is responsible for galectin-3 overexpression in well differentiated thyroid carcinomas. PLoS One 6:e20665
Soubeyran I, Mahouche I, Grigoletto A, Leste-Lasserre T, Drutel G, Rey C, Pedeboscq S, Blanchard F, Brouste V, Sabourin JC et al (2011) Tissue microarray cytometry reveals positive impact of homeodomain interacting protein kinase 2 in colon cancer survival irrespective of p53 function. Am J Pathol 178:1986–1998
Deshmukh H, Yeh TH, Yu J, Sharma MK, Perry A, Leonard JR, Watson MA, Gutmann DH, Nagarajan R (2008) High-resolution, dual-platform aCGH analysis reveals frequent HIPK2 amplification and increased expression in pilocytic astrocytomas. Oncogene 27:4745–4751
Yu J, Deshmukh H, Gutmann RJ, Emnett RJ, Rodriguez FJ, Watson MA, Nagarajan R, Gutmann DH (2009) Alterations of BRAF and HIPK2 loci predominate in sporadic pilocytic astrocytoma. Neurology 73:1526–1531
Cheng Y, Al-Beiti MA, Wang J, Wei G, Li J, Liang S, Lu X (2012) Correlation between homeodomain-interacting protein kinase 2 and apoptosis in cervical cancer. Mol Med Report 5:1251–1255
Pierantoni GM, Rinaldo C, Mottolese M, Di Benedetto A, Esposito F, Soddu S, Fusco A (2007) High-mobility group A1 inhibits p53 by cytoplasmic relocalization of its proapoptotic activator HIPK2. J Clin Invest 117:693–702
Wee HJ, Voon DC, Bae SC, Ito Y (2008) PEBP2-beta/CBF-beta-dependent phosphorylation of RUNX1 and p300 by HIPK2: implications for leukemogenesis. Blood 112:3777–3787
Moehlenbrink J, Bitomsky N, Hofmann TG (2010) Hypoxia suppresses chemotherapeutic drug-induced p53 Serine 46 phosphorylation by triggering HIPK2 degradation. Cancer Lett 292:119–124
Acknowledgments
This work was supported by grants from the German Research Foundation projects SCHM 1417/4-2, SCHM 1417/7-1, SCHM 1417/8-1, GRK 1566/1, SFB/TRR 81, the Excellence Cluster Cardio-Pulmonary System (ECCPS), German Academic Exchange Service (A/08/98404) and the LOEWE/UGMLC program.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Saul, V.V., Schmitz, M.L. Posttranslational modifications regulate HIPK2, a driver of proliferative diseases. J Mol Med 91, 1051–1058 (2013). https://doi.org/10.1007/s00109-013-1042-0
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00109-013-1042-0