Abstract
With the groundbreaking work of Takahashi and Yamanaka, induced pluripotent stem cells (iPSCs) have taken the stage of international stem cell research as a novel source of pluripotent cells and an alternative to embryonic stem cells (ESCs). Apart from their enormous potential as a starting source for the generation of patient-specific cell therapy products, iPSCs also highlight the power of artificially modulating transcriptional networks to induce dramatic changes of cell specification. Since small non-coding RNAs play important roles in the modulation and fine-tuning of transcriptional networks, microRNAs also exhibit important functions in directing cell fate decisions. In this review, we will discuss the role of microRNAs in pluripotent stem cells and their impact on the induction of pluripotency during reprogramming of somatic cells.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Pluripotent stem cells
Pluripotent stem cells (PSCs) are defined by their ability to differentiate into cells of all three germ layers and thus have the capacity to generate all somatic cells of a given organism. In addition, PSCs exhibit a seemingly unrestricted self-renewal potential. These unique features are enabled by a global epigenetic signature facilitating expression of a highly complex and intertwined set of factors, which suppress differentiation and allow for rapid cell cycling. These transcription factors act in a partially concerted manner and often co-occupy genomic loci. Moreover, the three main players of this network, Oct4, Sox2, and Nanog, collaboratively induce their own expression in an autoregulatory positive feedback loop, thereby mutually sustaining their expression [1].
Given their enormous differentiation as well as proliferation potential, PSCs hold a great promise for regenerative medicine, since they potentially can replenish all cells which are lost during acute or chronic diseases associated with profound cell death such as myocardial infarction, stroke, or hepatic failure; autoimmune disorders like diabetes type I or multiple sclerosis; or neurodegenerative conditions like Parkinson's and Huntington's disease. Likewise, PSCs represent a highly attractive target population for gene therapy approaches, as the genetic repair of initially only few cells would allow the generation of large quantities of gene-corrected differentiated progeny. This strategy currently is explored for a number of congenital defects including muscular dystrophies, hemoglobinopathies, lipid and glycogen storage diseases, or immunodeficiencies [2, 3].
Pluripotent stem cells originally were obtained from pre- or early post-implantation blastocysts as embryonic stem cells (ESCs), and under appropriate conditions, they can be propagated in vitro without losing their differentiation potential. Although first-in-human clinical trials involving human ESCs have already been initiated, the development of ESC-based therapies faces a number of ethical as well as biological problems. First, ESC generation depends on the destruction of surplus embryos—a fact that is diversely discussed with regard to ethical reasons. Furthermore, hESCs cannot be derived in a patient-specific manner and thus may evoke immunogenic reactions upon transplantation into a host [4]. In this context, induced pluripotent stem cells (iPSCs) appear as a promising alternative to ESCs as they can easily be derived in a patient-specific manner and suitable target cells can be procured easily. The generation of iPSCs through transcription factor-mediated reprogramming of somatic cells originally was performed from fibroblast cells with the four factors Oct4, Sox2, Klf4, and c-Myc (OSKM) expressed from γ-retroviral vectors [5]. Meanwhile, alternative donor cell sources have been identified, such as neural stem cells [6], keratinocytes, or hematopoietic progenitor cells [7]. Moreover, reprogramming factor delivery was modified, and iPSC generation nowadays is possible with various techniques, such as transposon-based vectors, non-integrating viruses, or direct protein transduction [8], thereby reducing the risk of insertional mutagenesis and insufficiently silenced reprogramming factor expression associated with integrating gene delivery systems. Furthermore, substantially modified reprogramming factor cocktails have been evaluated, and iPSC generation has been demonstrated with three (OSK), two (OS), and even one (O) factor cocktails [9]. In addition, alternative reprogramming factors such as Lin28 or Nanog, or small molecules such as valproic acid or inhibitors of transforming growth factor beta (TGFβ) signaling, have been introduced to the field, and reprogramming systems allowing optimal stoichiometric ratios of the reprogramming factors were shown to enhance reprogramming efficiencies [10, 11].
miRNA function in embryonic stem cells
MicroRNAs (miRNAs) are 21–25 nucleotide (nt) long small non-coding RNAs, which are able to bind complementary mRNA sequences and repress their expression through mRNA cleavage or inhibition of protein translation [12]. miRNAs are transcribed from either independent genes or introns of other genes in a predominantly RNA polymerase II-dependent manner and their processing involves the RNase III family member Drosha and the DiGeorge syndrome critical region 8 protein (Dgcr8). After Exportin-5-mediated export into the cytosol, precursor miRNAs (pre-miRNAs) are cleaved into ~20 nt long miRNA/miRNA* duplexes by the double strand-specific ribonuclease mDCR-1 (Dicer) [13]. Upon incorporation into the miRNA-induced silencing complex, miRNAs then bind to target mRNAs depending on their seed sequence (nucleotides 2–8 of the miRNA) and inhibit protein translation. In cases of extensive base-pair matching, miRNAs can even induce degradation of their target mRNAs [14].
The importance of miRNAs for ESCs was demonstrated in Dicer and Dgcr8 knockout mouse models. Both Dicer- and Dgcr8-deficient ESCs display pronounced differentiation defects and also show markedly delayed cell cycle progression, indicating that miRNAs control both hallmarks of ESCs, self-renewal, and pluripotency. Moreover, Dicer and Dgcr8 knockout ESCs also fail to contribute to chimera formation upon blastocyst injection [15, 16]. Interestingly, ESCs only transcribe a limited number of miRNAs, some of which are exclusively expressed in the pluripotent state and rapidly decrease upon differentiation stimuli [17]. The two most abundant miRNA families in ESCs are the 290 cluster (miR-290, miR-291a, miR-291b, miR-292, miR-293, miR-294, and miR-295) and the 302–367 cluster (miR-302a, miR-302b, miR-302c, miR-302d, and miR-367) in mice, and the miR-371 (consisting of the miR-290 homologues miR-371, miR-372, and miR-373) and miR-302 families in humans [18]. Importantly, the notion of a predominant expression of relatively few miRNA families in ESCs was given further evidence by Leung and colleagues who performed a photo-cross-linking-based immunoprecipitation of Argonaute 2 followed by deep sequencing of coprecipitated RNAs [19]. Intriguingly, it was demonstrated that also pluripotency-associated transcription factors induce expression of specific miRNAs. In this regard, the core pluripotency factors Oct4, Sox2, and Nanog were shown to co-occupy the promoters of the miR-290/371 or the miR-302–367 cluster [18], and recent work demonstrated that the H3K26 demethylase Jhdm1b cooperates with Oct4 in the induction of the miR-302–367 cluster, indicating that expression of these pluripotency-associated miRNAs is a complex and tightly regulated process [20].
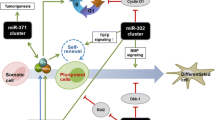
While miRNAs usually have hundreds of potential downstream targets, one of the main effects of the miR-290 cluster is to facilitate cell cycling and progression through the G1-S transition, since reintroduction of members of these miRNAs could at least partially rescue the proliferation defects in Dicer and Dgcr8 knockout models [21, 22]. Similarly, it has been shown that members of the miR-302 cluster, which are direct transcriptional targets of Oct4 and Sox2, accelerate cell cycling in human ESCs through repression of cyclin D1 and shortening of the G1 phase [23], although the role of cyclin D1 in pluripotent stem cells is not fully understood and cyclin D1 overexpression was demonstrated to enhance the generation of hiPSCs [24]. Consequently, the miR-290 and miR-302 clusters are also referred to as embryonic stem cell-specific cell cycle-regulating (ESCC) miRNAs [25] (Fig. 1a). Nevertheless, also repression of additional signaling pathways such as the canonical NFκB pathway has recently been suggested as potential cellular targets of the miR-290 family [26].
MicroRNA-mediated effects during iPSC generation. a The establishment and maintenance of an ESC-like self-renewal program is supported by different miRNAs. Members of the miR-290 and miR-302 cluster repress the cyclin-dependent kinase inhibitor Cdkn1a/p21, which under normal conditions would inhibit activity of the cyclin E/Cdk2 dimer, thereby preventing inactivation of p27 and arresting the cell in G1 phase. Furthermore, miR-130b, -301b, and -721 have been demonstrated to repress Meox2, which may lead to reduced Cdkn2a and Cdkn1a levels and helps in facilitating proliferation. b During iPSC generation, somatic cells with mesenchymal origin need to undergo an MET. Several miRNAs have been documented to facilitate this process, which allows expression of characteristic epithelial markers such as E-cadherin or Epcam. Members of the miR-106a–363 and miR-302–367 cluster as well as miR-93 have been shown to repress TGFβR2 thereby inhibiting anti-epithelial stimuli. In addition, the miR-106a–363 and miR-302–367 cluster induce expression of the epithelial surface marker E-cadherin, which is also supported by members of the miR-200 cluster and miR-205, which repress the E-cadherin antagonists Zeb1/2
However, miRNAs also regulate differentiation processes in pluripotent cells. In this context, members of the let-7 family were demonstrated to rescue the differentiation defect in Dgcr8-deficient murine ESCs, probably through antagonizing transcriptional targets of the major pluripotency factors Oct4, Sox2, and Nanog, such as Lin28, Sall4, and c-Myc. Interestingly, members of the let-7 family and Lin28 mutually repress each other: In the undifferentiated state, the RNA-binding protein Lin28 prevents let-7 from being processed, whereas upon differentiation, rising let-7 levels allow downregulation of Lin28 and promotion of the differentiation process [27]. In addition, miR-134, -296, and -470, as well as miR-145 and miR-21, expression of which is upregulated upon differentiation stimuli, were demonstrated to directly target Nanog, Oct4, and Sox2, thereby suppressing pluripotency and promoting differentiation in murine and human pluripotent stem cells [28, 29]. Recently, the miR-125 and miR-181 families were also reported to facilitate differentiation of murine ESCs through repression of the Polycomb ortholog Cbx7, which supports self-renewal and pluripotency in these cells [30]. Apart from downregulation of target mRNAs, miRNAs might also be directly involved in the transcriptional control of target genes, as recent data suggest a global association of Dicer 2 and Argonaute 2 with transcriptionally active loci in Drosophila [31]. This might add another level of complexity to the pleiotropic effects miRNAs exert to regulate crucial cell fate decisions.
miRNAs and cellular reprogramming
Although substantially improved protocols for iPSC generation are available today, transcription factor-mediated reprogramming has remained a rather slow and inefficient process as the generation of iPSCs still requires several rounds of cell divisions and usually takes 8–14 days depending on the starting cell source and the type of vector employed. This observation is in sharp contrast to cellular reprogramming by somatic cell nuclear transfer, which is completed within the first cell divisions. This discrepancy clearly calls for experiments deciphering the molecular mechanisms underlying this difference in order to enhance and accelerate transcription factor-mediated reprogramming.
In addition to their role in promoting pluripotency and self-renewal in ESCs, miRNAs have repeatedly been demonstrated to enhance the generation of iPSCs. The supportive effect of miRNAs on iPSC generation was initially demonstrated by Blelloch and colleagues, who observed a marked increase in reprogramming efficiencies upon overexpression of members of the ESCC miR-290 and miR-302 clusters, which was mainly caused through cell cycle promotion [32]. Apart from facilitating proliferation, the induction of pluripotency in somatic cells also involves pronounced phenotypic changes severely affecting cell morphology and architecture. Since embryonic and induced pluripotent stem cells exhibit characteristic features of epithelial cells and many strategies to obtain iPSCs employ mesenchymal cells, such as adult or embryonic fibroblasts, these cells need to undergo a mesenchymal-to-epithelial transition (MET) while being reprogrammed. This process has convincingly been shown to be promoted by several miRNAs, such as miR-205 and members of the miR-200 family, which repress the E-cadherin antagonists zinc finger E-box-binding homeobox 1/2 (Zeb1 and Zeb2) and thereby elevate E-cadherin levels during iPSC generation [33, 34]. Accordingly, the miRNA clusters miR-106a–363 and 302–367 were demonstrated to facilitate MET through the induction of E-cadherin and inhibition of TGFβ receptor 2 [35], repression of which could also be shown by miR-93 [36] (Fig. 1b). However, miRNA-mediated effects during the induction of pluripotency certainly are pleiotropic since targets of miR-302 and miR-372 may also be involved in other cellular processes, such as epigenetic regulation or vesicular transport [37]. Noteworthy, Morrisey and colleagues could successfully generate iPSCs from both human and murine somatic cells only by overexpression of the miR-302/367 cluster and in the presence of the histone deacytelase 2 inhibitor valproic acid [38]. However, miRNAs were constitutively expressed from integrating viral vectors in this study. Meanwhile, also this limitation has been overcome and a more recent report demonstrated iPSC generation after repetitive transient delivery of mature miRNAs miR-200c, -302a/b/c/d, -369-3p, and 369-5p [39].
However, more miRNAs may have a role during the generation of iPSCs than the ones that—mostly based on educated guesses—have been described so far. In order to identify novel miRNAs, which facilitate the generation of iPSCs, it is necessary to screen larger numbers of potential candidates in an experimental setup faithfully indicating reprogramming events. To this end, we have recently performed a full miRNA library screen using murine embryonic fibroblasts derived from Oct4-eGFP reporter mice and employing a polycistronic lentiviral vector expressing Oct4, Klf4, and Sox2 from a spleen focus-forming virus-derived promoter/enhancer [40] (Fig. 2). By applying this experimental strategy, it was possible to reproducibly generate eGFP+ iPSC colonies as early as day 7 after lentiviral transduction, and several miRNAs potently enhancing the appearance of Oct4-GFP+ colonies could be identified. These included an miRNA family consisting of miRNAs miR-130b, -301b, and -721, which share the transcription factor mesenchymal homeobox protein 2 (Meox2) as a common downstream target. The specific effect of this miRNA family was confirmed by Meox2 knockdown experiments, which recapitulated the miRNA-mediated effects [41]. Mechanistically, Meox2 might impede iPSC generation through upregulation of the cyclin-dependent kinase inhibitor 2a (Cdkn2a/p16) and the tumor suppressor protein Cdkn1a/p21, which were both suggested as downstream targets of Meox2 [42, 43] (Fig. 1a). Indeed, mRNA levels of Cdkn2a/p16 and Cdkn1a/p21 were considerably reduced upon siRNA-mediated Meox2 knockdown (unpublished data). In addition, our experimental system also appears suited to screen for miRNAs, which repress the generation of iPSCs. Also, these studies may substantially increase our understanding of the molecular mechanisms underlying iPSC generation as it was recently demonstrated for miRNAs miR-21 and miR-29 or miR-34 [44, 45].
A screening assay allowing for the identification of novel small interfering RNAs modulating iPSC generation: Transduction of Oct4-GFP reporter fibroblasts with a polycistronic lentiviral vector encoding for Oct4, Klf4, and Sox2 leads to robust and reproducible emergence of GFP+ iPSC colonies. In a multi-well setup and by simple transfection, library screens can be performed to identify novel miRNAs enhancing or repressing iPSC generation
In conclusion, miRNAs play important roles not only in maintaining pluripotency and self-renewal programs in pluripotent stem cells but also in the induction of pluripotency in somatic cells. Thus, screening for miRNAs that modulate iPSC generation clearly is a powerful tool to further elucidate the molecular mechanisms underlying transcription factor-mediated reprogramming. This strategy also should help to reveal novel downstream factors of miRNAs and thereby will enhance the generation of iPSCs. Optimized iPSC generation protocols might also decrease the variability among iPSC lines, which is an important step towards the generation of clinical-grade differentiated cell therapy products derived of iPSCs. In this regard, modulating miRNA networks might also be a powerful tool to optimize the specification of distinct somatic lineages during in vitro differentiation of iPSCs. Eventually, microRNA-based cell fate induction might support strategies of “in vivo reprogramming,” where cell fate decisions of somatic stem/progenitor cells are manipulated to enhance endogenous regeneration or to functionally replace scar tissue.
References
Boyer LA, Lee TI, Cole MF, Johnstone SE, Levine SS, Zucker JP, Guenther MG, Kumar RM, Murray HL, Jenner RG et al (2005) Core transcriptional regulatory circuitry in human embryonic stem cells. Cell 122:947–956
Sancho-Martinez I, Li M, Izpisua Belmonte JC (2011) Disease correction the iPSC way: advances in iPSC-based therapy. Clin Pharmacol Ther 89:746–749
Wu SM, Hochedlinger K (2011) Harnessing the potential of induced pluripotent stem cells for regenerative medicine. Nat Cell Biol 13:497–505
Drukker M, Benvenisty N (2004) The immunogenicity of human embryonic stem-derived cells. Trends Biotechnol 22:136–141
Takahashi K, Yamanaka S (2006) Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 126:663–676
Kim JB, Sebastiano V, Wu G, Arauzo-Bravo MJ, Sasse P, Gentile L, Ko K, Ruau D, Ehrich M, van den Boom D et al (2009) Oct4-induced pluripotency in adult neural stem cells. Cell 136:411–419
Pfaff N, Lachmann N, Kohlscheen S, Sgodda M, Arauzo-Bravo MJ, Greber B, Kues W, Glage S, Baum C, Niemann H et al (2011) Efficient hematopoietic redifferentiation of induced pluripotent stem cells derived from primitive murine bone marrow cells. Stem Cells Dev 21(5):689–701
Schambach A, Cantz T, Baum C, Cathomen T (2010) Generation and genetic modification of induced pluripotent stem cells. Expert Opin Biol Ther 10:1089–1103
Kim JB, Greber B, Arauzo-Bravo MJ, Meyer J, Park KI, Zaehres H, Schöler HR (2009) Direct reprogramming of human neural stem cells by OCT4. Nature 461:649–653
Papapetrou EP, Tomishima MJ, Chambers SM, Mica Y, Reed E, Menon J, Tabar V, Mo Q, Studer L, Sadelain M (2009) Stoichiometric and temporal requirements of Oct4, Sox2, Klf4, and c-Myc expression for efficient human iPSC induction and differentiation. Proc Natl Acad Sci USA 106:12759–12764
Tiemann U, Sgodda M, Warlich E, Ballmaier M, Schöler HR, Schambach A, Cantz T (2011) Optimal reprogramming factor stoichiometry increases colony numbers and affects molecular characteristics of murine induced pluripotent stem cells. Cytometry A 79A:426–435
Kim VN, Han J, Siomi MC (2009) Biogenesis of small RNAs in animals. Nat Rev Mol Cell Biol 10:126–139
Kim VN (2005) MicroRNA biogenesis: coordinated cropping and dicing. Nat Rev Mol Cell Biol 6:376–385
Krol J, Loedige I, Filipowicz W (2010) The widespread regulation of microRNA biogenesis, function and decay. Nat Rev Genet 11:597–610
Kanellopoulou C, Muljo SA, Kung AL, Ganesan S, Drapkin R, Jenuwein T, Livingston DM, Rajewsky K (2005) Dicer-deficient mouse embryonic stem cells are defective in differentiation and centromeric silencing. Genes Dev 19:489–501
Wang Y, Medvid R, Melton C, Jaenisch R, Blelloch R (2007) DGCR8 is essential for microRNA biogenesis and silencing of embryonic stem cell self-renewal. Nat Genet 39:380–385
Houbaviy HB, Murray MF, Sharp PA (2003) Embryonic stem cell-specific microRNAs. Dev Cell 5:351–358
Marson A, Levine SS, Cole MF, Frampton GM, Brambrink T, Johnstone S, Guenther MG, Johnston WK, Wernig M, Newman J et al (2008) Connecting microRNA genes to the core transcriptional regulatory circuitry of embryonic stem cells. Cell 134:521–533
Leung AK, Young AG, Bhutkar A, Zheng GX, Bosson AD, Nielsen CB, Sharp PA (2011) Genome-wide identification of Ago2 binding sites from mouse embryonic stem cells with and without mature microRNAs. Nat Struct Mol Biol 18:237–244
Wang T, Chen K, Zeng X, Yang J, Wu Y, Shi X, Qin B, Zeng L, Esteban MA, Pan G et al (2011) The histone demethylases Jhdm1a/1b enhance somatic cell reprogramming in a vitamin-C-dependent manner. Cell Stem Cell 9:575–587
Sinkkonen L, Hugenschmidt T, Berninger P, Gaidatzis D, Mohn F, Artus-Revel CG, Zavolan M, Svoboda P, Filipowicz W (2008) MicroRNAs control de novo DNA methylation through regulation of transcriptional repressors in mouse embryonic stem cells. Nat Struct Mol Biol 15:259–267
Wang Y, Baskerville S, Shenoy A, Babiarz JE, Baehner L, Blelloch R (2008) Embryonic stem cell-specific microRNAs regulate the G1-S transition and promote rapid proliferation. Nat Genet 40:1478–1483
Card DA, Hebbar PB, Li L, Trotter KW, Komatsu Y, Mishina Y, Archer TK (2008) Oct4/Sox2-regulated miR-302 targets cyclin D1 in human embryonic stem cells. Mol Cell Biol 28:6426–6438
Edel MJ, Menchon C, Menendez S, Consiglio A, Raya A, Izpisua Belmonte JC (2010) Rem2 GTPase maintains survival of human embryonic stem cells as well as enhancing reprogramming by regulating p53 and cyclin D1. Genes Dev 24:561–573
Martinez NJ, Gregory RI (2010) MicroRNA gene regulatory pathways in the establishment and maintenance of ESC identity. Cell Stem Cell 7:31–35
Luningschror P, Stocker B, Kaltschmidt B, Kaltschmidt C (2012) miR-290 cluster modulates pluripotency by repressing canonical NF-κB signaling. Stem Cells 30(4):655–664
Melton C, Judson RL, Blelloch R (2010) Opposing microRNA families regulate self-renewal in mouse embryonic stem cells. Nature 463:621–626
Tay Y, Zhang J, Thomson AM, Lim B, Rigoutsos I (2008) MicroRNAs to Nanog, Oct4 and Sox2 coding regions modulate embryonic stem cell differentiation. Nature 455:1124–1128
Xu N, Papagiannakopoulos T, Pan G, Thomson JA, Kosik KS (2009) MicroRNA-145 regulates OCT4, SOX2, and KLF4 and represses pluripotency in human embryonic stem cells. Cell 137:647–658
O'Loghlen A, Munoz-Cabello AM, Gaspar-Maia A, Wu HA, Banito A, Kunowska N, Racek T, Pemberton HN, Beolchi P, Lavial F et al (2012) MicroRNA regulation of Cbx7 mediates a switch of Polycomb orthologs during ESC differentiation. Cell Stem Cell 10:33–46
Cernilogar FM, Onorati MC, Kothe GO, Burroughs AM, Parsi KM, Breiling A, Lo Sardo F, Saxena A, Miyoshi K, Siomi H et al (2011) Chromatin-associated RNA interference components contribute to transcriptional regulation in Drosophila. Nature 480:391–395
Judson RL, Babiarz JE, Venere M, Blelloch R (2009) Embryonic stem cell-specific microRNAs promote induced pluripotency. Nat Biotechnol 27:459–461
Gregory PA, Bert AG, Paterson EL, Barry SC, Tsykin A, Farshid G, Vadas MA, Khew-Goodall Y, Goodall GJ (2008) The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nat Cell Biol 10:593–601
Samavarchi-Tehrani P, Golipour A, David L, Sung HK, Beyer TA, Datti A, Woltjen K, Nagy A, Wrana JL (2010) Functional genomics reveals a BMP-driven mesenchymal-to-epithelial transition in the initiation of somatic cell reprogramming. Cell Stem Cell 7:64–77
Liao B, Bao X, Liu L, Feng S, Zovoilis A, Liu W, Xue Y, Cai J, Guo X, Qin B et al (2011) MicroRNA cluster 302-367 enhances somatic cell reprogramming by accelerating a mesenchymal-to-epithelial transition. J Biol Chem 286:17359–17364
Li Z, Yang CS, Nakashima K, Rana TM (2011) Small RNA-mediated regulation of iPS cell generation. EMBO J 30:823–834
Subramanyam D, Lamouille S, Judson RL, Liu JY, Bucay N, Derynck R, Blelloch R (2011) Multiple targets of miR-302 and miR-372 promote reprogramming of human fibroblasts to induced pluripotent stem cells. Nat Biotechnol 29:443–448
Anokye-Danso F, Trivedi CM, Juhr D, Gupta M, Cui Z, Tian Y, Zhang Y, Yang W, Gruber PJ, Epstein JA et al (2011) Highly efficient miRNA-mediated reprogramming of mouse and human somatic cells to pluripotency. Cell Stem Cell 8:376–388
Miyoshi N, Ishii H, Nagano H, Haraguchi N, Dewi DL, Kano Y, Nishikawa S, Tanemura M, Mimori K, Tanaka F et al (2011) Reprogramming of mouse and human cells to pluripotency using mature microRNAs. Cell Stem Cell 8:633–638
Warlich E, Kuehle J, Cantz T, Brugman MH, Maetzig T, Galla M, Filipczyk AA, Halle S, Klump H, Schöler HR et al (2011) Lentiviral vector design and imaging approaches to visualize the early stages of cellular reprogramming. Mol Ther 19(4):782–789
Pfaff N, Fiedler J, Holzmann A, Schambach A, Moritz T, Cantz T, Thum T (2011) miRNA screening reveals a new miRNA family stimulating iPS cell generation via regulation of Meox2. EMBO Rep 12:1153–1159
Chen Y, Leal AD, Patel S, Gorski DH (2007) The homeobox gene GAX activates p21WAF1/CIP1 expression in vascular endothelial cells through direct interaction with upstream AT-rich sequences. J Biol Chem 282:507–517
Irelan JT, Gutierrez Del Arroyo A, Gutierrez A, Peters G, Quon KC, Miraglia L, Chanda SK (2009) A functional screen for regulators of CKDN2A reveals MEOX2 as a transcriptional activator of INK4a. PLoS One 4:e5067
Choi YJ, Lin CP, Ho JJ, He X, Okada N, Bu P, Zhong Y, Kim SY, Bennett MJ, Chen C et al (2011) miR-34 miRNAs provide a barrier for somatic cell reprogramming. Nat Cell Biol 13:1353–1360
Yang CS, Li Z, Rana TM (2011) microRNAs modulate iPS cell generation. RNA 17:1451–1460
Acknowledgments
The authors are grateful to Jan Fiedler and Amar Deep Sharma for fruitful discussions on microRNA biology. N.P., T.M., and T.C. received funding from the German Research Foundation through the cluster-of-excellence REBIRTH (EXC 62/1).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Pfaff, N., Moritz, T., Thum, T. et al. miRNAs involved in the generation, maintenance, and differentiation of pluripotent cells. J Mol Med 90, 747–752 (2012). https://doi.org/10.1007/s00109-012-0922-z
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00109-012-0922-z