Abstract
Several genome-wide transcriptome studies have shown that chronic inflammatory responses generally taking place in the inflamed tissue are also reflected at the level of peripheral blood leukocytes. Blood monocytes are highly sensitized cell type continuously activated under inflammatory conditions. For a better understanding of the transcriptional imprinting influenced by a multitude of pro- and anti-inflammatory mediators, we established a whole blood in vitro system to explore cell- and stimulus-specific gene expression signatures in peripheral monocytes. In an explorative study, whole blood from healthy donors was stimulated with tumour necrosis factor-alpha (TNF-α) or lipopolysaccharide (LPS) for 1.5 h. Subsequently, monocytes were isolated with a purity of >99% by high-speed fluorescence activated cell sorting. Transcriptional changes were explored by whole genome Affymetrix arrays using highly validated filtering algorithm to identify differentially expressed genes. In vitro stimulation of whole blood samples with TNF-α and LPS resulted in 4,529 and 5,036 differentially expressed genes, respectively. Although both stimuli induced similar inflammatory profiles in monocytes, TNF-α- or LPS-specific gene signatures were characterized. Functional classification identified significant numbers of differentially expressed cytokines, cytokine receptors and apoptosis-associated genes. To our knowledge, this is the first study presenting cell- and stimulus-specific gene expression signatures that can be used to decipher complex disease specific profiles of acute and chronic inflammation. Once a library of signatures from the most important inflammatory mediators is defined, it can be helpful to identify those signatures, which are predominantly driving the disease pathogenesis and which are of potential interest for a therapeutical intervention.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Inflammation that accompanies autoimmune diseases could affect different tissues and organs like skin, kidneys, central nervous system in systemic lupus erythematosus (SLE), joints, lungs, heart in rheumatoid arthritis (RA), brain and spinal cord in multiple sclerosis [1], skin and joint in psoriasis [2]. Although different organs are affected, peripheral blood as easily accessible sample material reflects acute and chronic inflammatory responses. Changes in the number of leukocytes or of specific leukocyte population, and elevated levels of blood markers like erythrocyte sedimentation rate, C-reactive protein, and fibrinogen are of relatively limited diagnostic importance, considering that these commonly used biological markers are more inflammation than diseases dependent. Nevertheless, some of inflammatory markers like anti-nuclear antibodies or anti-citrullinated peptide/protein antibodies and rheumatoid factor (RF) are more specific for SLE or RA, respectively. A possibility to improve diagnosis and to get a better insight into disease pathogenesis using blood as the sample material emerged with appearance of microarray technology. The DNA microarray technology, as a comprehensive approach for identifying thousands of differentially expressed genes at a given time, was used to reveal alterations in gene expression of peripheral blood cells (PBC) or peripheral blood mononuclear cells (PBMC) [3–8]. PBMCs have been used as the source for the establishment of disease-related profiles in patients with RA [3], psoriatic arthritis (PsA) [4, 5], dermatomyositis [6], SLE [7], type 1 diabetes [8] and multiple sclerosis (MS) [9]. However, changes in the fraction of leukocytes within PBMCs correlate with the alteration of overall PBMC's gene expression. In RA patients monocytes related gene expression profiles, monocytosis as well as activation of monocytes associated genes during relapse of disease have been described [3, 10, 11]. To circumvent the complexity and the cellular heterogeneity of PBCs or PBMCs, we have introduced an approach of cell-specific gene expression profiling [12]. Using this approach transcriptional patterns of monocytes have been established for discrimination between different rheumatic diseases like RA, SLE and ankylosing spondylitis (AS) from healthy donors [12]. To interpret disease-related changes of gene expression and to understand molecular pathomechanisms, it is essential to know response pattern of different cell types to specific triggers. For example, in vitro stimulation of PBMCs with interferons (IFNs) was the key experiment to identify the corresponding IFN type I gene expression signature in various diseases like SLE [7], dermatomyositis [6] and in a subset of RA patients [13]. Based on these findings we focused on the development of tumour necrosis factor-alpha (TNF-α) and lipopolysaccharide (LPS) signatures in human monocytes. TNF-α has been reported as one of the major inflammatory cytokine involved in septic shock, malaria, cancer, as well as in pathogenesis of many autoimmune diseases, like RA, PsA, AS and Crohn's disease [14, 15]. Produced predominantly by monocytes/macrophages but also by lymphocytes and NK cells, this potent cytokine acts through TNFR-1 (the p55 TNF receptor) and TNFR-2 (the p75 TNF receptor). These receptors are expressed on nearly all human cells and they are responsible for the pleiotropic effect of this cytokine [16]. On the other side, LPS as a component of the outer membrane of gram-negative bacteria is a powerful inducer of TNF-α production. As one of the strongest activators of the innate immune system including monocytes, the LPS-induced systemic inflammatory response is characterized by the release of other inflammatory mediators as well, such as IL-1α, IL-1β, IL-6, IL-8, CCL2, and CCL8. The relevance of monocytes/macrophages, as the main producers of TNF-α, as well as the significance of these cells in response to anti-TNF-α therapy [17], emphasize the importance of these cells in the development of autoimmune diseases such as RA [18] and AS [19]. Although response of all leukocytes and whole body in general is present under inflammatory condition, focus on a specific leukocyte population could improve understanding of altered circumstances, especially those where monocytes and their inflammatory mediators have a great impact like SLE, RA, sepsis and atherosclerosis.
Materials and methods
Blood collection and study design of in vitro signatures
A 200 ml of peripheral blood from four healthy donors was collected in Vacutainer Heparin tubes (Becton Dickinson, Heidelberg, Germany). Donors used in this study were Caucasian in origin and non-smoker; two donors were females (24 and 27 years old) and two were males (28 and 30 years old) and not under any medication. Samples that represent unstimulated controls were immediately processed. Other samples were incubated for 1.5 h at 37°C either without stimulus or were stimulated with 100 ng/ml TNF-α (Peprotech, London, UK) and 100 ng/ml LPS (O111:B4 E.coli, Sigma, Germany), respectively. After stimulation, blood samples were lysed by EL buffer (Qiagen, Hilden, Germany) at 4°C according to the instruction of the manufacturer. Subsequent depletion of granulocytes was performed using CD15-conjugated microbeads (MACS, Miltenyi Biotec, Bergisch Gladbach, Germany) and the automated separation system Auto-MACS (Miltenyi Biotec). CD15-depleted fraction was stained with CD14-fluorescein isothiocyanate (FITC) antibody (Becton Dickinson) to label monocytes and with propidium iodide to exclude dead cells. Using FACSAria cell sorter (Becton Dickinson) monocytes were isolated with purity and viability of >99%. Absolute numbers of monocytes were estimated after their separation, and comparable cell numbers were obtained from all donors: 6.3 ± 1.6 × 105 from 10 ml of blood (mean ± SD). The Ethics Committee of the Medical Faculty of Charité Universitätsmedizin Berlin approved the study.
RNA isolation, Affymetrix gene chip hybridization and quality checks
After sorting, monocytes were immediately lysed in RLT buffer (Qiagen), containing β-mercaptoethanol. Lysates were stored at −80°C until RNA isolation. RNA isolation, Affymetrix gene chip hybridization and quality checks were performed as already described [20]. Since not all 16 samples passed stringent quality criteria for a successfully hybridized microarray, recommended by Affymetrix, finally, 12 arrays were used for comparative data analysis. To obtain maximal statistical power as demonstrated elsewhere [21], we performed group-wise comparison analysis instead of donor-matched analysis. By this way, if comparing two groups of 2 and 3 arrays respectively, the results of six comparisons are still available, as compared with two comparisons only, when donor-matched analysis would be performed. The chips from four donors were labelled from 1 to 4 and numbers relate to the same donors in all four groups: unstimulated 0, unstimulated 1.5 h, TNF-α 1.5 h and LPS1.5 h. Each of the 3 chips of the TNF-α (TNF-1, TNF-3 and TNF-4) or the LPS groups (LPS-2, LPS-3, LPS-4) was compared with each of two chips of the unstimulated group “unstim 0 h” (UN0-1 and UN0-2). Thus, six comparisons were available for the determination of TNF-α and LPS profiles, respectively. However, if available, donor-matched analysis is also included as a part of group-wise comparison. For example one of six comparisons in TNF-α vs UN0 represents donor-matched analysis: TNF-1 vs UN0-1. All chips included in the analysis are also apparent in heat-maps of Figs. 1, 4, 5a, 6a and 8a. To reveal probe sets which behave sensitive to in vitro incubation (STIVI genes), four chips of the unstimulated group “unstim 1.5 h”, which represents samples that were incubated at 37°C for 1.5 h without any stimuli, were compared with two chips of the unstimulated group “unstim 0 h”, which represents samples of monocytes processed immediately after their isolation. Array data will be publically available in the BioRetis database (http://www.bioretis-analysis.de) and explanation of their selection is provided in Electronic supplementary File 1.
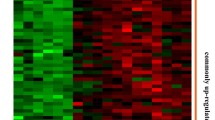
Gene expression profile of TNF-α- and LPS-stimulated monocytes for 1.5 h in whole blood. Hierarchical clustering of 11678 Affymetrix probe sets, which correspond to about 5896 known genes that distinguish stimulated monocytes from unstimulated cells. Each row represents one probe set and each column indicates one subject. Red indicates increased and green decreased expression compared with the mean. The colour intensity represents the magnitude of the deviation from the mean. Expression value intensities are depicted according to the colour-indicator key and range from -2 to 2
Chip data analysis and cluster algorithms
For chip data analysis we used all relevant data of the Affymetrix GCOS/MAS5 software, imported these into the BioRetis database (www.bioretis-analysis.de), performed group comparisons and filtered the significant differentially expressed probe sets as described before [21]. In detail, each chip was globally normalized and scaled to an average intensity of 150 (target value TGT = 150) to adjust for global differences in hybridization. All data of the GCOS software (expression heights, call for presence of transcript, p value for presence or absence of transcripts, log2 value of fold change and 95% confidence intervals of it, call for the significance of differential expression, the non-parametric p value for that call and additionally calculated strict Bonferroni corrected Welch t tests) were included in filtering significant probe sets (High Performance Chip Data Analysis, HPCDA; unpublished). Some of the data of all of these combined genes and probe sets are available in Electronic supplementary Table 1. For hierarchical cluster analysis (as Eisen plot), we used the programme Genes@Work [22]. It was used with gene vectors for normalization and Pearson w/mean for similarity measure. As cluster type, we used method centre of mass.
FACS staining and analysis
For intracellular staining monocytes were stimulated in the presence of 5 μg/ml Brefeldin A (Sigma). Cells were fixed with 3% paraformaldehyde, permeabilized with 0.5% saponine and labelled with human anti-IL-1β FITC (BD Bioscience), anti-IL-6 FITC (BD Bioscience), anti-IL-8 PE (BD Bioscience) and anti-TNF-α FITC (in-house generated) antibodies. The procedure for detection of NFκB phosphorylation was performed after 30 min stimulation. Fixed cells were permeabilized with 70% methanol and stained with human anti-pNFκB p65 FITC-labelled antibody (BD Bioscience). The experiments were repeated at least three times. Measurements were performed on a FACS Calibur (BD) and unstimulated monocytes immediately processed and unstimulated monocytes incubated for 1.5 h at 37°C were used as controls.
Microarray data from anti-TNF-α-treated RA patients
In the study of Stuhlmüller et al. [23] peripheral human monocytes were isolated from rheumatoid arthritis patients (RA) before and after treatment with the TNF-α-blocker adalimumab, labelled as RA1, RA2, RA3, RA5 and RA7, before Th and after therapy (anti-TNF). Array data were generated by using Affymetrix HG-U 133A gene chips.
Results
Defining genes induced by ex vivo incubation of monocytes for 1.5 h
To determine the genes that are sensitive to the in vitro manipulation of whole blood samples, we compared the samples of isolated monocytes after whole blood incubation for 1.5 h at 37°C with the samples of immediately isolated cells. The comparison identified 899 differentially expressed probe sets (correspond to 655 genes) in monocytes that were sensitive to in vitro incubation—STIVI (Electronic supplementary Table 1). These STIVI genes served as a technical control for the analysis of TNF-α and LPS effects in monocytes. Out of 899 STIVI probe sets, expression of 389 (270 genes) were increased and of 510 (385 genes) were decreased. Within this group of genes we identified up-regulation of TLR1 (FC = 1.22) and CCR5 (FC = 1.56), and down-regulation of CCR2 (FC = −1.56) and IL-6R (FC = −2.53). However, expression of the same genes was more affected by TNF-α and LPS stimulation, like down-regulation of CCR2 by FC = −13.93 and FC = −38.5, respectively.
TNF-α- and LPS-induced transcriptomes reveal common and stimulus-specific inflammatory profiles
To identify the response of monocytes to inflammatory conditions we studied gene expression profiles of isolated monocytes which were stimulated for 1.5 h in the whole blood by TNF-α- and LPS. Stimulated samples were compared with those that were unstimulated and immediately isolated, and which were used as a control group in chip data analysis. Considerable numbers of differentially expressed probe sets were identified, 8,321 after TNF-α and 9,711 after LPS stimulation (Fig. 1). Probe sets for uncharacterized genes were removed, and different probe sets of one gene were combined, revealing 4,529 and 5,036 genes differentially expressed by TNF-α and LPS, respectively. Both stimuli induced considerably overlapping expression profiles (Fig. 2). The differential regulation of 6,497 probe sets in common is consistent with the fact that both TNF-α and LPS activate signalling pathways, such as IKK-NFκB, and Jnk-AP1. Phosphorylation of NFκB, which we used as an indication of early monocytes activation and a control parameter for our experimental setup, was identified 30 min after TNF-α and LPS stimulation (Fig. 3). On the other hand, each stimulus also induced specific changes in expression, characterized with 1,703 probe sets (1,093 genes) which were TNF-α specific, while 3,151 probe sets (1,945 genes) were affected by LPS stimulation only (Fig. 4a, b). Down-regulation of TNFSF10, TLR8, EPOR, CD1A, CASP10 and up-regulation of FPR3, GPR35, CD22, CASP3 constitute part of the TNF-α-specific profile. Genes like TLR6, TNFSF14, TNFRSF1A as down-regulated and IFNB1, SOCS1, INHBA, CD200, IRF1, IRF7, IRF8 as up-regulated represent part of the LPS-specific pattern (Electronic supplementary Table 2). Noteworthy is the set of genes which were differentially expressed by TNF-α and LPS but in opposite direction (Fig. 4c). Remarkable numbers of these genes are considered as part of an IFN signature like IFIT1, IFIT3, IFIT5, IFI44, OAS1, OAS2 and XAF1. Our results revealed that LPS induced their expression as well, while these genes were suppressed by TNF-α (Electronic supplementary Table 2).
Overall distribution of differentially expressed probe-sets sensitive to in vitro incubation “STIVI”-, TNF-α- and LPS. Venn diagram represents 11,678 differentially expressed probe sets in TNF-α- and LPS-stimulated monocytes for 1.5 h in whole blood together with STIVI probe sets, whose expression was detected during incubation for 1.5 h. Values in parenthesis indicate the absolute number of differentially expressed probe sets in all three groups compared with unstimulated and immediately isolated monocytes
NFκB phosphorylation in monocytes detected 30 min after TNF-α and LPS stimulation in whole blood. After activation in whole blood monocytes were isolated using MACS technology, namely with CD15 magnetic beads granulocytes were removed and pure monocytes were obtained with additional MACS CD14 positive cell sorting. The alterations in phosphorylation of NFκB were measured by flow cytometry on a FACSCalibur. Histograms represent mean fluorescence intensity (MFI) of unstimulated and immediately processed monocytes (filled in gray), unstimulated monocytes incubated for 1.5 h (gray line), TNF-α stimulated (green line) and LPS stimulated (red line). Data show representative histogram from three independent experiments
Gene expression profiles that discriminate TNF-α- from LPS-stimulated monocytes. a Hierarchical clustering of 1,824 Affymetrix probe sets differentially expressed just by TNF-α. b Hierarchical clustering of 3,214 Affymetrix probe sets differentially expressed only by LPS. c Hierarchical clustering of 94 Affymetrix probe sets regulated by TNF-α and LPS that belong to common inflammatory profile but showed opposite direction of changes. Each row represents one probe set and each column indicates one subject. Red indicates increased and green decreased expression compared with the mean. The colour intensities are depicted to the deviation from the mean. Expression value intensities are depicted according to the colour-indicator key and ranged from −2 to 2
Functional classification of differentially expressed genes in monocytes stimulated by TNF-α and LPS
Database for Annotation, Visualization and Integrated Discovery [24] was used for functional classification and biological interpretation of differentially expressed genes. Within the group of up-regulated genes, those involved in signal transduction, apoptosis, and immune/inflammatory response were identified among the top most significant biological processes (Table 1). Genes involved in metabolic processes and genes encoding components of mitochondria were significantly down-regulated. “Metabolic process” was the functional group with the highest significance (Bonferroni corrected p values, p = 8.46 10−20 for TNF-α and p = 1.57 10-32 for LPS). We next evaluated all differentially expressed cytokines, cytokine receptors and apoptosis-associated genes using Gene Ontology annotations as selection criteria and described two profiles in monocytes in more detail: the cytokine–cytokine receptor profile (Fig. 5a) and the apoptosis-survival profile (Fig. 6a).
Cytokine–cytokine receptor gene expression profile with functional classification. a Hierarchical clustering of 120 differentially expressed cytokine and cytokine receptor genes that distinguish TNF-α- and LPS-stimulated monocytes from unstimulated samples. Blue bars of the dendrogram represent unstimulated samples, red bars TNF-α stimulated and black bars LPS stimulated. b Functional classification of cytokines and cytokine receptors which are differentially expressed by TNF-α and LPS stimulation. Differentially expressed genes in TNF-α- and LPS-stimulated monocytes compared with unstimulated samples can be classified into functional families. The cytokine–cytokine receptor profile is subdivided into the following families: Interleukinome (interleukins and interleukin receptors), Chemokinome (chemokines and chemokine receptors), TNF–TNFR family members and IFNs, growth-, differentiation- and proliferation-factors and their receptors. Bars present the log2 of the average fold change in gene expression level of STIVI- (yellow bars), TNF-α- (green bars) and/or LPS- (red bars) regulated genes compared with unstimulated samples
Hierarchical clustering of the apoptosis-survival gene expressional profile with functional classification of caspase- and BCL2-family members in monocytes following TNF-α and LPS stimulation for 1.5 h. a 240 pro- and anti-apoptotic genes showed altered expression by stimulation for 1.5 h with TNF-α and LPS compared with unstimulated samples. Blue bars of the individual dendrogram represent unstimulated samples, red bars TNF-α stimulated and black bars LPS stimulated. b Members of caspase- and BCL2-family, as a part of apoptosis-survival profile, that were differentially expressed in monocytes following TNF-α and LPS stimulation for 1.5 h. Genes from apoptosis-survival profile that are caspase or BCL2 family members were selected. Bars present log2 of the average fold change in gene expression level of STIVI (yellow bars), TNF-α (green bars) and LPS (red bars) genes compared with unstimulated ones
Cytokine–cytokine receptor profile is composed of 120 differentially regulated genes, and subdivided into interleukins and interleukin receptors named here as “Interleukinome”, chemokines and chemokine receptors, named as “Chemokinome”, TNF–TNFR family members, as well as interferons, growth-, differentiation- and proliferation-factors and their corresponding receptors (Fig. 5b, Electronic supplementary Table 3).
The Interleukinome consisted of 37 differentially expressed genes. Beside interleukins and interleukin receptors caspase 1, also known as IL-1β converting enzyme, was also included. This enzyme has a crucial role in processing of inactive IL-1 family members, such as IL-1β and IL-18 into their active forms [25], and was induced both by TNF-α and LPS. Up-regulation of IL-1α, IL-1β and IL-18, the classical IL-1 family members, was a common feature of both stimuli. However, up-regulation of IL-1F9, a recently identified member of this family [26], was specific for LPS.
On the protein level we also identified IL-1β production induced after 1.5 h stimulation by TNF-α and LPS (Fig. 7a) and in respect to cytokine production monocytes exhibited a stronger response to LPS as compared with TNF-α stimulation. In addition our results revealed production of other pro-inflammatory cytokines such as IL-6 and IL-8 (Fig. 7b) already 1.5 h after stimulation. The production of above mentioned cytokines correlated with their up-regulation detected within the Interleukinome.
Detection of cytokines production and expression of surface molecule CD83 in monocytes following TNF-α and LPS stimulation in whole blood. Whole blood was left unstimulated or was treated with TNF-α (100 ng/ml) and LPS (100 ng/ml) for 1.5 h in the presence of Brefeldin A for intracellular staining or without it for CD83 staining. After activation in the whole blood monocytes were isolated using MACS technology, namely with CD15 magnetic beads granulocytes were removed and pure monocytes were obtained with additional MACS CD14-positive sorting. The alterations in cytokine production, IL-1ß, IL-6 and IL8, and expression of CD83 were measured by flow cytometry on a FACSCalibur. Mean fluorescence intensities (MFI) for control samples, unstimulated and immediately processed (filled in gray) and unstimulated but incubated samples for 1.5 h (gray line), and for TNF-α (green line) and LPS (red line) stimulated cells are presented. Data show representative histograms from three independent experiments
The changes observed for the Interleukinome indicate that stimulation with TNF-α and LPS induces not only an inflammatory but an anti-inflammatory response as well, represented by down-regulation of IL-1RAP (IL-1 receptor accessory protein), and up-regulation of IL-1R2 (IL-1 receptor 2), IL-1RN (IL-1 receptor antagonist) and IL-10.
The Chemokinome of TNF-α- and LPS-stimulated monocytes contains 29 differentially expressed chemokines: ten belong to the CCL chemokine family, ten to the CXCL family and nine to the family of chemokine receptors (Fig. 5b). All differentially expressed CCL and CXCL genes were up-regulated. Up-regulation of CCL18, CCL19 and CXCL9 was specific for LPS. The expression of chemokine receptors is also important for the overall tuning of the chemokine-chemokine receptor system during inflammation. Our data revealed decreased expression of CCR2, CXCR2, CXCR4 and CX3CR1, while expression of CCRL2, CCR5 and CCR7 were increased. CCR1 was specifically up-regulated by TNF-α, whereas CXCR7, the recently identified chemokine receptor for CXCL11 and CXCL12 [27, 28], was exclusively up-regulated by LPS.
Profile of TNF and TNFR family members differentially expressed in TNF-α and LPS-stimulated monocytes includes ten ligands, 12 receptors and one enzyme “A disintegrin and metalloproteinase domain 17” (ADAM17, also known as TACE). The most of the TNF superfamily ligands were influenced both after TNF-α and LPS stimulation such as up-regulation of TNF-α itself, LTB, CD30L and VEGI and down-regulation of TWEAK and APRIL. TNF-α- and LPS-stimulated monocytes also showed the up-regulation of ADAM17, the enzyme that cleaves the membrane-bound form of TNF-α and generates its soluble form.
Profile of IFNs, growth-, differentiation- and proliferation-factor family members and their receptors contains 34 differentially expressed genes, such as IFNs (IFNβ and IFNε), growth factors like HGF, INHBA, INHBC, and their corresponding receptors IFNAR1, IFNGR1, IFNGR2, MET, ACVR1, ACVR1B, ACVR2, ACVR2A (Fig. 5b). Members of the IFN family such as IFNβ1 and IFNε1 were induced only by LPS. Also IFNGR1, receptor for IFN type II, was down-regulated with the same stimulus. IFN response triggered via TLR4 is part of inflammatory profile that is LPS specific.
Profile of “Apoptosis-Survival” genes in TNF-α- and LPS-activated monocytes exhibits 240 genes, both pro- and anti-apoptotic. 190 genes were regulated by TNF-α- and 213 by LPS. Out of these genes 163 were affected by both stimuli and belong to the common inflammatory profile in monocytes. Nevertheless, 27 genes were differentially expressed only by TNF-α, and 50 genes only by LPS (Electronic supplementary Table 4). Beside the overall response of monocytes regarding differential expression of all pro- and anti-apoptotic genes, we described in more detail specific profiles for differentially expressed BCL2- and caspase family members. The BCL2 family, which includes both pro- and anti-apoptotic members, represents a death-survival check-point in cells. Another major component of the cell death machinery is the family of caspases, which notably contributes to the regulation of apoptosis by its proteolytic activity.
The caspase family members that changed their gene expression level following stimulation were CASP1, CASP3, CASP4, CASP5, and CASP7 which were up-regulated and CASP2, 6, 8 and 10 which were down-regulated (Fig. 6b). Up-regulation of CASP3 and down-regulation of CASP10 were specific for TNF-α stimulation.
The BCL-2 family members are important regulators of the mitochondrial pathway of apoptosis. The members of this family are subdivided in three main groups based on the function and BCL2 homology region (BH domains). There are one anti-apoptotic and two pro-apoptotic groups. Members of the anti-apoptotic group were up-regulated, such as BCL2A1 and MCL1 by TNF-α and LPS. Also BCL2, as a member of the same anti-apoptotic group was up-regulated, but only by LPS. The members of the multidomain pro-apoptotic group BAX and BAK1 showed decreased expression. But the members of the BH3 pro-apoptotic group showed both down- and up-regulation, such as BIM, BAD, BMF, BNIP3 and BNIP3L which decreased their expression and BID, NOXA and BCL2L14 which displayed increased level in their expression.
Application of the in vitro TNF-α-gene signature to monitor anti-TNF-α-induced effects in monocytes of Adalimumab-treated RA patients
Based on gene expression data published very recently by Stuhlmüller et al., we have monitored the imprinting of our in vitro-generated TNF-α-signature in monocytes of RA patients before and after successful treatment with a TNF-α-blocking antibody (Adalimumab) [23]. Since different array types were used in our study (HG-U133 Plus 2.0 representing nearly 54.000 probe sets) and in that of Stuhlmüller et al. (HG-U133A representing only 22.000 probe sets), we could not perform direct group comparison analyses. Therefore, we have determined the overlap of TNF-α-regulated probe sets identified by our in vitro approach and the profile of differentially regulated genes before and after treatment. Thus, 179 out of 471 treatment-regulated probe sets (457 were up- and just 14 were down-regulated) were common in both gene expression profiles and used for hierarchical cluster analyses (Fig. 8 and listed in the Electronic supplementary Table 5). In Fig. 8a, it is shown that 174 out of these 179 common probe sets were down-regulated in vitro and allowed a clear classification of unstimulated and TNF-α-stimulated samples. Interestingly, the majority of these probe sets was reversely regulated in monocytes isolated from anti-TNF-α-treated samples, which have been compared with cells before treatment (Fig. 8b). TXNIP, HHEX, P2RY5, ZNF217, SAMD4A, HDAC9, APEX1 and LARP4 were identified as the strongest up-regulated genes by anti-TNF-α responders, while very few genes, like EREG, PDE4B, PLAUR, PFKFB3 were found to be down-regulated.
Gene expression profile of in vitro TNF signature in monocytes that overlapped with anti-TNF-α responders in RA patients but show opposite direction of changes. a Gene expression profile of TNF-α in vitro stimulated monocytes that show opposite direction of changes when compared with monocytes from RA patients before and after anti-TNF-α treatment. Hierarchical clustering of 179 Probe sets that are common for in vitro stimulated monocytes by TNF-α- and for anti-TNF-α-treated RA patients before and after therapy. These common probe sets show opposite direction of changes, 174 probe sets which were down-regulated by TNF-α were up-regulated after anti-TNF-α treatment and five in vitro TNF-α up-regulated were down-regulated in monocytes from patients that responded to anti-TNF-α therapy. For hierarchical clustering, gene vectors for normalization, Euclidian distance for similarity measure, and centre of mass as cluster type were used. b 179 Probe sets that show differential gene expression in monocytes from five RA responders before and after anti-TNF-α therapy and that overlapped with in vitro TNF-α signature. Altogether 471 Probe sets from anti-TNF-α responder were selected when 80% of pair-wise comparisons revealed significantly increased or decreased expression (Electronic supplementary Table 5), but 179 Probe sets were overlapped with in vitro TNF signature. Each row represents the average signal for one gene, while each column indicates one subject. Genes which are expressed at higher levels relative to the mean expression levels are presented in red, and genes that are expressed at lower levels relative to the mean expression levels are presented in green. The colour intensity represents the magnitude of the deviation from the mean. Expression value intensities are depicted according to the colour-indicator key and ranged from −2 to 2
Discussion
Our approach focuses on the generation of cell-specific gene expression profiles that can be used to unravel complexity of gene expression signatures underlying inflammatory diseases. Given that inflammatory responses are driven by more or less specific cytokines, such as type I IFN in SLE [7, 20] or TNF-α in RA and septicaemia [14], it would be very helpful for a better functional understanding of inflammatory diseases, to know which genes are specifically induced by a certain cytokine in a certain cell type. Although microarray technology has made serious progress with respect to robustness and reproducibility, a functional understanding of genes differentially expressed under pathophysiological conditions remains a major challenge for bioinformatics. Functional annotation software, such as Ingenuity, Panther or Kyoto Encyclopaedia of Genes and Genomes pathway (KEGG), are promising steps in the right direction, but often end up in a new level of confusion. In general, it has to be considered that gene expression studies possess the inherent limitation of not being able to unambiguously elucidate a molecular mechanism right to its last detail without doing additional functional studies. But this technology possesses the unique potency to reveal molecular alterations at the transcriptional level that can be assigned to a particular mechanism. Therefore, the knowledge about cytokine-specific gene signatures will help to strengthen the validity of gene expression profiles available from different cells or tissues, which are embedded in a cytokine network under inflammatory conditions.
In a first attempt, we have focused on human monocytes isolated from whole blood that have been stimulated for 1.5 h with TNF-α or bacterial LPS, respectively and which were analyzed by genome-wide expression arrays. Our method of whole blood stimulation has been designed as an approach, which resembles more physiological conditions for short-term cell stimulation as compared with approaches using cells or cell lines completely separated from their physiological milieu. Stimulation of cells in whole blood under continuous agitation and their subsequent isolation, which was performed continuously at 4°C, seems to be the most promising way to circumvent an artificial activation of monocytes, which is almost inevitably induced when these cells are first isolated and then cultured. The combined MACS/FACS procedure applied for the isolation of peripheral blood monocytes has been optimized to avoid almost any additional stimulation [29]. Therefore, we assume that the ex vivo generation of gene expressional profiles from monocytes stimulated in whole blood will provide more precisely insights into their transcriptome than any other in vitro model described in the literature [30]. Although we have focused on early TNF-α and LPS responses in monocytes to exclude indirect effects, some cytokines like IL-1β or IL-8 were already produced within 90 min of stimulation, which may have an additional impact on the monocytes. Otherwise, we are aware that short-term stimulation only partially reflects a chronic inflammatory response that is caused by a long-lasting inflammatory milieu. However, short-term exposure to TNF-α and LPS might be able to provide a stimulus-specific signature which resembles the initial phase of chronic inflammation.
Incubation of whole blood ex vivo for 1.5 h at 37°C without any stimulus was already associated with an altered gene transcription reflecting an early response to the artificial in vitro milieu. Although only minor changes occurred compared with the effects induced by TNF-α or LPS, these genes sensitive to in vitro stimulation (STIVI genes should be considered when performing in vitro manipulations of monocytes to distinguish artificially induced from pathophysiologically relevant profiles.
Stimulation with LPS was performed as a prototype of gram-negative bacterial infection that strongly activates monocytes. On the other side, cell activation by TNF-α, which reflects a result of already changed environment, was reported in pathogenesis of sepsis, atherosclerosis, RA, AS, Crohn's disease, and psoriatic arthritis [14, 31, 32].
We could identify several thousands of genes, which were differentially expressed in response to TNF-α and LPS, respectively. Considering that both stimuli are able to activate the same signalling pathways via their receptors, TNFR1 and TLR4 [33, 34], it was not surprising that their gene expression profiles showed large overlap. As a result most of the TNF-α- and LPS-regulated genes constitute a more common inflammatory signature, which is described by 6,497 probe sets that correspond to 3,735 genes. Besides TNF-α and LPS, stimuli-like IL-1 and other TLR agonist are also able to activate NFκB, and we were prompted to deduce that the convergence of different signalling pathways toward activation of the same transcription factors is relevant for the induction of a more general inflammatory response.
To describe TNF-α specific gene signature, we compared TNF-α induced genes with those of LPS and identified 1,824 probe sets that reflect 1,093 genes more specifically regulated by TNF-α. Genes like SYNGR3, GJA3, ARNTL2, CD22, SPP1, MAP3K7IP3, PPAP2A, GPR35 and FPR3 were identified as specifically TNF-α up-regulated. Specificity of LPS in comparison to TNF-α is displayed in 3,214 probe sets (1,945 genes), and among them are IFNB1, IFNE1, SOCS1, INHBA, DEFB1, CD200, CFB.
Moreover, profiling of TNF-α- and LPS-stimulated monocytes revealed relatively small but relevant group of genes, which belong to TNF-α and LPS common inflammatory profile but whose expression were changed in opposite direction, like OAS1, OAS2, IFI44, IFIT1, IFIT3, IFIT5 and STAT1, RSAD2 (CIG5). Namely, while TNF-α silenced, LPS promoted their expression. Interestingly, these genes are considered as the part of already described type I IFN signature. The evidence that part of LPS expressional pattern resemble with IFN signature is in addition supported with the group of genes that are specifically induced by LPS, such as ADAR, IFI35, IFI44L, IFI6, IFIT2, IFITM1, IFITM3, MX1, MX2, OAS3, SOCS1. The genes that have not been described as a part of IFN profile but which were up-regulated by LPS are IFNB1, IFNE1 themselves, and transcription factors IRF1, IRF7, and IRF8. The finding that a fraction of LPS-regulated genes resembles an IFN signature could be explained by the fact that a more general and comprehensive response is triggered by LPS than by IFN or TNF-α. In fact, two components of the TLR4 signalling pathway are activated by LPS, a Myd88-dependent and a Myd88-independent branch. The first one results in an activation of NFκB and thereby inducing the transcription of pro-inflammatory target genes, which results in similarity with TNF-α stimulation. The other one, a Myd88-independent, preferentially mediates induction of IFN-inducible genes via IRF3 and IRF7, that resemble the IFN stimulation [34].
An IFN type I signature is detected in SLE, dermatomyositis, subgroup of RA patients, but interestingly, it is also induced in Systemic Onset Juvenile Idiopathic Arthritis (SoJIA) patients following anti-TNF-α treatment [35]. A part of therapeutically induced type I IFN genes were identified as TNF-α down-regulated, like IFIT1, IFIT3, OAS1, RSAD2. Namely, these genes were suppressed by TNF-α stimulation. The identification of these genes supports the hypothesis regarding the dominance of either TNF-α or IFN type I as the key cytokines involved in driving autoimmune diseases like SLE or RA [36].
In addition, the pro-apoptotic genes XAF1 and TRAIL (TNFSF10) contribute to the discrimination of IFN type I and LPS common genes from one side and TNF-α regulated genes from the other side [37]. Up-regulated by type I IFN and LPS, but down-regulated by TNF-α, they seem to be responsible for inflammation-dependent apoptotic processes. The final outcome of apoptotic response which includes both pro- and anti-apoptotic genes seems to be finely tuned, especially considering that monocytes apoptotic behaviour could be beneficial or detrimental depending of the overall inflammatory context.
Our results showed remarkable numbers of pro- and anti-apoptotic genes in monocytes affected by TNF-α and LPS stimulation that we used to describe the apoptosis-survival profile. BCL2 family members showed a more anti-apoptotic programme considering that all anti-apoptotic genes, BCL2, BCL2A1 and MCL1 were up-regulated and most of the pro-apoptotic genes were down-regulated. Part of the apoptosis-survival profile also constitutes caspases, proteolitic enzymes whose activation is responsible for cell death. Beside their role in apoptosis some of them, like CASP1, CASP4, CASP5, and CASP12 are involved in cytokine processing and associate inflammation and apoptosis [38].
To show the applicability of in vitro-generated cytokine signatures in practice, we used the TNF-α-signature to monitor RA patients before and after treatment with Adalimumab, a TNF-α-blocking antibody [23]. Thus, we could identify a profile of 174 probe sets that was obviously silenced by TNF-α, since these genes were found to be down-regulated in response to TNF-α in vitro and up-regulated in successfully treated RA patients if compared with monocytes analyzed before treatment. Nearly 30% of this 174 probe sets were not only suppressed in vitro, but were also identified as being suppressed in vivo when untreated RA samples were compared with a group of monocytes isolated from healthy donors (data not shown). Thus, reversed changes in gene expression detected in monocytes stimulated with TNF-α from the one side and monocytes from RA patients, which were treated with TNF-α blockers from the other side, make it promising to assume that our concept of using in vitro-generated gene signatures in a cell-specific manner will be helpful for following and predicting therapeutic responses.
Altogether, to our knowledge, the present study demonstrates for the first time a challenging opportunity to analyze cytokine-specific gene expression profiles in a cell-specific manner. The development of additional cytokine-specific signatures in monocytes, such as IFN type I, IFN type II, IL-1, IL-6 will be necessary to improve specificity of already determined signatures. This complex knowledge seems to be not only indispensable to understand complexity of acute and chronic inflammatory diseases, but is also fundamental in the field of systems biology exploring physiological and pathophysiological dysregulations mainly triggered by pro- and anti-inflammatory cytokine responses.
References
Marrack P, Kappler J, Kotzin BL (2001) Autoimmune disease: why and where it occurs. Nat Med 7:899–905. doi:10.1038/90935
Veale DJ, Ritchlin C, FitzGerald O (2005) Immunopathology of psoriasis and psoriatic arthritis. Ann Rheum Dis 64(Suppl 2):ii26–ii29. doi:10.1136/ard.2004.031740
Batliwalla FM, Baechler EC, Xiao X, Li W, Balasubramanian S, Khalili H, Damle A, Ortmann WA, Perrone A, Kantor AB, Gulko PS, Kern M, Furie R, Behrens TW, Gregersen PK (2005) Peripheral blood gene expression profiling in rheumatoid arthritis. Genes Immun 6:388–397
Batliwalla FM, Li W, Ritchlin CT, Xiao X, Brenner M, Laragione T, Shao T, Durham R, Kemshetti S, Schwarz E, Coe R, Kern M, Baechler EC, Behrens TW, Gregersen PK, Gulko PS (2005) Microarray analyses of peripheral blood cells identifies unique gene expression signature in psoriatic arthritis. Mol Med 11:21–29
Stoeckman AK, Baechler EC, Ortmann WA, Behrens TW, Michet CJ, Peterson EJ (2006) A distinct inflammatory gene expression profile in patients with psoriatic arthritis. Genes Immun 7:583–591. doi:10.1038/sj.gene.6364334
Baechler EC, Bauer JW, Slattery CA, Ortmann WA, Espe KJ, Novitzke J, Ytterberg SR, Gregersen PK, Behrens TW, Reed AM (2007) An interferon signature in the peripheral blood of dermatomyositis patients is associated with disease activity. Mol Med 13:59–68
Baechler EC, Batliwalla FM, Karypis G, Gaffney PM, Ortmann WA, Espe KJ, Shark KB, Grande WJ, Hughes KM, Kapur V, Gregersen PK, Behrens TW (2003) Interferon-inducible gene expression signature in peripheral blood cells of patients with severe lupus. Proc Natl Acad Sci USA 100:2610–2615
Maas K, Chan S, Parker J, Slater A, Moore J, Olsen N, Aune TM (2002) Cutting edge: molecular portrait of human autoimmune disease. J Immunol 169:5–9
Lock C, Hermans G, Pedotti R, Brendolan A, Schadt E, Garren H, Langer-Gould A, Strober S, Cannella B, Allard J, Klonowski P, Austin A, Lad N, Kaminski N, Galli SJ, Oksenberg JR, Raine CS, Heller R, Steinman L (2002) Gene-microarray analysis of multiple sclerosis lesions yields new targets validated in autoimmune encephalomyelitis. Nat Med 8:500–508. doi:10.1038/nm0502-500
Häupl T, Ostensen M, Grützkau A, Burmester GR, Villiger PM (2008) Interaction between rheumatoid arthritis and pregnancy: correlation of molecular data with clinical disease activity measures. Rheumatology Oxford 47(Suppl 3):iii19–iii22. doi:10.1093/rheumatology/ken157
Häupl T, Ostensen M, Grützkau A, Radbruch A, Burmester GR, Villiger PM (2008) Reactivation of rheumatoid arthritis after pregnancy: increased phagocyte and recurring lymphocyte gene activity. Arthritis Rheum 58:2981–2992. doi:10.1002/art.23907
Grützkau A, Grün J, Häupl T, Burmester GR, Radbruch A (2007) Gene expression in inflammatory rheumatic diseases. Dtsch Med Wochenschr 132:1888–1891
van der Pouw Kraan TC, Wijbrandts CA, van Baarsen LG, Voskuyl AE, Rustenburg F, Baggen JM, Ibrahim SM, Fero M, Dijkmans BA, Tak PP, Verweij CL (2007) Rheumatoid arthritis subtypes identified by genomic profiling of peripheral blood cells: assignment of a type I interferon signature in a subpopulation of patients. Ann Rheum Dis 66:1008–1014. doi:10.1136/ard.2006.063412
Clark IA (2007) How TNF was recognized as a key mechanism of disease. Cytokine Growth Factor Rev 18:335–343. doi:10.1016/j.cytogfr.2007.04.002
McInnes IB, Schett G (2007) Cytokines in the pathogenesis of rheumatoid arthritis. Nat Rev Immunol 7:429–442
Rigby WF (2007) Drug insight: different mechanisms of action of tumor necrosis factor antagonists-passive-aggressive behavior? Nat Clin Pract Rheumatol 3:227–233
Catrina AI, Trollmo C, af Klint E, Engstrom M, Lampa J, Hermansson Y, Klareskog L, Ulfgren AK (2005) Evidence that anti-tumor necrosis factor therapy with both etanercept and infliximab induces apoptosis in macrophages, but not lymphocytes, in rheumatoid arthritis joints: extended report. Arthritis Rheum 52:61–72. doi:10.1002/art.20764
Kinne RW, Brauer R, Stuhlmuller B, Palombo-Kinne E, Burmester GR (2000) Macrophages in rheumatoid arthritis. Arthritis Res 2:189–202
Taylor PC, Williams RO, Feldmann M (2004) Tumour necrosis factor alpha as a therapeutic target for immune-mediated inflammatory diseases. Curr Opin Biotechnol 15:557–563. doi:10.1016/j.copbio.2004.09.005
Biesen R, Demir C, Barkhudarova F, Grün JR, Steinbrich-Zollner M, Backhaus M, Häupl T, Rudwaleit M, Riemekasten G, Radbruch A, Hiepe F, Burmester GR, Grützkau A (2008) Sialic acid-binding Ig-like lectin 1 expression in inflammatory and resident monocytes is a potential biomarker for monitoring disease activity and success of therapy in systemic lupus erythematosus. Arthritis Rheum 58:1136–1145. doi:10.1002/art.23404
Menßen A, Edinger G, Grün J, Haase U, Baumgrass R, Grützkau A, Radbruch A, Burmester G, Häupl T (2009) SiPaGene: a new repository for instant online retrieval, sharing and meta-analyses of GeneChip® expression data. BMC Genomics 10:98
Califano A, Stolovitzky G, Tu Y (2000) Analysis of gene expression microarrays for phenotype classification. Proc Int Conf Intell Syst Mol Biol 8:75–85
Stühlmuller B, Häupl T, Hernandez MM, Grützkau A, Kuban RJ, Tandon N, Voss JW, Salfeld J, Kinne RW, Burmester GR (2010) CD11c as a transcriptional biomarker to predict response to anti-TNF monotherapy with adalimumab in patients with rheumatoid arthritis. Clin Pharmacol Ther 87:311–321. doi:10.1038/clpt.2009.244
Dennis G Jr, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, Lempicki RA (2003) DAVID: database for annotation, visualization, and integrated discovery. Genome Biol 4:P3
Dinarello CA (1998) Interleukin-1 beta, interleukin-18, and the interleukin-1 beta converting enzyme. Ann NY Acad Sci 856:1–11
Barksby HE, Lea SR, Preshaw PM, Taylor JJ (2007) The expanding family of interleukin-1 cytokines and their role in destructive inflammatory disorders. Clin Exp Immunol 149:217–225. doi:10.1111/j.1365-2249.2007.03441.x
Balabanian K, Lagane B, Infantino S, Chow KY, Harriague J, Moepps B, Arenzana-Seisdedos F, Thelen M, Bachelerie F (2005) The chemokine SDF-1/CXCL12 binds to and signals through the orphan receptor RDC1 in T lymphocytes. J Biol Chem 280:35760–35766. doi:10.1074/jbc.M508234200
Zlotnik A, Yoshie O, Nomiyama H (2006) The chemokine and chemokine receptor superfamilies and their molecular evolution. Genome Biol 7:243
Grützkau A, Radbruch A (2008) Separation of whole blood cells and its impact on gene expression. In: GB BA (ed) Microarrays in inflammation. Basel, Birkhaeuser Verlag AG, pp 31–40
Wahl LM, Wahl SM, Smythies LE, Smith PD (2006) Isolation of human monocyte populations. Curr Protoc Immunol chapter 7: Unit 7 6A. doi 10.1002/0471142735.im0706as70
Tracey D, Klareskog L, Sasso EH, Salfeld JG, Tak PP (2008) Tumor necrosis factor antagonist mechanisms of action: a comprehensive review. Pharmacol Ther 117:244–279. doi:10.1016/j.pharmthera.2007.10.001
Ulloa L, Tracey KJ (2005) The “cytokine profile”: a code for sepsis. Trends Mol Med 11:56–63. doi:10.1016/j.molmed.2004.12.007
Baud V, Karin M (2001) Signal transduction by tumor necrosis factor and its relatives. Trends Cell Biol 11:372–377.
Baccala R, Hoebe K, Kono DH, Beutler B, Theofilopoulos AN (2007) TLR-dependent and TLR-independent pathways of type I interferon induction in systemic autoimmunity. Nat Med 13:543–551. doi:10.1038/nm1590
Palucka AK, Blanck JP, Bennett L, Pascual V, Banchereau J (2005) Cross-regulation of TNF and IFN-alpha in autoimmune diseases. Proc Natl Acad Sci USA 102:3372–3377. doi:10.1073/pnas.0408506102
Banchereau J, Pascual V (2006) Type I interferon in systemic lupus erythematosus and other autoimmune diseases. Immunity 25:383–392. doi:10.1016/j.immuni.2006.08.010
Bennett L, Palucka AK, Arce E, Cantrell V, Borvak J, Banchereau J, Pascual V (2003) Interferon and granulopoiesis signatures in systemic lupus erythematosus blood. J Exp Med 197:711–723
Hotchkiss RS, Nicholson DW (2006) Apoptosis and caspases regulate death and inflammation in sepsis. Nat Rev Immunol 6:813–822. doi:10.1038/nri1943
Acknowledgments
We are grateful to Heidi Schliemann responsible for generating gene expression data. The work was supported by the German Ministry of Education and Research (BMBF) within the National Genome Research Network NGFN (01GS0413) and by the European Union’s Sixth Framework Programme (project AutoCure; LSHB-CT-2006-018861).
Disclosure of potential conflict of interests
The authors declare no conflict of interests related to this study.
Author information
Authors and Affiliations
Corresponding author
Additional information
Author Contributions
Conceived and designed the experiments: AG, GRB, RB, AR. Performed the experiments: BSm, AG. Analyzed the data: BSm, AG, JG, TH, BSt, MSZ. Wrote the paper: BSm, AG, RB, JG.
Andreas Grützkau and Ria Baumgrass contributed equally to this work
Supported by the German Federal Ministry of Education and Research (BMBF) through the National Genome Research Network (Infection and Inflammation Network SIPAGE, grant 01GS0413) and by the European Union's Sixth Framework Programme (project AutoCure; LSHB-CT-2006-018861)
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplementary File 1
This file includes a description how to get access to the publically available array data using BioRetis database. Furthermore, filtering criteria are described for the selection of differentially expressed genes. (PDF 15 kb)
Supplementary Table 1
with sheet 1-9. These tables include Signal values, p values, Signal Log Ratio (SLR), Fold Change (FC) of differentially expressed probe sets that distinguish unstimulated monocytes but incubated, TNF-α- and LPS-stimulated monocytes from Unstimulated and immediately processed monocytes. (XLS 8343 kb)
Supplementary Table 2
with sheet 1-3. These tables include the probe sets that discriminate TNF-α- from LPS-stimulated monocytes. Beside 1,824 probe sets that are TNF-α specific and 3,214 probe sets specific for LPS, there are 94 probe sets that are common for TNF and LPS stimulation but they are regulated in opposite direction by these two stimuli. (XLS 789 kb)
Supplementary Table 3
with sheet 1-2. These tables include the probe sets that describe the cytokine–cytokine receptor profile in monocytes. Altogether 182 probe sets are differentially expressed in monocytes stimulated with TNF-α or LPS for 1.5 h compared with unstimulated cells. 120 genes out of 182 probe sets that are differentially expressed are presented with average signals and average fold changes. (XLS 80 kb)
Supplementary Table 4
with sheet 1-2. These tables include the probe sets of the genes that describe the apoptosis-survival profile in monocytes. Altogether 419 probe sets are differentially expressed and involved in apoptosis and survival in monocytes after 1.5 h of incubation in the presence of TNF-α or LPS compared with unstimulated cells. 240 genes out of 419 differentially expressed probe sets were presented with average signals and average fold changes. (XLS 163 kb)
Supplementary Table 5
with sheet 1-3. These tables include Signal values, p values, Signal Log Ratio (SLR), Fold Change (FC) of 471 differentially expressed probe sets that distinguish monocytes from RA patients that responded to anti-TNF-α therapy, before and after treatment (sheet 1 and 2). These probe sets were revealed as significantly increased or decreased in 80% of pair-wise comparisons. Sheet 3 includes 179 probe sets with FC that are common for in vitro stimulated monocytes by TNF-α and monocytes from RA patients treated with anti-TNF-α, and which showed opposite direction of changes. Affymetrix arrays used for analysis of in vitro stimulated monocytes belong to HG-U133 Plus 2.0 platform and for analysis of anti-TNF treated RA patients to HG-U133A 2.0. (XLS 287 kb)
Rights and permissions
About this article
Cite this article
Smiljanovic, B., Grün, J.R., Steinbrich-Zöllner, M. et al. Defining TNF-α- and LPS-induced gene signatures in monocytes to unravel the complexity of peripheral blood transcriptomes in health and disease. J Mol Med 88, 1065–1079 (2010). https://doi.org/10.1007/s00109-010-0648-8
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00109-010-0648-8